Interaction between Duodenase, a Proteinase with Dual Specificity, and Soybean Inhibitors of Bowman--Birk and Kunitz Type
I. P. Gladysheva1*, T. S. Zamolodchikova2, E. A. Sokolova2, and N. I. Larionova1
1School of Chemistry, Lomonosov Moscow State University, Moscow, 119899 Russia; fax: (095) 939-5417; E-mail: lar_lab@enzyme.chem.msu.ru2Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, ul. Miklukho-Maklaya 16/10, Moscow, 117871 Russia; fax: (095) 310-7007
* To whom correspondence should be addressed.
Received April 29, 1999; Revision received June 24, 1999
The interaction between duodenase, which belongs to a group of Janus-faced proteinases, and classical Bowman--Birk (BBI) and Kunitz (STI) type inhibitors from soybean was investigated. Duodenase was shown to interact only with the antichymotrypsin site (Leu-Ser) of BBI, whereas the antitrypsin site (Lys-Ser) of the inhibitor appeared to be vacant and capable of interaction with trypsin. The inhibition constants of duodenase by BBI, the BBI--trypsin complex, and STI were 4, 400, and 40 nM, respectively.
KEY WORDS: duodenase, Bowman--Birk inhibitor, Kunitz inhibitor, specificity, inhibition constant
Duodenase, a serine proteinase from bovine duodenal glands, is a member of a small group of so-called Janus-faced proteinases with dual substrate specificity, combining trypsin- and chymotrypsin-like properties [1]. Cathepsin G of human leukocytes [2] and chymases of sheep and goat mast cells [3, 4] also belong to this group. Clearly expressed structural homology of these enzymes (more than 60% identical residues) and other serine proteinases of hemopoietic cells (rat and human chymases) provides evidence for their close evolutionary relationship [5-8]. Dual specificity of proteinases of this group is mainly a consequence of changes in the structure of their S1 site as compared to trypsin-like proteinases. The S1 site binds to the side chain of the P1 residue of the substrate; the carboxylic group of the latter forms the hydrolyzed bond (classification according to Schechter and Berger [9]). X-Ray structure analysis of cathepsin G [2] showed that its S1 site is divided into two compartments by the side chain of the Glu-226 residue (from here on the residues are number as for chymotrypsinogen A), corresponding to the Asp-226 residue of duodenase [7] and sheep chymase [8]. Such an unusual substrate-binding “pocket” can efficiently interact not only with the positively charged residues Lys and Arg, but also with the aromatic residue Phe; in the latter case electrostatic rather than hydrophobic interactions are the main binding factors [2]. Thus, in Janus-faced proteinases including duodenase, the charged residue which determines the enzyme specificity occupies position 226, whereas in trypsin and trypsin-like proteinases the Asp-189 residue is crucial for specificity [10].
The goal of this work was to study the interaction between duodenase as a representative of the group of Janus-faced proteinases and Bowman--Birk type inhibitor (BBI) from soybean, capable of simultaneous and independent binding to trypsin by the Lys-Ser site and to alpha-chymotrypsin and chymotrypsin-like proteinases by the Leu-Ser site [11], and also Kunitz type inhibitor from soybean [12].
MATERIALS AND METHODS
The following reagents were used in this study: bovine alpha-chymotrypsin and Tris-HCl from the factory of chemical reagents in Olaine (Latvia); ethyl ester of N-benzoyl-L-tyrosine (Bz-Tyr-EE), ethyl ester of N-benzoyl-L-arginine (Bz-Arg-EE), Suc-Ala-Ala-Pro-Phe-pNA, Bz-Arg-pNA, Suc-Val-Pro-Phe-pNA, and bovine trypsin from Sigma (USA); STI from Reanal (Hungary).
alpha-Chymotrypsin was purified on a Toyopearl HW-50 column (eluent, dilute HCl, pH 3). The content of the active enzyme was 90% [13], and that in the trypsin preparation was 63% [14].
Classical BBI (2-IV) was isolated from soybean strain VNIIS-2 according to Odani and Ikenaka [15] but with some modifications [16]. The concentration of the active inhibitor in BBI and STI preparations was assayed by titration with trypsin of known active site concentration. The BBI (2-IV) preparation contained 100% concentration of the active inhibitor against the protein.
Duodenase was isolated from bovine duodenal glands by affinity chromatography on a column with STI-Sepharose 4B [1]. Protein concentration in duodenase preparations was assayed according to Lowry [17]. The active site concentration in duodenase was assayed by several methods: titration with BBI with the known activity, titration with labeled diisopropyl fluorophosphate, and titration with p-nitrophenyl-p´-guanidine-benzoate [14].
Determination of activity of proteinases and soybean inhibitors of Bowman--Birk and Kunitz type. The enzymatic activity of duodenase, alpha-chymotrypsin, and trypsin was estimated in 0.05 M Tris-HCl buffer (pH 8.0) at 25°C (for trypsin, the buffer contained 20 mM CaCl2) using a Shimadzu 265 FW spectrophotometer (Japan) with a thermostatted cuvette. Suc-Val-Pro-Phe-pNA (1.0 mM) and Suc-Ala-Ala-Pro-Phe-pNA (1.0 mM) were used as duodenase substrates, Bz-Arg-pNA (3.0 mM) was used as trypsin substrate, and Bz-Tyr-EE (0.125 mM) and Bz-Arg-EE (0.15 mM) were used as alpha-chymotrypsin substrates. The concentration of nitroaniline formed during the reaction was monitored by spectrophotometry at 405 nm (molar extinction epsilon = 9800 M-1·cm-1). During hydrolysis of Bz-Arg-EE and Bz-Tyr-EE, the concentrations of the products were monitored at 253 and 256 nm, respectively.
The inhibitory activities of BBI (2-IV) and STI were estimated by their suppression of the amidase activity of duodenase and trypsin and esterase activity of alpha-chymotrypsin. The enzyme--inhibitor mixture was incubated in the working buffer for 10-15 min, and then the reaction was initiated by addition of the substrate solution.
Determination of the inhibition constants of duodenase by STI, BBI, and the BBI--trypsin complex. The inhibition constants of duodenase by BBI, STI, and the BBI--trypsin complex were determined according to Bieth and Frechin [18]. The essence of this method is in the estimation of remaining enzymatic activity (a) at various concentrations of the inhibitor and constant concentration of the enzyme, when the reaction is initiated by the addition of the substrate into the equilibrium mixture of the enzyme and inhibitor.
The apparent Ki was determined graphically using the equation:
where Ki(app) is the dissociation constant of the EI complex without accounting for the influence of the substrate, [I]0 is the total inhibitor concentration in the reaction mixture, and [E]0 is the total enzyme concentration.
The true value of the inhibition constant was calculated using the equation:
For competitive inhibition, when [S] = Km, the inhibition constant is calculated with rather large error using Eq. (3) [19]:
To attain equilibrium with inhibition of duodenase by native BBI, the mixture was incubated for 10 min.
To study the inhibition of duodenase by the BBI--trypsin complex, the latter was obtained by incubation of equimolar quantities of the native BBI and trypsin for 10 min. It was demonstrated in a separate experiment that the BBI--trypsin complex does not hydrolyze trypsin substrate (3.0 mM). The BBI--trypsin complex was incubated with duodenase for 40 min. It was demonstrated in a separate experiment that the native duodenase does not lose its activity during the experiment.
Competition of duodenase and trypsin for binding to BBI. An equimolar mixture of BBI and trypsin was incubated for 10 min to attain the equilibrium complex. Aliquots were taken for estimation of the trypsin activity (0.125 µM in the cuvette). An equimolar quantity of duodenase was added to the reaction mixture, and aliquots for estimation of the remaining activity of duodenase (0.125 µM in the cuvette) against Suc-Ala-Ala-Pro-Phe-pNA were taken after 2, 5, 10, 15, and 20 min. The activity of trypsin was estimated simultaneously.
In other experiment the BBI--duodenase complex was first formed, its activity was estimated after 7-10 min, and then an equimolar quantity of trypsin was added. The remaining activity was estimated as described above. It was shown that the native duodenase does not lose its activity during the experiment.
Determination of the association rate constant of duodenase with the BBI--trypsin complex. The association of proteinases with protein inhibitors during the initial period of time can be considered as second-order irreversible reactions. If equal concentrations of the enzyme and the BBI--trypsin complex participate in the reaction, kass can be determined by Eq. (4) [20]:
where [E]0 is the initial enzyme concentration and [E] is the enzyme concentration at the moment t.
Mixtures of the enzyme with the BBI--trypsin complex were incubated for various time intervals. The reaction was initiated by addition of the substrate to the reaction mixture.
Competition of duodenase and alpha-chymotrypsin for binding to BBI. An equimolar mixture of BBI (0.2 µM) and alpha-chymotrypsin was incubated for 10 min to form a complex. Aliquots were taken for estimation of the remaining activity of alpha-chymotrypsin against Bz-Tyr-EE (0.125 mM). An equimolar quantity of duodenase was added to the reaction mixture, and then the remaining activity of duodenase against Suc-Val-Pro-Phe-pNA was estimated in 5, 10, 15, and 20 min. Simultaneously, the remaining activity of alpha-chymotrypsin was estimated against Bz-Tyr-EE.
Study of inhibition of alpha-chymotrypsin by the BBI--duodenase complex. A constant concentration (1 µM) of alpha-chymotrypsin and various concentrations of the BBI--duodenase complex (0.097-1.45 µM) were incubated for 50 min. The reaction was initiated by addition of Bz-Arg-EE substrate (0.15 mM).
RESULTS AND DISCUSSION
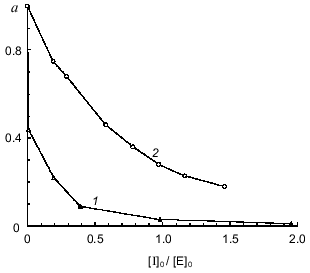
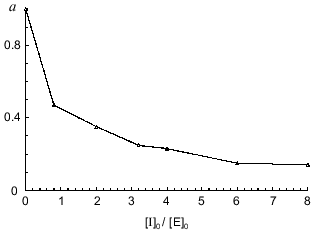
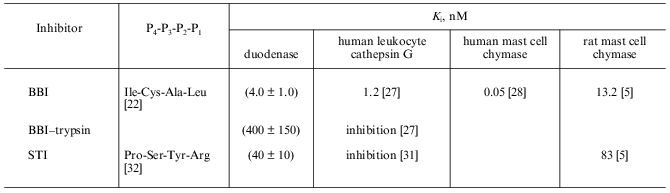
To quantitatively characterize the interaction of Bowman--Birk and Kunitz type soybean inhibitors with duodenase, we estimated and calculated the inhibition constants of duodenase by these inhibitors. The titration curves of duodenase with BBI, the BBI--trypsin complex, and STI are presented in Figs. 1 and 2. In a preliminary experiment we demonstrated that at the ratio [E]0/Ki = 100-1000, one BBI molecule binds to one duodenase molecule. Consequently, only one of two centers of the BBI inhibits the enzyme. The values of Ki(app) were determined from Eq. (1) as the slope of the linear plot of [I]0/(1 - a) versus 1/a. The true inhibition constants obtained using Eq. (2) are summarized in the table. The Ki values correspond to the values obtained from Eq. (3). It can be seen from these data that the classical soybean BBI binds to duodenase one order of magnitude more efficiently than STI.
Fig. 1. Remaining activity of duodenase versus concentration of BBI (0.05 µM duodenase) (1) and concentration of the BBI--trypsin complex (0.5 µM duodenase) (2); 1.0 mM Suc-Val-Pro-Phe-pNA, 0.05 M Tris-HCl, and 20 mM CaCl2 (pH 8.0).
Inhibition constants of serine proteinases by soybean inhibitorsFig. 2. Remaining activity of duodenase versus STI concentration; 0.1 µM duodenase, 1.0 mM Suc-Val-Pro-Phe-pNA, and 0.05 M Tris-HCl (pH 8.0).
A previous study of the substrate specificity of duodenase [21] showed that it has a dual nature: a trypsin-like type of substrate hydrolysis (Lys or Arg in the P1 position) is typical of this enzyme; also, similarly to alpha-chymotrypsin, duodenase efficiently hydrolyses substrates with hydrophobic residues (Phe, Tyr, or Leu) in the P1 position. However, the tryptic substrate specificity of duodenase manifests itself more clearly. For duodenase, the preference in the P1 position decreases in the sequence Lys > Arg > Phe > Leu. Thus, Suc-Thr-Pro-Lys-pNA is hydrolyzed by duodenase 20 times more efficiently than Suc-Val-Pro-Phe-pNA [21].
Duodenase is also shown to be more active against synthetic substrates with extended peptide chains than against “short” substrates. This suggests the existence of an extended substrate-binding site in duodenase [21].
Thus, the Bowman--Birk type soybean inhibitor is able to simultaneously and independently bind to trypsin-like (Lys-Ser site) and to chymotrypsin-like (Leu-Ser site) serine proteinases [22]; it is thus an ideal protein analog of a substrate for subsequent studies of duodenase specificity. Based on the data on the substrate specificity, Lys in the P1 position of the BBI trypsin domain appears to be more preferential for duodenase than Leu in the P1 position of the chymotrypsin domain of this protein inhibitor. However, as will be demonstrated below, the results we obtained disprove this conclusion quite clearly.
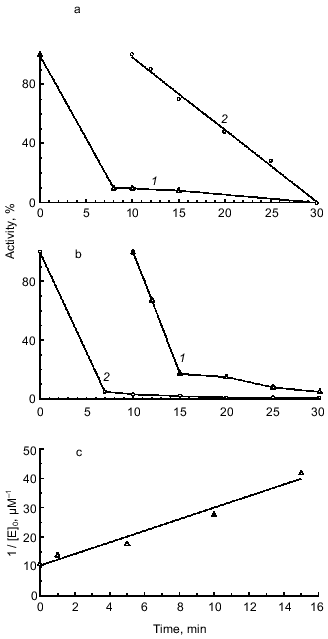
In the next step of our studies, we performed experiments indicating which of the two BBI sites binds to duodenase. As seen in Fig. 3a, duodenase is completely inhibited by the BBI--trypsin complex. Trypsin is not displaced from the complex because the activity of the reaction mixture against Bz-Arg-pNA is zero (a separate experiment showed that duodenase does not hydrolyze Bz-Arg-pNA). However, the time needed for essentially complete inhibition of duodenase (t99%) increases. At the enzyme and inhibitor concentrations 0.125 µM, the BBI--duodenase complex is formed in 10 min, and the trypsin--BBI--duodenase complex is formed in 20 min. The value of t99% is inversely proportional to the association rate constant of the enzyme--inhibitor complex (kass). We calculated by linear anamorphosis (Eq. (4)) of the experimental data presented in Fig. 3a that kass of duodenase with the trypsin--duodenase complex is 2.7·104 M-1·sec-1 (Fig. 3c). Also, while inhibiting duodenase by the BBI--trypsin complex, the Ki increases two orders of magnitude compared to the native inhibitor. This phenomenon cannot be rationalized by steric hindrances arising from the interaction of duodenase with the antichymotrypsin site of BBI because of formation of the BBI--trypsin complex. According to the 3D structure of BBI in solution obtained by NMR, the antitryptic and antichymotryptic sites are far removed from each other to opposite ends of the molecule [23]. Increase in the inhibition constant is most probably related to the fact that BBI binding to trypsin can to some extent break the contacts between the antitryptic and antichymotryptic domains of the BBI molecule. As mentioned earlier, these interdomain interactions promote stabilization of such structural organization of the antichymotrypsin domain of BBI which provides efficient inhibition of chymotrypsin-like proteinases [24, 25].
We also showed that the BBI--duodenase complex retains capacity for complete inhibition of trypsin without appearance of duodenase activity (Fig. 3b). Thus, we conclude that BBI, the double-site classical soybean inhibitor of Bowman--Birk type, binds to bovine duodenase by its antichymotrypsin site (Leu-Ser) and does not use its antitrypsin site (Lys-Ser).Fig. 3. Remaining activity of duodenase versus time of interaction with the BBI--trypsin complex (a); remaining activity of trypsin versus time of interaction with the BBI--duodenase complex (b); 0.125 µM duodenase, trypsin, and BBI, 1.0 mM Suc-Ala-Ala-Pro-Phe-pNA, 3.0 mM Bz-Arg-pNA, 0.05 M Tris-HCl, 20 mM CaCl2 (pH 8.0): 1) active trypsin; 2) active duodenase. c) Anamorphosis of curve 2 from Fig. 3a in coordinates of Eq. (4).
We performed a separate experiment on the formation of the triple complex with the simultaneous presence of equimolar quantities of trypsin, BBI, and duodenase in the reaction mixture. The mixture was incubated for 30 min, and the remaining activity of trypsin and duodenase was estimated. Under these experimental conditions, trypsin is 100% inhibited, and duodenase is 50% inhibited. This can be rationalized by higher affinity of BBI to trypsin (Ki = 0.14 nM [16]) as compared with duodenase (Ki = 2.7 nM). Also, BBI binds first to trypsin (kass = 1.1·106 M-1·sec-1 [26]), and duodenase interacts not with the native BBI, but with its complex. When duodenase is inhibited by the BBI--trypsin complex, Ki increases two orders of magnitude as compared with the native inhibitor. Consequently, the [E]0/Ki ratio decreases by two orders of magnitude, and 100% inhibition cannot be attained at the given concentration of duodenase.
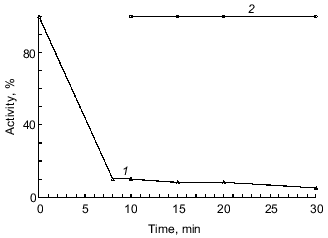
The experiment on inhibition of duodenase by the BBI--chymotrypsin complex demonstrated that when the antichymotrypsin site of BBI (Leu-Ser) is occupied by another enzyme (in this case, alpha-chymotrypsin), BBI does not bind to duodenase (Fig. 4). Thus, we confirmed once again that BBI binds to duodenase by the antichymotrypsin site.
We also demonstrated that alpha-chymotrypsin is unable to displace duodenase from the BBI--duodenase complex at concentration ten times higher than that of the complex. To determine such a high concentration of alpha-chymotrypsin (1 µM), we used Bz-Arg-EE as substrate (0.15 mM). In a separate experiment we demonstrated that duodenase does not hydrolyze Bz-Arg-EE. alpha-Chymotrypsin activity at increased concentrations of the BBI--duodenase complex remains 100%.Fig. 4. Inhibition of duodenase by the BBI--chymotrypsin complex. Remaining activity of duodenase and alpha-chymotrypsin versus time of interaction with BBI; 0.2 µM duodenase, alpha-chymotrypsin, and BBI, 1.0 mM Suc-Val-Pro-Phe-pNA, 0.125 mM Bz-Tyr-EE, 0.05 M Tris-HCl (pH 8.0): 1) active chymotrypsin; 2) active duodenase.
These results suggest that in spite of the clearly manifested trypsin specificity against synthetic substrates, duodenase binds to the antichymotrypsin site of the protein inhibitor BBI, similar to cathepsin G of human leukocytes [27] and chymase of mast cells of human skin [28].
Judging from the amino acid sequence [7], duodenase is homologous to other serine proteinases such as cathepsin G and mast cell chymases. Duodenase also has some overlap in the substrate specificity with the mentioned proteinases.
Study of a series of peptide 4-nitroanilide substrates [29] showed that cathepsin G prefers bulky aliphatic or aromatic amino acids in the P1 position of the reactive site. Suc-Val-Pro-Phe-pNA is the best substrate for cathepsin G (kcat/Km = 6.9·103 M-1·sec-1). Chymases of human and rat mast cells also prefer the Phe residue in the P1 position of the substrate [5, 6].
Among hydrophobic substrates of duodenase with Phe in the P1 position, Suc-Val-Pro-Phe-pNA is the most reactive one, similar to cathepsin G. However, for duodenase kcat/Km is sixfold lower than that for cathepsin G [21]. Also, analogously to other cathepsin G-like serine proteinases [6], duodenase most actively cleaves hydrophobic substrates which have Pro in the P2 position [21].
Duodenase also demonstrates its chymotrypsin-like specificity, interacting with chymostatin: Ki = 60-67 nM (unpublished data). Chymostatin inhibits chymotrypsin-like enzymes with inhibition constant ranging from 17 nM (human mast cell chymase) to 280 nM (human leukocyte cathepsin G) [6]. Chymostatin does not inhibit trypsin-like enzymes including thrombin, plasmin, kallikrein, and trypsin [30].
The Ki values for bovine duodenase, human leukocyte cathepsin G, and human mast cell chymase by soybean inhibitors are presented in the table. BBI is shown to be as highly efficient inhibitor of duodenase as the other cathepsin G-like proteinases. As described above, all these proteinases bind to BBI by its antichymotrypsin site (Leu-Ser), whereas its antitrypsin site (Lys-Ser) does not bind to any chymotrypsin-like serine proteinases.
The basic pancreatic trypsin inhibitor (BPTI), having Lys in the P1 position of the active site [32], does not inhibit duodenase even in concentrations two orders of magnitude higher than the enzyme concentration at [E]0 = 2 µM and on incubation of the enzyme with inhibitor for 30 min. BPTI behaves analogously in relation to chymase [28], and it only slightly binds to human leukocyte cathepsin G (Ki = 1 µM [33]). However, it is interesting to note that BPTI-Sepharose can be a useful affinity sorbent for isolation of both cathepsin G and duodenase [1, 34, 35].
Soybean trypsin inhibitor of Kunitz type (STI) has Arg in the P1 position of the active site and inhibits duodenase and rat mast cell chymase approximately an order of magnitude weaker than BBI.
So, we conclude that duodenase efficiently interacts with a protein inhibitor having Leu in the P1 position of its active site. The efficiency of inhibition lowers when Arg occupies this position. In the case when this position is occupied by Lys, duodenase is not inhibited. Thus, interacting with protein inhibitors, duodenase essentially behaves as a chymotrypsin-like serine proteinase.
This study was financially supported by the Russian Foundation for Basic Research (grant No. 99-04-48135).
REFERENCES
1.Zamolodchikova, T. S., Vorotyntseva, T. I., and
Antonov, V. K. (1995) Eur. J. Biochem., 227, 866-872.
2.Hof, P., Mayr, I., Huber, R., Korzus, E., Potempa,
J., Travis, J., Powers, G. C., and Bode, W. (1996) EMBO J.,
15, 5481-5491.
3.Pemberton, A. D., Huntley, J. F., and Miller, H. R.
P. (1997) Biochem. J., 321, 665-670.
4.Macaldowie, C. N., Mackellar, A., and Huntley, J.
F. (1998) Res. Vet. Sci., 64, 17-24.
5.Katunuma, T., and Kido, H. (1988) J. Cell.
Biochem., 38, 291-301.
6.Powers, I. C., Tanaka, T., Harper, J. W.,
Minematsu, Y., Barker, L., Lincoln, D., Crumley, K. V., Fraki, J. E.,
Schechter, N. M., Lazarus, G. G., Nakajima, K., Nakashino, K., Neurath,
H., and Woodbury, R. G. (1985) Biochemistry, 24,
2048-2058.
7.Zamolodchikova, T. S., Vorotyntseva, T. I.,
Najimov, T. V., and Grishina, G. A. (1995) Eur. J. Biochem.,
227, 873-879.
8.McAleese, S. M., Pemberton, A. D., McGrath, M. E.,
Huntley, J. F., and Miller, H. R. P. (1998) Biochem. J.,
333, 801-809.
9.Schechter, J., and Berger, A. (1967) Biochem.
Biophys. Res. Commun., 27, 157-162.
10.Perona, J. J., and Craik, C. S. (1995) Prot.
Sci., 4, 337-360.
11.Birk, Y. (1976) Meth. Enzymol., 45,
695-739.
12.Kunitz, M. (1947) J. Gen. Physiol.,
30, 311-320.
13.Shonbaum, J. R., Zerner, B., and Bender, M.
(1961) J. Biol. Chem., 236, 2930-2935.
14.Chase, T., and Shaw, E. (1970) Meth.
Enzymol., 19, 20-27.
15.Odani, S., and Ikenaka, T. (1977) J.
Biochem., 82, 1513-1522.
16.Larionova, N. I., Gladysheva, I. P., Tikhonova,
T. V., and Kazanskaya, N. F. (1993) Biochemistry (Moscow),
58, 1046-1052.
17.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and
Randall, R. J. (1951) J. Biol. Chem., 193, 265-275.
18.Bieth, J. (1974) Bayer-Symposium V
“Proteinase Inhibitors”, Springer-Verlag, Berlin, pp.
463-469.
19.Brandt, R. B., Laux, J. E., and Yates, S. W.
(1987) Biochem. Med. Metabol. Biol., 37, 344-349.
20.Larionova, N. I., Kazanskaya, N. F., and
Yakubovskaya, R. I. (1979) Biokhimiya, 44, 3-17.
21.Zamolodchikova, T. S., Sokolova, E. A.,
Alexandrov, S. I., Mikhaleva, I. I., Prudchenko, I. A., Morozov, I. A.,
Kononenko, N. V., Mirgorodskaya, O. A., Da, U., Larionova, N. I.,
Pozdnev, V. F., Ghosh, D., Duax, W. L., and Vorotyntseva, T. I. (1997)
Eur. J. Biochem., 249, 612-621.
22.Odani, S., and Ikenaka, T. (1972) J.
Biochem., 71, 839-848.
23.Werner, M. H., and Wemmer, D. E. (1992)
Biochemistry, 31, 999-1010.
24.Ascenzi, P., Amiconi, G., Bolognesi, M.,
Menegatti, E., and Guarneri, M. (1990) J. Mol. Recognition,
3, 192-196.
25.Odani, S., and Ikenaka, T. (1978) J.
Biochem., 83, 747-753.
26.Gladysheva, I. P. (1994) Author’s abstract
of Candidate’s dissertation [in Russian], Moscow State
University.
27.Gladysheva, I. P., Larionova, N. I., Gladyshev,
D. P., Tikhonova, T. V., and Kazanskaya, N. F. (1994) Biochemistry
(Moscow), 59, 371-374.
28.Ware, J. H., Wan, X. S., Rubin, H., Schechter, N.
M., and Kennedy, A. R. (1997) Arch. Biochem. Biophys.,
344, 133-138.
29.Yoshida, Y., Everitt, M. T., Neurath, H.,
Woodburg, R. G., and Powers, J. C. (1980) Biochemistry,
19, 5799-5804.
30.Umezawa, H., Aoyagi, T., Morishima, H., Kunimoto,
S., Matsuzaki, M., Hamad, M., and Takenchi, T. (1970) J.
Antibiot., 23, 425-430.
31.Virca, G. D., Metz, G., and Schnebli, H. P.
(1984) Eur. J. Biochem., 114, 1-9.
32.Kassel, B., and Laskowski, M. (1965) Biochem.
Biophys. Res. Commun., 20, 463-468.
33.Starkey, P. M., and Barrett, A. J. (1976)
Biochem. J., 155, 255-263.
34.Saklatvala, J., and Barrett, A. J. (1980)
Biochim. Biophys. Acta, 615, 167-177.
35.Tikhonova, T. V., Gladysheva, I. P., and
Larionova, N. I. (1995) FEBS Lett., 362, 225-228.