A New Subtilisin-Like Proteinase from Roots of the Dandelion Taraxacum officinale Webb S. L.
A. M. Bogacheva1*, G. N. Rudenskaya1, A. Preusser1, I. O. Tchikileva1, Ya. E. Dunaevsky2, B. N. Golovkin3, and V. M. Stepanov1
1Department of Natural Compounds, School of Chemistry, Lomonosov Moscow State University, Moscow, 119899 Russia; fax: (7-095) 939-3181; E-mail: annamb@genebee.msu.su2Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow, 119899 Russia; fax: (7-095) 939-3181
3Department of Tropical Plants, Botanical Garden, Russian Academy of Sciences, Moscow, 127276 Russia
* To whom correspondence should be addressed.
Received December 1, 1998; Revision received May 11, 1999
A serine proteinase from roots of Taraxacum officinale Webb S. L. was isolated by affinity chromatography and gel-filtration on Superose 6R using FPLC. The enzyme is a 67-kD glycoprotein containing 54% carbohydrate which we have named taraxalisin. The substrate specificity of taraxalisin toward synthetic peptides and oxidized insulin B-chain is comparable with that of cucumisin from Cucumis melo and the subtilisin-like serine proteinase macluralisin from Maclura pomifera. The proteinase is inactivated by DFP and PMSF. Taraxalisin exhibits maximal activity at pH 8.0. The pH range for stability of the enzyme is narrow--6.0-9.0. The temperature optimum for the subtilisin-like activity is 40°C. The N-terminal sequence of taraxalisin has 40% of its residues identical to those of subtilisin Carlsberg. Thus, the serine proteinase from dandelion roots is a member of the subtilisin family, which is evidently widespread in the plant kingdom.
KEY WORDS: proteinases, subtilisins, taraxalisin, dandelion roots, isolation, properties
Abbreviations: pNA) p-nitroanilide; Glp) pyroglutamyl; DFP) diisopropyl fluorophosphate; PMSF) phenylmethylsulfonyl fluoride; DMF) dimethylformamide; mrp) morpholine residue; STI) soybean trypsin inhibitor.
Proteinases play a prominent role in plant physiology. They are
catalysts of important processes like degradation of defective
proteins, activation of proenzymes, hydrolysis of storage proteins
during seed germination, etc. The proteolytic enzymes of seeds and
fruits have been most studied; those from leaves have been much less
studied. Proteinases from roots are almost unknown except for MRP--a
serine proteinase from maize roots [1]. MRP is
reported to be an elastase-like enzyme based on its inhibition pattern
and substrate specificity [1].
The latexes of tropical plant genuses (Hevea, Ficus, Euphorbia, and Parthenium) are known to be rich in proteolytic enzymes. A subtilisin-like serine proteinase present in the latex of Maclura pomifera from the family Moraceae was isolated in our laboratory earlier [2]. Analysis of information about latex-forming plants has shown that the Compositae (Asteraceae) family also includes a number of plants forming latex. We have chosen the dandelion Taraxacum officinale Webb S. L., a plant widely spread in Central Russia, as the object of further search for proteolytic enzymes. Preliminary experiments have shown that the latex effluent from the roots of this plant contains a proteinase that hydrolyzes Glp-Ala-Ala-Leu-pNA--a typical substrate of serine proteinases from the chymotrypsin or subtilisin families [3]. The isolation and molecular and functional characteristics of this serine proteinase, which we have named taraxalisin, are described in this article.
MATERIALS AND METHODS
Isolation of taraxalisin. Taraxalisin is isolated as described in the following typical experiment. Dandelion roots (70 g) were harvested in Moscow in May 1996, washed, and frozen to -20°C. The frozen roots were chopped with a knife, then homogenized in 100 ml of 50 mM Tris-HCl buffer (pH 8.0) with 5 mM (CH3COO)2Ca at room temperature using a Waring Blendor-like homogenizer (Mechanika Precyzyjna Type 362, Poland). The homogenate was filtered using a water-driven vacuum pump. Protein in the filtrate (103 ml, supplemented with 8 ml of 1 M Tris-HCl buffer, pH 8.0) was precipitated by adding dry (NH4)2SO4 to 80% saturation, and the sample was kept overnight at 5°C for the formation of the protein precipitate. The protein precipitate was collected by centrifugation at 40,000g for 45 min at 5°C using a Beckman JA-20 centrifuge (Austria). The pellet was dissolved in 50 ml of 50 mM Tris-HCl buffer (pH 8.0) containing 5 mM (CH3COO)2Ca, and the solution was desalted using an Amicon (USA) ultrafiltration cell with a YM-10 membrane.
Affinity chromatography. The protein fraction in 50 ml of 50 mM Tris-HCl buffer (pH 8.0) with 5 mM (CH3COO)2Ca was subjected to affinity chromatography on a 2.5 × 45-cm Sepharose-AALmrp column [4] preequilibrated with the same buffer. After washing the column with the same buffer, the enzyme was eluted with 50 ml of 25% ethanol (v/v) in 50 mM Tris-HCl buffer (pH 8.0) containing 5 mM (CH3COO)2Ca and 1 M NaCl. The eluate containing the enzyme active against Glp-Ala-Ala-Leu-pNA was desalted and concentrated to 15 ml using an Amicon ultrafiltration cell with a YM-10 membrane.
FPLC gel-filtration. The preparation was lyophilized and then dissolved in a minimal volume of 50 mM Tris-HCl buffer (pH 8.0) containing 0.5 M NaCl. Using FPLC, the sample (100 µl) was gel-filtered on a 1 × 30-cm Superose 6R column equilibrated with the same buffer. Proteins were fractionated by molecular mass on the column using the same buffer under pressure providing a flow rate of 0.5 ml/min. The column was calibrated with the following molecular mass markers from Bio-Rad (USA): cytochrome c (12.6 kD), ribonuclease (14 kD), trypsin inhibitor (22 kD), ovalbumin (44 kD), BSA (65 kD), ovalbumin-dimer (88 kD), BSA dimer (130 kD).
Isolation of taraxalisin substrate. Proteins were extracted and precipitated as described for the proteinase. The protein fraction was then desalted by gel-filtration on Sephadex G-25 and lyophilized.
Gel-filtration. The lyophilized protein fraction (15 mg of protein) was dissolved in 500 µl of 50 mM Tris-HCl buffer (pH 8.0) containing 0.5 M NaCl and subjected to gel-filtration on a 1.6 × 80-cm Sephadex G-75 column using the same buffer.
Chromatofocusing. This was performed on a 0.5 × 1-5-cm PBE-94 chromatography column (Pharmacia, Sweden). The column was equilibrated with 15 ml of 25 mM histidine-HCl buffer, pH 7.4 (starting buffer). PB-74-polybuffer (diluted 8-fold), pH 4.0, was used as the eluent (gradient from pH 7.4 to 4.0). The protein sample was dissolved in 500 µl of the eluting polybuffer.
Study of the possible natural substrates of taraxalisin. The analyzed protein was dissolved in 50 mM Tris-HCl buffer (pH 8.0) containing 5 mM (CH3COO)2Ca and taraxalisin was added protein/proteinase ratio 100:1. The sample was incubated at 37° for 24 h. The resulting hydrolysis was studied by SDS-PAGE (10%).
Protein concentration. Protein concentrations in solution were determined by a modification of the Bradford method [5].
Proteolytic activity. The proteolytic activity of taraxalisin was studied by its cleavage of synthetic chromogenic substrates. The substrates (Glp-Ala-Ala-Leu-pNA, Glp-Phe-Ala-pNA, Glp-Ala-Ala-Phe-Leu-pNA, and Bz-Arg-pNA) were dissolved in DMF. One unit of activity was defined as the amount of enzyme that cleaves 1 µmole of substrate in 1 h [3]. The reaction mixtures contained 50 µl of the enzyme, 50 µl of substrate (5 mg/ml in DMF), and 2.5 ml 50 mM Tris-HCl buffer (pH 8.0). The samples were incubated in a thermostat at 37°C until yellow coloration appeared. The reaction was stopped with 200 µl of 50% acetic acid.
The proteolytic activity of taraxalisin towards 0.5% solutions of BSA (Diam, Russia), ovalbumin (Reakhim, Russia), and azocasein and thrombin (Ferment, Lithuania) was studied. The protein solutions (1 ml) were incubated with the enzyme at 37°C for 24 h. The unit of activity was defined as the amount of enzyme that formed hydrolysis products corresponding to 1 milliequivalent of tyrosine in 1 h. Activity against azocasein was assessed as described in [6].
Hydrolysis of bovine insulin B-chain. The hydrolysis oxidized bovine insulin B-chain (0.7 mg in 250 ml 50 mM Tris-HCl buffer, pH 8.0) by taraxalisin (6 µg) was studied. After 3.5 h at 37°C, the hydrolysis was stopped by rapid freezing of the sample. The hydrolysate was separated on a 4.6 × 200-mm Ultrasphere-Octyl column using a Beckman-344 liquid chromatograph (Beckman, Austria). The column was eluted with 0.1% trifluoroacetic acid with a linear acetonitrile gradient from 0 to 50%. The isolated peptides were hydrolyzed with 5.7 M HCl and trifluoroacetic acid (2:1) containing 0.1% beta-mercaptoethanol at 160°C. Amino acid compositions were determined with a Hitachi-835 amino acid analyzer (Hitachi, Japan).
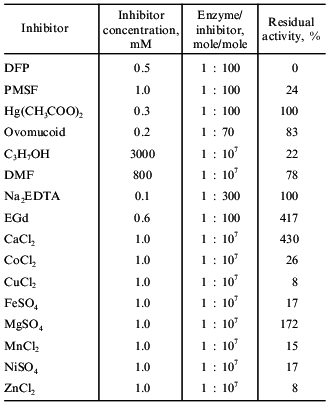
Inhibition of taraxalisin. Inhibition of the enzyme by 0.5 mM DFP and 0.5 mM PMSF (Serva, Germany), 0.5 mM Na2EDTA (Reanal, Hungary), 0.2 mM ovomucoid (Sigma, USA), 0.3 mM Hg(CH3COO)2 and 0.6 mM EGd (Boehringer Mannheim, Germany), 25% C3H7OH, and 5% DMF was determined. The enzyme was incubated in 50 mM Tris-HCl buffer (pH 8.0) for 4 h at 20°C. Residual activities were determined against the synthetic substrate Glp-Ala-Ala-Leu-pNA and are given in percent of initial activity.
Influence metal ions on the activity of taraxalisin was studied in 50 mM Tris-HCl buffer (pH 8.0) using 1 mM salt solutions: CaCl2, CoCl2, CuCl2, FeSO4, MgSO4, MnCl2, NiSO4, ZnCl2. The enzyme was incubated in the solutions for 1 h at 20°C, then the residual activity was measured with Glp-Ala-Ala-Leu-pNA and expressed in percent of initial activity.
Determination of Km and kcat. The values were determined in 2.5 ml of 50 mM Tris-HCl buffer (pH 8.0) containing 50 µl enzyme solution (1.57 mg/ml). Solutions of the synthetic chromogenic substrate Glp-Ala-Ala-Leu-pNA in DMF at 0.83, 1.11, 1.65, 2.22, 3.33, 4.95, 7.43, 9.90, 14.85, 19.81, and 29.70 mg/ml were added, the final concentration of the substrate varying from 0.033 to 1.016 mM. The samples were incubated at 37°C for 12 h. The values of Km and kcat were calculated from 1/v versus 1/S double reciprocal plots using the program Enzfitter.
Stability of taraxalisin versus pH. The stability of the enzyme at 25°C was determined in buffer solutions with different pH values (1 M acetate buffer at pH 4.0, 4.5, and 5.0; 50 mM Mes at pH 5.5, 6.0, and 6.5; 50 mM Tris-HCl at pH 7.0, 7.5, 8.0, and 8.5; 50 mM borate buffer at pH 9.0, 9.5, 10.0, and 10.5) by hydrolysis of Glp-Ala-Ala-Leu-pNA as described above. Enzyme solution (100 µl; 0.15 mg/ml) was added to 450 µl of the buffer solutions and 100 µl aliquots were taken to measure the activity. The solutions were incubated for 4 and 23.5 h at 25°C, and residual activity was expressed as percent of the control.
Activity of taraxalisin versus temperature. The activity of the enzyme with Glp-Ala-Ala-Leu-pNA in 50 mM Tris-HCl buffer (pH 8.0) was measured at temperatures in the interval 20-55°C.
Isoelectrofocusing. The isoelectric point of taraxalisin was determined by isoelectrofocusing in 5% polyacrylamide gel in the presence of 2% ampholines Biolyte 3/10 with a Bio-Rad mini IEF-Cell. The following proteins of known pI were used for calibration: cytochrome c (10.65), ribonuclease (9.45), goat myoglobin (8.55), horse myoglobin (7.3), beta-lactalbumin (5.34), BSA (4.7), ferritin (4.4), aminoglycosidase (3.5) (Serva).
Electrophoresis. SDS-PAGE (in 12.5 and 10% gels) was performed using a Bio-Rad mini-systems cell at 100 V and 20 mA (concentrating gel) and 250 V and 40 mA (separating gel) for 40 min by the Laemmli method [7]. The sample was preliminarily processed with 7% trichloroacetic acid for denaturation and sedimentation of the protein.
Phosphorylase B from rabbit muscle (94 kD), BSA (67 kD), ovalbumin (43 kD), carbonic anhydrase (30.1 kD), soybean trypsin inhibitor (20.1 kD), and alpha-lactalbumin (14.4 kD) (Bio-Rad) were used as markers.
Ultracentrifugation. Protein samples in 50 mM Tris-HCl buffer (pH 8.0) containing 0.5 M NaCl were analyzed by non-equilibrium sedimentation at 5°C for 1 h at 180,000g using a Spinco model E ultracentrifuge (Beckman, Austria). The protein was monitored at 280 nm. The molecular mass was computed from the equation: logM = 1.5·logS + 3.925.
Determination of carbohydrate content. A taraxalisin sample (111 µg) was denatured in 300 µl of 0.5% SDS and 1% beta-mercaptoethanol at 100°C for 10 min and then dialyzed against water. A portion of the denatured enzyme (37 µg) in 100 µl of 50 mM sodium citrate (pH 5.5) was incubated with 9.1 µg of recombinant endoglycosidase (Endo Hf) from Streptomyces plicatus [8] (Biolabs, USA) at 37°C for 14.5 and 21 h. A sample without Endo Hf was used as a control. The resulting hydrolysates were studied by SDS-PAGE (12.5%).
For qualitative carbohydrate analysis, 20 µg of taraxalisin was hydrolyzed in 500 µl of anhydrous hydrazine in an atmosphere of nitrogen at 100°C for 10 h. The residue was analyzed [9] using a Beckman Gold 126 HPLC instrument (Beckman, Austria) with a 4.6 × 150-mm Phenomenex C18 analytical column and fluorometric determination of molar ratios on a Gilson 121 flow-type fluorometer (365/440 nm) (Gilson, France). N-Mannosamine was used as an internal standard.
Determination of amino acid composition. Samples of the enzyme were hydrolyzed in tubes sealed under reduced pressure with 5.7 M HCl for 48 h at 105°C. Methionine sulfone and cysteic acid contents were assayed after oxidation of the sample with performic acid. Tryptophan content was determined after 24 h hydrolysis with 4 M methanesulfonic acid in the presence of 0.2% tryptamine. The amino acid content was determined with a Hitachi-835 amino acid analyzer (Japan).
N-Terminal sequencing. The N-terminal sequence of taraxalisin was determined by Edman degradation. The enzyme (0.53 mg) dissolved in 3 ml of 0.05 M Tris-HCl buffer (pH 8.0) was inactivated for 30 min with DFP at 25°C, dialyzed against water, and lyophilized. The enzyme was further purified by HPLC on a 4.6 × 100-mm Aquapore column using a concentration gradient (15-60% acetonitrile in 0.1% trifluoroacetic acid). The protein was sequenced using a gaseous-phase sequencer Knauer m. d. 120A PIH ANALISER one Line (Applied Biosystems, USA).
RESULTS AND DISCUSSION
The dandelion plant accumulates in its roots nutrients for winter: inulin (up to 24%), rubber (3%), triterpenes, and fatty acids and their glycerides [10]. All these substances hamper the proteinase isolation and complicate its chromatography. Protein precipitation with ammonium sulfate was used to remove inulin and rubber (Table 1). The precipitation of protein must be done at the reagent ratios described in "Materials and Methods" because from very diluted extract even saturated ammonium sulfate (100%) does not precipitate the bulk of the enzyme, which remains in the supernatant. Also, the pH of the solution must be maintained at 7.0-8.0; the proteinase is unstable to even brief exposure to lower pH values.
Table 1. Isolation of the serine proteinase
from dandelion roots
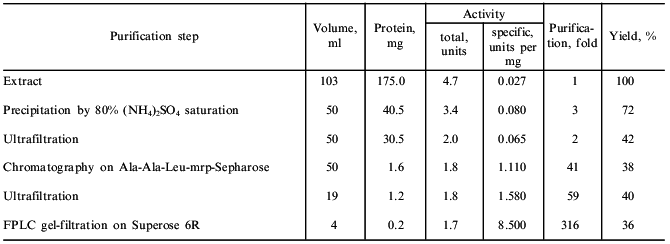
The enzyme preparation can concentrated and desalted between purification steps by ultrafiltration.
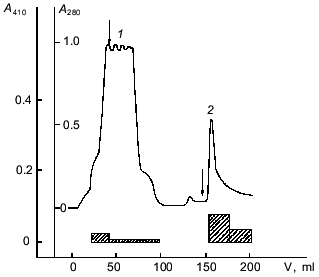
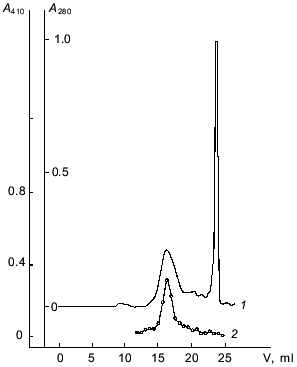
Affinity chromatography was used for further separation of the enzyme from pigments and other admixtures. It should be noted that a well-known affinity sorbent for isolation of proteinases, bacitracin-Sepharose [11], was ineffective for the isolation of taraxalisin. However, a sorbent recently synthesized in our laboratory containing as a ligand the substrate analog Ala-Ala-Leu-mrp attached via p-benzoquinone to AH-Sepharose [4] was effective. The proteinase from dandelion roots has high affinity to this sorbent, and no loss of the enzyme was observed during the chromatography (Fig. 1). Final separation of a pigment was achieved by FPLC gel-filtration on a Superose 6R column (Fig. 2). The proteinase acting on Glp-Ala-Ala-Leu-pNA appears in the first major peak and corresponds to a 71-kD protein. The enzyme taraxalisin was purified 316-fold with a yield of 36%.
Fig. 1. Affinity chromatography of taraxalisin on Ala-Ala-Leu-mrp-Sepharose: 1) unsorbed fraction; 2) sorbed fraction containing taraxalisin; A410, activity against Glp-Ala-Ala-Leu-pNA (hatched bars).
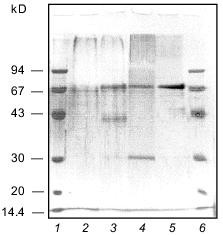
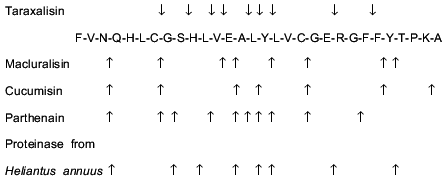
The homogeneity of the taraxalisin preparation was confirmed by SDS-PAGE (12.5% gel, Fig. 3) and the finding of a single N-terminal amino acid sequence (Fig. 4).Fig. 2. FPLC gel-filtration of taraxalisin on Superose 6R: 1) A280; 2) taraxalisin activity (A410).
Fig. 3. SDS-PAGE analysis of taraxalisin: 1, 6) marker proteins; 2) taraxalisin; 3) taraxalisin + Endo Hf (after 14.5-h incubation); 4) taraxalisin + Endo Hf (after 21-h incubation); 5) Endo Hf.
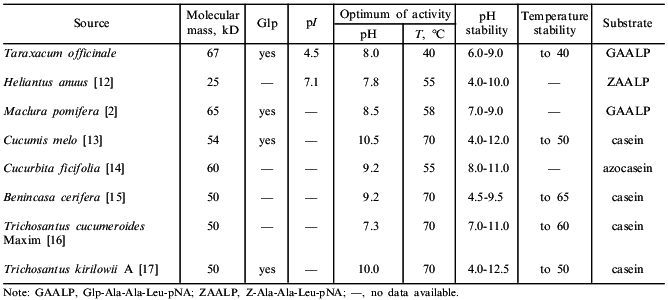
Some enzymatic and physicochemical characteristics of taraxalisin and of some other subtilisin-like proteinases of plants are presented in Table 2.Fig. 4. N-Terminal sequences of taraxalisin and subtilisin Carlsberg.
Table 2. Characteristics of taraxalisin and
other subtilisin-like proteinases of plants
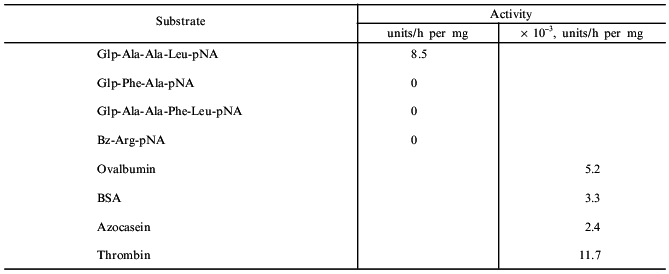
Data on the substrate specificity of taraxalisin are presented in Table 3. It should be noted that taraxalisin is rapidly inactivated in solutions containing more than 2% organic solvents such as dimethylformamide and isopropyl alcohol. This limited the selection of specific substrates for testing of enzyme activity. Instead of the hydrophobic chromogenic substrate Z-Ala-Ala-Leu-pNA commonly used for assessing the activity of bacterial subtilisins, we used the more hydrophilic substrate Glp-Ala-Ala-Leu-pNA; the latter required 10-fold lower percent of dimethylformamide for its dissolution. This substrate turned out to be one of the best of the studied substrates. The Km determined for Glp-Ala-Ala-Leu-pNA was 0.37 ± 0.06 mM, an order of magnitude higher than that for the subtilisin-like enzyme from Maclura pomifera fruits [2] and 2.5-fold higher than that found for subtilisin BPN´ [3]. These data show the rather high affinity of taraxalisin to this substrate; however, the low value of kcat (0.3 ± 0.02 sec-1) suggests that the cleavage of the enzyme--substrate complex is quite slow. Taraxalisin splits, although relatively slowly, various protein and peptide substrates (Table 3). The possible natural substrates of the taraxalisin are latex proteins with molecular mass (by ultracentrifugation) 148 kD (S = 6.78 ± 1.14) and 132 kD (S = 6.26 ± 0.17). The isoelectric points of these proteins are 6.7 and 5.6, respectively.
Table 3. Activity of taraxalisin with
various peptide and protein substrates
The substrate specificity of taraxalisin was tested by action on oxidized insulin B-chain (Fig. 5). Taraxalisin showed rather broad substrate specificity, splitting peptide bonds formed by the carboxyl group of hydrophobic amino acids such as Leu, Val, Tyr, and Phe, which is characteristic for subtilisin-like plant proteinases, as well as those of glutamic and cysteic acid. The serine proteinase from maize roots splits peptide substrates only after alanine residues [18].
Table 4 demonstrates that taraxalisin is inhibited by specific inhibitors of serine proteinases, DFP and PMSF. This confirms that the dandelion root proteinase belongs to the serine proteinase family.Fig. 5. Hydrolysis of oxidized bovine insulin B-chain by taraxalisin and some other serine proteinases.
Table 4. Effects of inhibitors on the
activity of taraxalisin
The molecular mass of taraxalisin by SDS-PAGE (12.5% gel) was 67 kD, similar to that by gel-filtration. This is similar to the molecular masses of subtilisin-like proteinases from plant fruits. After cleavage of the carbohydrate component with the recombinant endoglycosidase Endo Hf from Streptomyces plicatus, which cleaves N-acetylglycoside bonds [8], the molecular mass taraxalisin is 31 kD (Fig. 3). This is comparable with the molecular masses of bacterial subtilisins.
Reversed-phase HPLC of the carbohydrate part of taraxalisin suggests that is contains about 42% carbohydrate (not counting glucose) and includes 12% galactose, 8% mannose, 12% arabinose, 13% N-acetylglucosamine, 2% fructose, and 6% N-acetylgalactosamine.
The isoelectric point of taraxalisin is at pH 4.5. Subtilisin-like enzymes usually have isoelectric points in the alkaline pH region, but "acidic" subtilisins are also known. Thus, the subtilisin-like proteinase I (pI . 10) was isolated from Ac. chrysogenum together with the subtilisin-like proteinase II (pI 4.0) [19].
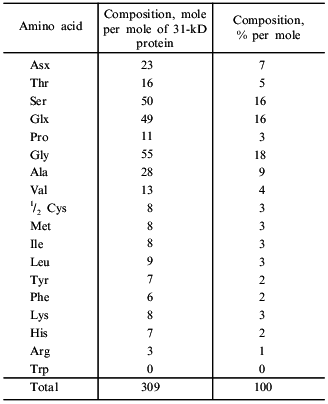
The amino acid composition of taraxalisin presented in Table 5 differs substantially from those of subtilisins from fruits of southern plants. There are no other data on the amino acid composition of serine proteinases from roots in the literature. The most characteristic feature of taraxalisin is its significant Cys content.
Table 5. Amino acid composition of
taraxalisin
Comparison of the aligned N-terminal sequences of subtilisin Carlsberg and taraxalisin traced for 15 amino acid residues revealed 40% identity (Fig. 4).
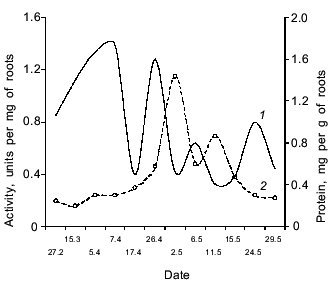
The dandelion plant growing in Central Russia is a biennial or perennial herb which forms a rosette of leaves during the first season and forms flower buds while still under snow. It blooms after the snow melts and ripens from the middle of May to early June. The dynamics of the accumulation of the enzyme with proteolytic activity against Glp-Ala-Ala-Leu-pNA in dandelion roots depend on the phase of seasonal development of the second year plant. Figure 6 shows that the maximum of the proteinase activity was observed in the root latex at the end of April, soon after snow melting. Later, the enzyme activity in the roots decreased during the peduncle growth. A second peak of the proteinase activity was observed from May 10 to June 10, during the blooming and seed formation of the plant. Simultaneously, the active proteinase appeared also in the latex and penetrated into the peduncles. After ripening of the seeds, which usually finishes before the first week of June, the enzyme activity in the roots decreases rapidly and was not detected in July-September. Hence, the proteinase is especially active during bud, flower, and seed formation and presumably plays an important role in the reproduction of the plant.
The authors are grateful to V. V. Nasonov for the qualitative carbohydrate analysis.Fig. 6. Dynamics of proteinase activity against Glp-Ala-Ala-Leu-pNA and soluble protein content in dandelion roots in different physiological phases of the plant: 1) protein; 2) activity.
This work was supported in part by a grant from the Russian Foundation for Basic Research (No. 96-04-49904).
REFERENCES
1.Batt, R., and Wallace, W. (1989) Biochim.
Biophys. Acta, 990, 109-112.
2.Rudenskaya, G. N., Bogdanova, E. A., Revina, L. P.,
Golovkin, B. N., and Stepanov, V. M. (1996) Planta, 196,
174-179.
3.Lublinskaya, L. A., Haidu, I., Balandina, G. N.,
Filippova, I. Yu., Markaryan, A. N., Lysogorskaya, E. N., Oksenoit, E.
S., and Stepanov, V. M. (1987) Bioorg. Khim., 13,
748-753.
4.Kuznetsova, A. V., Bogacheva, A. M., Rudenskaya, G.
N., and Stepanov, V. M. (1997) Chromatography, 45,
44-48.
5.Spector, T. (1978) Analyt. Biochem.,
86, 142-146.
6.Hencke, R. M., and Smith, T. L. (1967) Biochim.
Biophys. Acta, 139, 405-420.
7.Laemmli, U. K. (1970) Nature, 227,
680-685.
8.Robbins, P. (1984) J. Biol. Chem.,
259, 7577-7583.
9.Horlin, A. Ya., Shiyan, S. D., Nasonov, V. V., and
Markin, V. A. (1987) Bioorg. Khim., 13, 1266-1274.
10.Vavilova, N. M. (1994) Homeopathic
Pharmacodynamics [in Russian], Everest, Moscow.
11.Stepanov, V. M., and Rudenskaya, G. N. (1983)
J. Appl. Biochem., 5, 420-428.
12.Rudenskaya, G. N., Stepanov, V. M., Zakharova,
Yu. A., Revina, L. P., and Khodova, O. M. (1987) Biokhimiya,
52, 1753-1755.
13.Kaneda, M., and Tominago, N. (1975) J.
Biochem., 78, 1287-1296.
14.Curotto, E., and Gonzales, G. (1989) FEBS
Lett., 243, 363-365.
15.Kaneda, M., and Tominago, N. (1977)
Phytochemistry, 16, 345-346.
16.Kaneda, M., Sobue, A., Eida, S., and Tominago, N.
(1986) J. Biochem., 99, 569-577.
17.Uchikoba, T., Horita, H., and Kaneda, M. (1990)
Phytochemistry, 29, 1979-1981.
18.Goodfellow, V. J., Solomonson, L. P., and Oaks,
A. (1993) Plant Physiol., 101, 415-419.
19.Stepanov, V. M., Rudenskaya, G. N., Vasilyeva, L.
I., Krestyanova, I. N., Khodova, O. M., and Bartoshevitch, Yu. E.
(1986) Int. J. Biochem., 18, 369-375.