REVIEW: UGA: a Dual Signal for 'Stop' and for Recoding in Protein Synthesis
W. P. Tate*, J. B. Mansell, S. A. Mannering, J. H. Irvine, L. L. Major, and D. N. Wilson
Department of Biochemistry and Centre for Gene Research, University of Otago, P.O. Box 56, Dunedin, New Zealand; fax: +64 3 479-7866; E-mail: warren.tate@stonebow.otago.ac.nz* To whom correspondence should be addressed.
Received July 7, 1999
UGA remains an enigma as a signal in protein synthesis. Long recognized as a stop signal that is prone to failure when under competition from near cognate events, there was growing belief that there might be functional significance in the production of small amounts of extended proteins. This view has been reinforced with the discovery that UGA is found at some recoding sites where frameshifting occurs as a regulatory mechanism for controlling the gene expression of specific proteins, and it also serves as the code for selenocysteine (Sec), the 21st amino acid. Why does UGA among the stop signals play this role specifically, and how does it escape being used to stop protein synthesis efficiently at recoding sites involving Sec incorporation or shifts to a new translational frame? These issues concerning the UGA stop signals are discussed in this review.
KEY WORDS: recoding, selenocysteine, UGA, frameshifting, protein synthesis, Sec, translation termination, antizyme, RF2, release factor, genetic code, stop signal
UGA CODES FOR STOP AND FOR AMINO ACIDS
The universal genetic code identified three codons as specifying stop in protein synthesis--UAA, UGA, and UAG. Since these were shown to be used in bacteria, amphibians, and mammals [1] the concept of universality was established. However, in part this was directed by the small number of organisms that were amenable for study at that time. As organisms from different environments have come under scrutiny and organelles have been studied, it has become evident that the code is not universal and that the stop signals are among the most variable aspects of these alternative variant codes. For example, Mycoplasma species use UGA as a code for tryptophan [2], and most mitochondria also have captured this signal for this amino acid. Does this mean UGA is the least desirable of stop signals and is dispensed with when some evolutionary pressure arises? It seems not. Other organisms such as Tetrahymena species have retained UGA for stop but instead use UAG and UAA for glutamine. Rather, a particular organism can tailor its codon usage according to other factors impinging on it, such as a drift in the base composition of its DNA ((G + C)-content). With an (A + T)-rich genome in mitochondria one can understand why UGA rather than UGG is the predominant codon for tryptophan.
There are indications that organisms not only accommodate UGA as a mainstream signal, but also suggestions that UGA has special characteristics to provide these organisms flexibility to respond physiologically to various environments. For example, in Escherichia coli UGA is used at natural termination sites in approximately one out of every three genes, as compared with UAA (one in two) and UAG (only one in ten genes). In mammals UGA is the most common stop signal, although there is a more even distribution of the use of the three signals at gene termination sites in these organisms [3]. The discovery of UGA as a code for selenocysteine in a wide range of organisms suggests that this is an ancient mechanism, perhaps when pre-oxygen environments were more accommodating to the selenol group at the active centers of proteins. In modern environments the selenol group is susceptible to oxidation and may therefore be gradually disappearing. Selenium, whilst stable in the +4 oxidation state under environmental conditions, exists in the -2 oxidation state in biological macromolecules. The selenol group (pKa = 5.2) is fully ionized at physiological conditions and therefore fully active as a powerful nucleophile. In contrast, the substitute thiol of most modern enzymes is not fully ionized in the physiological pH range. To achieve comparable activity, a thiol (pKa = 8 or greater) must attain an unusually low pKa by virtue of its specific local environment. Does this mean we are seeing a stop codon takeover of what was once an important codon for an amino acid, rather than observing a steady state where UGA has been captured for specific instances to encode the amino acid?
We may well have now attained a steady state where the selenoenzymes have ecological niches, and the UGA encoding two distinct events endures within a genetic system, even though this may have been part of a takeover process. Relevant to this discussion is how an organism can accommodate UGA as a dual signal when clearly there will be competition between the separate decoding processes. Similarly UGA is found at recoding sites not only where selenocysteine is incorporated but where an alternative genetic event such as misincorporation of an amino acid or translational frameshifting occurs. Here the UGA stop signal fails, sometimes so badly that one out of every two ribosomal passages will take the alternative route.
A SPECIAL FACTOR FOR DECODING UGA IN BACTERIA
One puzzling feature in bacteria is the retention or acquisition of a special factor to decode UGA as a stop signal. Mammals and other eukaryotes have one release factor, eRF1, to decode all three stop signals, and a single site on the factor recognizes them [4]. In contrast, bacteria have a release factor to recognize UAA and UAG, namely RF1, but an unique factor RF2 for UGA, although this factor can also recognize UAA. Indeed genetic selectionhas derived a factor variant that can recognize all three codons [5], and the yeast mitochondrial factor mtRF1 can recognize UGA on bacterial ribosomes [6]. Clearly then it is not difficult to derive a single factor that could recognize all three stop signals in bacteria. This suggests that there been some selective pressure to retain a separate factor for the decoding of UGA. The expression and the activity of the UGA decoding factor RF2 is controlled in a highly sophisticated way in contrast to the other factor, RF1. It has one of the now classic recoding sites in its gene, providing an exquisite mechanism to control its synthesis [7], and this seems to have been conserved in most of the prokaryotes species examined to date. In addition, the activity of the factor is also apparently controlled by a conformational switch linking two functional domains for decoding UGA and for hydrolysis of the completed polypeptide [8]. Interestingly this region has now been found to contain a motif that is highly conserved between all known decoding release factors [9]. Bacterial RF1 seems not to be so sensitive to this conformational switch and thereby subject to activity regulation. These observations seem to indicate that bacteria have undergone positive selection to retain a special factor for UGA and have evolved special mechanisms to regulate its activity. It may mean that there are aspects to the role of the UGA signal that are beyond our understanding at present.
THE STOP SIGNAL IS A SEQUENCE ELEMENT
Why is UGA, rather than the other stop codons, used as a dual signal in protein synthesis for stop or another event? The answer to this seems to be found when we examine its role as a stop signal. The key here is the realization that the signal for stop in protein synthesis has elements upstream and downstream that strongly influence its strength. We have shown that the base following the stop codon (+4) is a strong determinant in the efficiency by which a stop signal is decoded [10, 11]. Moreover, the +4, +5, and +6 bases following UGA will cross-link to the decoding factor, RF2, from a zero length thio-U moiety in the mRNA [12, 13], in addition to the first position of the stop codon (thio-UGA) [14]; and the +6 base can influence the efficiency of decoding by the factor [15]. This implies a contact between the factor and at least 6 bases of the mRNA during the decoding process. Isaksson and coworkers have shown the last two codons before the stop codon influence near cognate misincorporation of amino acids at UGA stop codons. The critical parameter is the coding potential of the two codons, that is the nature of the amino acids they encode and, in some cases, the isoaccepting species of tRNA at the -1 codon [16, 17]. The implication of these observations is that the decoding factor RF2 enters the A site of the active center of the ribosome, spanning the decoding site of the small subunit and the peptidyltransferase center of the large subunit, and it makes contact with the last amino acids of the completed polypeptide chain and the ultimate tRNA. Whether these are specific interactions between the release factor and the other ligands in the center, or whether there are some combinations of these ligands that allow a lower rate of association of the factor with the site just from steric hindrance, remains to be determined. However, the two upstream codons have the net effect of contributing to the strength of the termination signal. We envisage that bacteria have a twelve base sequence element influencing signal strength with a core of four bases, the codon and the base following. Are these parameters specific to stop signals containing only UGA or relevant to all three stop codons? While these considerations are indeed relevant to all signals, the UGA-containing signals seem to be more sensitive to the parameters that determine the strength of the signal. This may be the reason why UGA is able to act as a dual signal, where there is strong competition between the stop mechanism and an alternative event, and why UGA superficially appears to be a poor choice for a mainstream stop signal.
SENSITIVITY OF UGA TO THE STRENGTH OF THE STOP SIGNAL SEQUENCE
ELEMENT
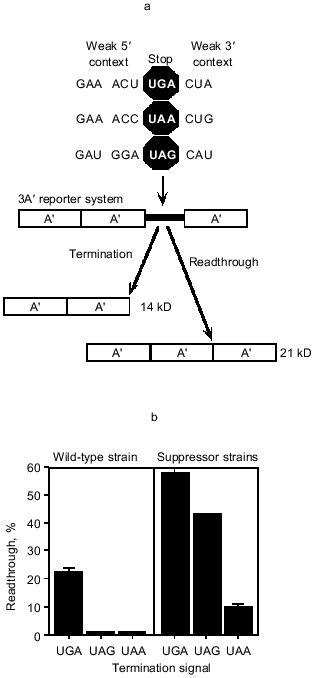
One of the implications of a sequence element, rather than a codon, determining the strength of the termination signal is that once the important parameters are understood it is possible to design signals of varying strengths. We have used our current knowledge to design the weakest possible signals for each of the three stop codons. These have been placed as the termination site between repeats of the gene sequence for the immunoglobulin-binding domain of Staphylococcus aureus protein A, in a three-domain reporter system. If the stop signals act efficiently, then a two-domain 14 kD protein is produced, whereas failure of the stop signal allows an additional domain to be translatedand a 21 kD protein is synthesized. This is illustrated in Fig. 1a. We have compared how release factor decoding of each of the three 'weak' signals competes with near cognate decoding by aminoacyl-tRNAs. Essentially the kinetics of decoding the signals as stop will influence whether a near cognate event can occur. In addition to measuring these effects in wild-type strains, where only near cognate coding will be competing with the stop signal decoding, the experiments have also been carried out in strains carrying specific suppressors for each stop signal, where there is direct competition between two cognate decoding events, stop and amino acid incorporation. A comparison of the three signals shows clearly that UGA is much more sensitive to the weakening of the signal than the other two signals. Against near cognate events, the UAA and UAG signals fail in only one or two passages per hundred passages of ribosomes, while the UGA signal fails about one in four times. Under more direct competition with suppressor tRNAs, the failure rates increase with UGA now failing in six out of each ten passages, UAG in four out of ten, and UAA in one out of ten. This is illustrated in Fig. 1b. This study gives an indication as to why UGA might be the preferred signal to feature at sites where an alternative translation event is important. UGA-containing stop signals seem particularly sensitive to the strength of the stop sequence element, and that may reflect how the RF2 protein fits into the active center and is stabilized within it compared with RF1, which has the capacity to recognize the other two signals. Indeed, it has been claimed that in E. coli most UAA signals are recognized by RF1 [18]. 10Sa RNA marks incompletely synthesized proteins for degradation by attaching a short peptide tag to the C terminus [19]. The 10Sa RNA acts as the first tRNA as well as the mRNA for the peptide tag. UGA is the stop codon for 10Sa RNA in only two of the fifty-six organisms available for analysis [20], suggesting UGA may be selected against as the stop signal.
Fig. 1. Expression of weak termination signals in the 3A´ reporter system. a) The 3A´ reporter system was used to investigate termination signal strength. Sequences that were predicted to be poor for termination for specific stop codons were placed upstream and downstream of that codon for each of UGA, UAG, and UAA. The termination signals were cloned between the 2nd and 3rd A´ domains in the 3A´ reporter plasmid. In vivo expression from this plasmid results in two products, a 14 kD termination product and a 21 kD readthrough product. The efficiency of the competing termination and readthrough events determines the molar ratio of the two products. b) Expression of the 3A´ reporter system containing weak UGA, UAG, and UAA termination signals (as presented in Fig. 1a) in wild-type (left) and suppressor strains (right). Error bars are the standard error of the mean for each construct.
UGA AND TRANSLATIONAL FRAMESHIFTING
The first observations of unusual, non-linear translation during the synthesis of phage MS2 and T7 proteins were reported two decades ago [21, 22]. An enormous range and number of programmed frameshift sites have now been documented and characterized, and while most have been associated with viruses, there are several noteworthy cases within essential genes in the prokaryotic and eukaryotic genomes.
Regulation of expression of the bacterial RF2 gene at the frameshifting site. The first frameshift site to be discovered in an essential cellular gene was that in bacterial RF2 [7] and it was notable because the key initial feature recognized was an in-frame UGA codon, early in the coding region of the protein. This was discovered when the gene was first cloned and the open reading frame was insufficient to encode a protein of the size of the factor. Sequencing the then precious RF2 protein, purified from bacteria, provided the N terminal forty-four amino acids. Remarkably this revealed that the start of the RF2 gene was upstream of an in-frame UGA, and that the code for the protein sequence continued in the +1 frame from this UGA until the end of the reading frame. Since the recombinant clone gave 50-fold enhanced activity of the factor, and the expected size of expressed protein, it was clear that this was not simply an aberrant clone. Once the trivial explanation of a sequencing error had been eliminated, an exquisite mechanism for how the gene was regulating its own expression unfolded. RF2 recognized UGA stop signals and here was an internal in-frame UGA in its own mRNA. Faithful decoding of this signal by existing RF2 would prematurely terminate the synthesis of new release factor, and a translational frameshift event in the forward direction (+1) would be required to complete the synthesis of a functional protein [7]. The implication was that the kinetics of recoding the internal UGA stop signal determined the subsequent fate of the translation of the RF2; fast kinetics for recognition of the UGA as stop would commit the site to a termination pathway, while slower kinetics would increase the probability for the alternative to occur. Subsequent studies of the site indicated that this mechanism of self regulation by RF2 was indeed occurring [23, 24], and that remarkably the stop signal was failing every second or third ribosomal passage of the mRNA. UGA stop signals at the end of genes were not known to be inefficient to that degree, despite occasional reports of small amounts of readthrough products at UGA signals [25]. Why was UGA failing so badly then at the RF2 frameshift site?
Initial focus was on the identification of cis elements in the mRNA as facilitators of the alternative frameshift pathway, and little attention was given to the possibility that the UGA might itself be a significant contributor to the regulation. The discovery of a Shine-Dalgarno sequence upstream from the site at an unusual spacing identified such a cis determinant and the nature of the codon at position 25 (immediately before the stop codon) was also important [26]. There is no doubt that these elements set up the frameshift event at this site in a way that is not found at UGA termination sites. Changes to the sequence or spacing of the Shine-Dalgarno element or the 25th codon all significantly reduced the competitiveness of the frameshift event against the stop at the UGA signal [26]. This means these elements allow slippage of the decoding complexes, at the active center of the ribosome, one base forward on the mRNA so as to take the UGA signal out of frame. It could occur while the A site was still either empty awaiting the decoding RF2, or filled but not in a competent state for the release reaction. Indeed, increasing the concentration of the cellular RF2 can dramatically lower the competitiveness of the frameshifting event so that it is undetectable. Conversely, a disabled RF2 which can bind to the ribosomal site but not mediate release allows 100% frameshifting [24]. This gives credence to the kinetic argument that it is the rate of commitment to the termination pathway that determines if frameshifting occurs, despite the site being so carefully crafted for the frameshift event.
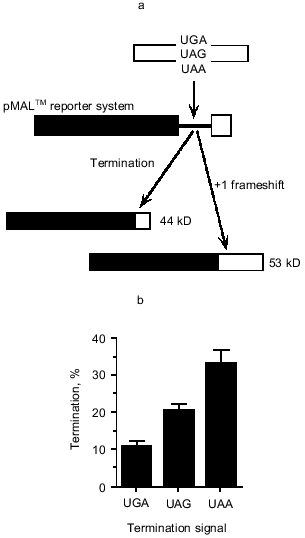
Such an argument brings attention back onto the UGA termination signal because it implies that the efficiency of reading this signal by the release factor is a critical determinant. At the time of these studies it was not appreciated that the termination signal might be an extended element, rather than just a codon somewhat influenced by context. However, even focussing on the codon alone meant UGA could be compared with the other two stop codons in their ability to function at the site. We used a malE reporter gene with an excellent immunological detection system for the maltose-binding protein to determine whether UGA was especially important for regulation at the site. This is illustrated in Fig. 2a. Termination at the site gave a 44 kD protein, but if the +1 frameshifting event occurred then a 53 kD protein was produced. These products were well separated on SDS-PAGE and the subsequent Western Blot gave beautifully clean specific banding patterns that were easily quantitated. In this system, with the RF2 frameshift site cloned behind the malE gene and expressed in vivo at a high rate under the control of a strong promoter, the competitiveness of the frameshifting event was significantly enhanced over that of the natural site in the RF2 gene. Here as seen in Fig. 2b, frameshifting occurred at ~90% of ribosomal passages along the mRNA (UGA; 10% termination) [10], as compared with 30-50% of passages when the RF2 site is expressed with its own promoter. We believe this can be explained simply by the demand placed on the RF2 protein and its local concentration at the frameshift site when the mRNA is present in such high amounts due to the strong promoter. Hence we are simulating a situation of high apparent need for the RF2 protein. What happens when the UGA is replaced by UAG and UAA? Clearly the termination event is more competitive; these codons reduce frameshifting from ~90 to ~67% with UAA and ~80% with UAG (Fig. 2b) [10]. The rate of decoding the stop signals in these cases must be higher and this may reflect a higher efficiency of RF1 to commit to the termination pathway than RF2. Both release factors are able to decode the UAA signal, so the concentration of the decoding ligands (RF1 and RF2) at the site might also be the important determinant.
Now it is known that the stop signal is an extended element and that upstream and downstream elements contribute to its efficiency of decoding; how does this affect the regulation at the RF2 frameshift site? The codon immediately upstream of the stop codon (CUU) is an important part of the cis element facilitating frameshifting and so there may be little leeway here to evolve to an amino acid contributing to optimum termination efficiency of the UGA. CUU has not been examined, but the CUC leucine codon is a good context for termination efficiency of a following UGA [16]. The -2 codon in the frameshift site provides only the spacing to the Shine-Dalgarno sequence and therefore its coding potential could theoretically evolve to be a regulator of the termination efficiency of the downstream UGA. Indeed among 20 different organisms the amino acid at this site is variable, in contrast to the encoded leucine at the -1 codon position [27]. A tyrosine codon (UAU) is found at the -2 codon position in the E. coli RF2 frameshift site; this amino acid, unlike the charged amino acids in this position, does not particularly favor or disfavor efficient stopping at downstream UGAs [17]. This indicates that perhaps the upstream sequences to the UGA at the frameshift site are not having a major influence on the efficiency of UGA decoding as stop, in that they are providing neither a particularly strong nor particularly weak element for the signal.Fig. 2. The pMAL-RF2 frameshift window termination assay. a) In the pMAL-RF2 frameshift window termination assay, protein synthesis termination is in competition with a +1 frameshift event. The gene encoding maltose-binding protein is followed by the RF-2 frameshift window which contains the termination context of interest. Termination will result in a 44 kD protein, while frameshifting produces a 53 kD protein; these products can be detected by SDS-PAGE followed by immunoblotting and anti-MBP detection. The termination efficiency of the signal in competition with +1 frameshifting can then be determined. b) Expression of the pMAL-RF2 frameshift window termination system containing UGAcua, UAGcua, and UAAcua termination signals. Error bars are the standard error of the mean for each construct. These experiments were performed in E. coli strain FJU112, which contains wild-type ribosomes and no suppressor tRNA species.
The same is certainly not true however, for the downstream part of the signal. Consider the strength of the stop signal just from the perspective of the +4 base, the position that provides a significant contribution to the efficiency of decoding stop signals, and shows the strongest bias (outside of the codon itself) in the bacterial stop signals at natural termination sites (U is highly favored and C is highly disfavored). UGAC signals are the weakest of the twelve possible, if a four base stop signal is considered. Hence the weakest context is found at the RF2 frameshift site, contributing to the poor performance as a stop signal. We know that the RF2 factor makes contact with the bases up to +6, but apparently not beyond (thio-U in positions +7 to +10 of the mRNA do not cross-link to RF2), and so we examined the frequency of the +4 to +6 bases of the frameshift site to see whether they were also found at natural termination sites after a UGA and whether they conferred a particularly weak signature to the UGA stop signal. A clear indication came from an examination of the frequency of the native (RF2 frameshift) UGACUA sequence at the ends of genes in the E. coli genome. Only three such sequences are found at the end of putative genes in the genome; but all are open reading frames yet to be confirmed as functional genes and so this number could be as low as zero. This is by far the lowest frequency of occurrence of any UGANNN sequence at natural termination sites and indicates there has been a strong selection against this sequence. We were not surprised therefore to find that UGACUA was the weakest of the set of UGACNA and UGACUN stop signals. The +5 position seems relatively neutral with respect to the efficiency of the UGA in committing to the termination pathway but the A in the +6 position confers a significant weakening of the signal to commit to this pathway [15].
What does this mean for the role of UGA at the RF2 frameshift site? Perhaps it is not just a passive part of the site, with the facilitators of frameshifting alone determining the regulatory mechanism. Rather a frameshifting mechanism has arisen because of the cis elements at the site, but the critical key to the regulation of RF2 production is the efficiency of decoding of the stop signal at the 26th codon position. If this decoding efficiency is high because the interacting ligands are upregulated or if the stop signal itself is not a particularly weak signal, then the positive facilitators of frameshifting become particularly ineffective. This raises the interesting question as to whether the frameshifting elements arose around a particularly weak stop signal, or whether the element arose first and then a stop signal was added at a later date to provide more precise regulation. Both mechanisms are possible, but the protein may initially have been shorter at its N terminus. The evolution of cis elements around a weak stop signal not part of an existing gene would then have allowed an N terminal extension of the protein and a specific regulatory mechanism for its synthesis. This must have conveyed some advantage to the cell, indeed overexpression of RF2 is toxic and rapidly leads to the excess RF2 being in an inactive conformation. To date, the known functional sites of RF2 seem to be in the middle of the protein or at its C terminus, for example at nucleotide positions 170-210 for the anticodon domain [28], and positions 240-280 for the ribosomal binding domain and potential peptidyl-tRNA hydrolysis domain [8]. The N terminal region of RF2 from different bacteria is the most variable part of the protein. It is interesting that this frameshifting regulatory mechanism seems to be of ancient origin, and while it has been retained in many subsequent lineages it has been lost in three independent lineages [27].
Clearly this is an example of how UGA is doing more than just stopping protein synthesis and an insight into how this stop codon has taken on a greater role in cellular physiology is exquisitely revealed. This is by far the best studied example of UGA functioning at a site other than a natural termination site, at least from the perspective of the stop signal itself.
Regulation of expression of the ornithine decarboxylase antizyme gene in eukaryotes. There is an example of UGA functioning at a frameshifting site in eukaryotes which has analogies to the RF2 bacterial site in that it is a +1 shift, and it serves to regulate the expression of the protein, ornithine decarboxylase antizyme, in whose mRNA the site is found. Where the bacterial example was a protein regulating its own synthesis, in this case an essential mediator of cell growth and differentiation, polyamine, is a key regulator of the site, and the antizyme protein produced is a regulator of the polyamine synthesis pathway [29].
We are not so advanced in our understanding of the stop signal in eukaryotes and therefore it is not possible to undertake such a detailed and critical analysis of the role of the UGA at this site. However some useful conclusions can be made. Firstly, the same factor is involved in decoding the stop signal no matter whether UGA, UAA or UAG is within the signal at the site (eukaryotic eRF1 recognizes all three codons [4]), in contrast to the situation at the RF2 frameshift site. Therefore this feature cannot explain any sensitivity of UGA to competition, compared with the other two codons. Nevertheless, there are indications that UGA-containing signals in mammalian contexts are subject to failure [25]. Hence UGA-containing signals may be more sensitive to upstream and downstream contexts just like the prokaryotic signals. A detailed comparison of the mechanism of how the mammalian cis elements of the antizyme mRNA function in mammals, and in the fission and budding yeasts, have revealed what is important for the frameshifting event. The stop codon is critically important for the frameshift event and UGA allows more frameshifting than UAG or UAA [30]. Some sense codons (mainly rare codons) can partially substitute for stop codons in supporting frameshifting and these differ between the mammalian and yeast systems. Here this may reflect a slow rate of commitment to the canonical decoding pathways (either termination with UGA or amino acid incorporation with rare sense codons) thereby allowing the alternative frameshifting event to occur.
The cis stimulatory elements identified at the antizyme mRNA in addition to the UGA stop signal include a pseudoknot beginning four bases downstream [31]. This may be the equivalent to the Shine-Dalgarno element in the RF2 site that distorts the orientation of the mRNA at the A site of the decoding center in the bacterial ribosomal active center. In addition, an upstream element has been identified that has a comparable effect on frameshifting efficiency to the pseudoknot in mammals and in the budding yeast Schizosaccharomyces pombe. However, the pseudoknot in the genetic background of the fission yeast Saccharomyces cerevisiae is more effective in promoting frameshifting and the upstream element has no effect [31]. The sequence immediately upstream of the UGA stop codon in the antizyme site is conserved between mammals and Drosophila [32]. What does our current understanding of the mammalian or yeast termination signals tell us about the possible involvement and importance of the UGA-containing signal at the site? Despite an earlier belief that the nature of the stop signal was perhaps not really an important discriminator for this frameshift site, a conclusion derived from in vitro experiments [30], Atkins and colleagues now point to the fact that all eukaryotic antizymes identified to date have retained UGA at the site [32]. Clearly there may be an advantage in vivo of UGA over the other stop codons for fine modulation of the translational events at the site, thereby providing the antizyme protein in amounts that allow for optimum polyamine concentrations in eukaryotic cells.
Are there any clues to the strength of the UGA signal within the antizyme site? In stark contrast to prokaryotes, a U in the base following the stop codon of mammalian signals has a strong negative effect on the efficiency of termination [11]. Either of the two purine nucleotides in this +4 position is a strong positive modulator of the efficiency, while C, as in prokaryotes, weakens the signal. What nucleotide is found at the antizyme site in this position? In all cases U is found at +4 giving a definite signature of a weak stop signal. Moreover, the sequences downstream of this position (+6 to +10) are strongly pyrimidine rich, and we have recently found from in vitro experiments that such a string of pyrimidines also significantly weakens further the UGA stop signal (McCaughan and Tate, unpublished). These observations support the concept that a particularly weak UGA-containing stop signal is functioning at the antizyme frameshifting site, and is presumably contributing to a slow rate of commitment to the termination pathway. This provides the window of opportunity for the alternative frameshifting event to occur. While there is less compelling data to date that the -1 and -2 codons have major effects on termination in eukaryotes compared with the prokaryotic studies, it is of interest that one of the major stimulatory elements for antizyme frameshifting encompasses the -1 to -3 codons. Despite the fact that it appears that the nucleotide sequence and not the coding potential is important, the effect of this element may also relate to it being the upstream part of the termination signal. This frameshifting site may assist in the characterization of the mammalian sequence element, and at the very least it provides a strong impetus to uncover further subtleties of the composition of the eukaryotic termination signal.
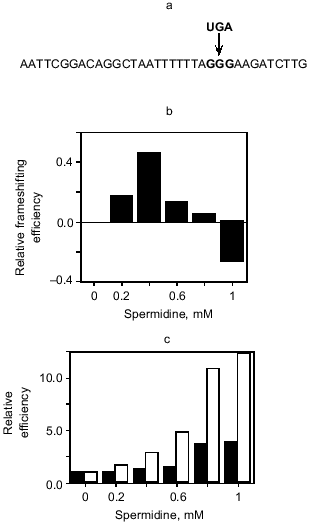
Could polyamines affect the eRF1-mediated decoding of the UGA stop signal at the antizyme frameshifting site? We have investigated this indirectly by substituting the equivalent GGG codon of the HIV-1 gag-pol frameshifting site with UGA as it would be in the antizyme site (Fig. 3a). HIV-1 has a -1 frameshifting mechanism and polyamine does not affect frameshifting with the native sequence. Remarkably, polyamine now has a modest effect on frameshifting efficiency in the HIV UGA construct, initially stimulatory, and then inhibitory at higher concentrations (Fig. 3b) [33]. This gives an indication that polyamines may be affecting negatively the eRF1-mediated decoding of the UGA signal at the antizyme site, thereby slowing the commitment to the termination pathway and giving greater opportunity for frameshifting to occur. Moreover, investigation of the bacterial RF2 frameshift site in rabbit reticulocyte lysate also revealed a polyamine effect on the eRF1-mediated decoding of the UGA signal at that site. Both readthrough of the stop codon, as a result of misincorporation of an amino acid through a near cognate decoding event, and +1 frameshifting were stimulated (Fig. 3c). These are clear signatures of the failure of the stop signal and give further credence to the idea that the polyamine is affecting the commitment to the termination event at the frameshift sites.
Fig. 3. The effect of spermidine on frameshift efficiency. a) The HIV gag-pol -1 frameshift site used for translational assays in rabbit reticulocyte lysate. Proteins were translated in the presence of [35S]methionine and were detected using a phosphoimager after separation by SDS-PAGE. Two constructs were used, one with the HIV GGG codon at the site of frameshifting and another where the native GGG was replaced with UGA. Frameshifting at UGA or GGG was measured with respect to total translation of the construct; e.g., for the UGA construct this is (frameshift product)/(termination + frameshift products). The proportion of frameshift product was then measured relative to the frameshifting observed at 0 mM spermidine. b) This graph illustrates the difference in spermidine effect upon frameshifting at UGA and GGG frameshift sites. The values are for (relative frameshifting at UGA) - (relative frameshifting at GGG). c) Events at the RF2 +1 frameshift site as measured in rabbit reticulocyte lysate translation assays. Frameshift, readthrough and termination products were detectable, and frameshift and readthrough products were measured relative to termination. The efficiency of these events at different concentrations of spermidine is presented relative to that measured at 0 mM spermidine. The black bars are (frameshift product)/(termination product) while the white bars are (readthrough product)/(termination product) for the same translation reaction.
UGA AND SEC INCORPORATION
Competition for decoding UGA as stop or Sec during synthesis of bacterial formate dehydrogenase H. Selenocysteine (Sec) is a cysteine in which the thiol group is replaced by a selenol group and is now regarded as the 21st amino acid [34]. A number of enzymes and proteins across the prokaryotic, eukaryotic, and archaebacterial kingdoms contain selenocysteine. Just over 10 years ago a startling observation was made that the code for Sec in the E. coli formate dehydrogenase H gene, fdhF, was an in-frame TGA [35], as was the Sec codon in the mammalian glutathione peroxidase gene [36]. It was convincing in that the TGA codon in the gene sequence for glutathione peroxidase was colinear with Sec and the surrounding amino acid sequence in the active site of the enzyme. There were several important aspects to this discovery. Firstly there must be a specific cellular machinery for Sec biosynthesis and its co-translational incorporation. The Böck laboratory has almost single handedly elucidated this mechanism in bacteria [37]. The other major point of interest, and relevant to this discussion, is how UGA can encode both Sec and stop within the same cellular milieu.
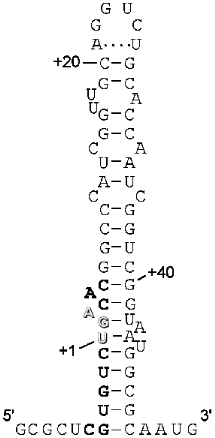
The key point to this dual signalling is whether there is competition between the stopping mechanism and the Sec incorporation mechanism, or whether somehow the UGA at this site is precluded from acting as a termination signal. In a sense this is similar to competition between decoding release factors and suppressor tRNAs in a normal suppression scenario. However, there are some special features of this competition. Böck and colleagues have shown there is a special prokaryotic elongation factor, SELB, equivalent to elongation factor EF-Tu, and this factor binds to a special structural element (stem loop) just downstream from the in-frame UGA in the fdhF mRNA, along with the Sec-specific tRNA (SELC) which has an anticodon recognizing the UGA codon. Tethered to the incoming mRNA, this machinery has an enormous advantage over the decoding release factor, RF2, because it ensures the interaction of SELB·Sec-tRNA with the mRNA at the A site of the ribosome. The structure of the stem loop has been determined by chemical and enzymatic probing and these studies suggest a tertiary structure consisting of two domains [38]. The lower domain involves the UGA codon in a distorted double stranded region (Fig. 4). It has been suggested that this functions to entrap the UGA codon [38] and 5´ and 3´ elements of the stop signal to prevent its recognition by RF2, although it is difficult to envisage how this might occur since the secondary structure must be disrupted when the mRNA is in its channel in the decoding site. Certainly when this lower helical region is disrupted by mutagenesis Sec incorporation does decrease modestly, consistent with RF2 having better access to the codon [39]. The spacing of the upper domain of the stem loop which binds SELB is also critical to Sec incorporation.
Hüttenhofer and Böck have proposed a model for the interaction of the SELB·GTP·Sec-tRNA complex with the ribosome. The ternary complex is proposed to be in a pre-competent state; binding to the stem loop of the mRNA induces a conformational change in SELB, which enables a productive interaction with the ribosome. The formation of the cognate codon--anticodon interaction would then promote Sec incorporation. Experimental evidence supports the formation of the quaternary complex with mRNA off the ribosome [40]. This presents a daunting task for the competing release factor to access the UGA codon and decode it as a stop signal. The expectation was that Sec incorporation would be very efficient and termination at the site would be at best modest, if functioning at all. It was a surprise to us in our initial studies that we could only measure termination at the site and Sec incorporation was almost undetectable (Mansell and Tate, unpublished data). Clearly, despite the impressive mechanism of the Sec incorporation system it was not completely occluding RF2 decoding at the site. Decoding of the codon immediately upstream of the UGA requires the lower domain of the stem loop to be unwound, and of course decoding of the UGA would also necessitate this codon being single stranded. This suggests the UGA codon and larger signal is not involved in any secondary structure at, or immediately prior to entering, the A site of the ribosomal active center. Non-specific suppression at the site is prevented in the absence of selenium, SELB, or Sec-tRNA. This suggests that RF2 is able to recognize the UGA efficiently if one component of the Sec incorporation machinery is lacking. It may be that a good competition is only set up when the UGA of the Sec incorporation site in the mRNA arrives at the decoding site with the quaternary complex firmly established in its competent state.Fig. 4. Secondary structure of the fdhF mRNA stem loop. The secondary structure of the fdhF stem loop as predicted by Hüttenhofer et al. (1996), based on chemical and enzymatic protection data.
Do the sequences of prokaryotic Sec insertion sites give any indication of the strengths of the stop signals at the sites? Taking three E. coli examples, the fdhF, fdoG, and fdnG genes, the twelve-base sequence element contributing to the termination signal is highly conserved at the Sec incorporation sites (CGT/C GTC TGA CAC). For the downstream CAC, C in the +4 position is selected against at natural termination sites and contributes significantly to weakening the signal, as explained for the RF2 frameshift site. However, the CAC codon is not selected against following UGA at natural termination sites. The upstream sequence (CGT GTC) is not found at any natural termination sites of the 1256 UGA-terminating E. coli genes, although statistically this is not surprising. CGC GTC is found at one natural termination site and that is what would be expected. If these two codons are considered separately then there is also no obvious bias against their use at natural termination sites. In fact these codons encode Arg-Val which favor efficient termination at UGA stop codons [16, 17]. Of course selenoenzymes have the Sec at the active center and there may be severe restrictions on what amino acids can be encoded in the flanking positions to maintain a viable active center, not allowing therefore subtle selection for better translational regulation at the sites. Alternatively, a strong upstream context for termination may be desirable for the requirements of the cell in regulating the synthesis of these proteins. The key contributor then to lowering the strength of the stop signal appears to be the +4 base weakening its coding efficiency.
Test systems to measure competition between Sec incorporation and termination at Sec sites have been established in several reporter systems. Initially, we have used an expression system under a strong promoter and, while being able to detect Sec incorporation, the efficiency was in the range of only 0.1-0.5% [41]. Using a comparable expression system, Suppmann et al. determined the efficiency of Sec incorporation during translation of fdhF mRNA as 4-5% [42]. However these systems produce abundant mRNA and if the Sec incorporation machinery is limiting, most mRNAs being translated may not be in the 'competent translation complex' for Sec incorporation. Indeed it has been shown that expression of a mRNA stem loop to act as a competitor for the mRNAs dramatically reduced Sec incorporation, consistent with this explanation [40]. Hence one difficulty of these studies is to ensure that the Sec incorporation machinery is not separated into unproductive complexes. Despite this, reduction in expression of these initial systems did not decrease the competitiveness of the termination event.
A two gene reporter system based on an upstream lacZ gene and a downstream luc+ gene (with the fdhF UGA insertion site cloned between the two) has been more successful in the study of the competition between termination and Sec incorporation at this site, as Sec incorporation is much more significant (20-40%). With this system, the strength of the stop signal could be modified to modulate significantly the competitiveness of termination. Overproduction of the decoding RF2 reduced Sec incorporation to a low level, whereas overexpression of SELC resulted in increased Sec incorporation. This indicated how the ligands were competing for decoding the UGA at the Sec site. This competition varies with growth rate of the organism. At low growth rates the Sec incorporation is favored [41] and, consistent with this observation, there is evidence that the number of RF2 molecules is much lower under these conditions [43].
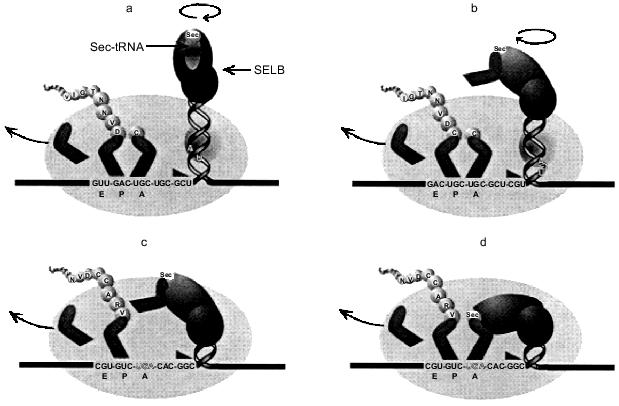
The SELB quaternary complex bound to the apical loop of the fdhF stem loop would be envisaged to hinder unwinding of the stem loop structure. For such a complex to remain bound at the apex of the stem loop, it must rotate about the axis of the helical stem as the secondary structure unwinds. This may increase the torsional load experienced by the advancing ribosome, and in a fashion analogous to a pseudoknot, increase the likelihood of the ribosomal pause required for its own productive interaction with the ribosome. Recent cryoelectron-microscopy studies show the mRNA track may in places resemble a tunnel rather than a trough, suggesting a conformational change after the initial binding of the mRNA. There is the possibility for further conformational changes and possible interactions with ribosomal RNA when the ribosome encounters structures like the fdhF stem loop structure. Mansell (1999) has proposed a 'helical approach' model to explain how the Sec-tRNA could be delivered to the ribosomal A site, with a ribosomal pause partially generated by the imposed torsional stress of the unwinding hairpin and the rotating bound SELB·Sec-tRNA complex (shown in part in Fig. 5) [41]. The rate-limiting step during Sec incorporation is the interaction of the SELB·GTP·Sec-tRNA complex with the A site and, as shown in Fig. 5b, there would still be potential competition with the decoding RF2. The efficiency of Sec incorporation will therefore depend on the competing efficiencies of the two decoding ligands (SELB·GTP·Sec-tRNA and RF2), such that a decrease in the rate of RF2 selection would increase the opportunity of the SELB·Sec-tRNA to decode the UGA codon and promote Sec incorporation.
Sec incorporation into mammalian proteins. UGA also acts as the code for Sec in mammalian cells. These selenoproteins, such as glutathione peroxidase and the thyroid processing enzyme, type I 5´-deiodinase, play key roles in cellular metabolism and are essential for the survival of the organism. They must be produced at all times although selenium levels in an organism can determine how much is synthesized. The competition between UGA as a stop signal and as the code for Sec has been somewhat easier to document than for the bacterial proteins. Transfection into animal cells of the deiodinase gene and its subsequent expression have shown that both the termination product (14 kD) and the complete iodinase protein can be detected (28 kD) [11]. The relative amounts of these products can vary with cell type and expression systems, reflecting a competition between the two UGA decoding mechanisms.Fig. 5. The 'helical approach' model of Mansell (1999) [41] for selenocysteine incorporation in E. coli. a) As the translating ribosome (oval) advances toward the fdhF hairpin and bound SELB·Sec-tRNASec·GTP complex, (b) the ribosomal mRNA melting impetus (triangle) encounters the hairpin and begins unwinding the secondary structure, drawing the SELB complex toward the ribosomal SELB-binding site as UGA approaches the A-site. c) As the UGA enters the A-site, the SELB complex interacts with the ribosome, which induces a conformational transition to the (d) A-site-interactive state. A cognate anticodon--codon interaction occurs, allowing the incorporation of selenocysteine into the nascent polypeptide chain.
A sequence element (SECIS) essential for Sec incorporation is found 1.2 kb downstream from the UGA codon in the untranslated region of the deiodinase mRNA, and this seems to be the equivalent of the stem loop immediately behind the UGA in bacterial selenoprotein mRNAs [44]. This distant element may also act as a delivery mechanism for a specialized elongation factor complex, equivalent to prokaryotic SELB. Functionally interchangeable cis acting SECIS structures have been found in the mRNAs of 5´-deiodinase and SelP [44], the latter containing ten internal in-frame UGAs [45].
How is the competition at UGA regulated to allow synthesis of the active enzymes and complete selenoproteins? There are features of the stop signal which suggest it is weak. For example, deiodinase mRNA has a +4 base C immediately following the UGA, and this gives one of the weakest of the twelve possible four base signals. In addition there is a pyrimidine rich sequence in the downstream region beyond the +4 base and this also has been shown in vitro to be a particularly weak context for a stop signal (McCaughan and Tate, unpublished). Hence all indications are that the site, at least in the deiodinase mRNA, is crafted so that the competition for the stop signal is minimized. It is of interest that most of the UGAs in the rodent SelP mRNA have weak termination contexts, otherwise it would be difficult to understand how the protein could ever be synthesized; at each UGA there would be a high chance of premature termination of synthesis. Certainly a premature chain termination product has been detected, corresponding to one UGA site in this mRNA where the +4 base is a purine contributing to a stronger stop signal [46].
UGA AND THE HISTORY OF THE GENETIC CODE
The origins and forces that shaped the universal genetic code and the nuclear and mitochondrial variant codes are still the subject of much debate. The simple and elegant idea of a 'frozen accident' or the alternative more likely 'adaptive', 'historical', and 'chemical' alternatives speak to what generated the last common ancestor but not necessarily the more recent changes [47]. Where does UGA fit in these arguments? The implication that the universal code is that of the last common ancestor and the variant codes are more recent variations suggests that UGA as a stop signal is quite ancient. How does UGA encoding Sec fit into this scenario? This could be a case of more recent codon swapping, as perhaps occurs with the use of UGA for Trp in most mitochondrial variant codes, and for Trp and Cys in some nuclear variant codes.
UGA no longer encoding Sec except in rare cases might be an example of codon loss as a result of the arrival of the oxygen environment. Osawa and Jukes invoke that codons vanish because of some mutational pressure such as changing G + C content of genomes [48]. It would then be important to use the codon for another amino acid so that translation would not be inhibited if there was a drift back in the genome base composition; hence a codon reassignment would have occurred. How does UGA as stop fit into this picture of the universal code for the last common ancestor? Unless UGA was relatively rarely used for Sec, even in ancient times, and thus could easily be taken over as a stop codon, the coding regions of genes would need to have a drift away from UGA perhaps to UGG or UGC. Alternatively, perhaps UGA was initially stop in a precursor to the common ancestor but was taken over by Sec to varying degrees during the evolutionary period to the common ancestor (Sec is found encoded by UGA in the three kingdoms of organisms).
We still see UGA being used for Sec in modern organisms where the selenoproteins seem to have established in important physiological niches. Are these examples where the codon takeover has been incomplete, and where there has been adaptation, so that organisms can accommodate a codon representing two completely different events? As described above, the examples we now understand in some detail support this contention. In addition, in UGA at frameshifting sites, which are used as an elegant control mechanism for gene expression, we see this codon playing a further adaptive role apparently of some importance to the organisms in which these mechanisms are found. UGA is indeed an enigma as a code signal in the modern organism, not only retaining a prime position for its mainstream role as signalling stop in protein synthesis, but also engaged in niche roles for the production of specialized Sec-containing proteins, and at frameshift sites for the production of the right amounts of key proteins.
The work from our own laboratory described here has been supported by a Howard Hughes Medical Institute International Investigator award to WPT, a Human Frontiers of Science Program grant to WPT (awarded with Y. Nakamura, L. Kisselev, and M. Philippe), and grants from the Marsden Fund and the Health Research Council from New Zealand.
REFERENCES
1.Marshall, R. E., Caskey, C. T., and Nirenberg, M.
(1967) Science, 155, 820-826.
2.Barrell, G., Bankier, A. T., and Drouin, J. (1979)
Nature, 282, 189-194.
3.Tate, W. P., Dalphin, M. E., Pel, H. J., and
Mannering, S. A. (1996) Gen. Eng.,18, 157-182.
4.Tate, W. P., Beaudet, A. L., and Caskey, C. T.
(1973) Proc. Natl. Acad. Sci. USA, 70, 2350-2352.
5.Ito, K., Uno, M., and Nakamura, Y. (1998) Proc.
Natl. Acad. Sci. USA, 95, 8165-8169.
6.Askarian-Amiri, E. (1999) M.Sc. Thesis, University
of Otago, New Zealand.
7.Craigen, W. J., Cook, R. G., Tate, W. P., and
Caskey, C. T. (1985) Proc. Natl. Acad. Sci. USA, 82,
3616-3620.
8.Wilson, D. N. (1999) Ph.D. Thesis, University of
Otago, New Zealand.
9.Frolova, L. Y., Tsivkovskii, R. Y., Sivolobova, G.
F., Oparina, N. Y., Serpinsky, O. I., Blinov, V. M., Tatkov, S. I., and
Kisselev, L. L. (1999) RNA, 5.
10.Poole, E. S., Brown, C. M., and Tate, W. P.
(1995) EMBO J., 14, 151-158.
11.McCaughan, K. K., Brown, C. M., Dalphin, M. E.,
Berry, M. J., and Tate, W. P. (1995) Proc. Natl. Acad. Sci.
USA,92, 5431-5435.
12.Poole, E. S., Brimacombe, R., and Tate, W. P.
(1997) RNA, 3, 974-982.
13.Poole, E. S., Major, L. L., Mannering, S. A., and
Tate, W. P. (1998) Nucleic Acids Res.,26, 954-960.
14.Brown, C. M., and Tate, W. P. (1994) J. Biol.
Chem.,269, 33164-33170.
15.Major, L. L., Poole, E. S., Dalphin, M. E.,
Mannering, S. A., and Tate, W. P. (1996) Nucleic Acids
Res.,24, 2673-2678.
16.Björnsson, A., Mottagui-Tabar, S., and
Isaksson, L. A. (1996) EMBO J., 15, 1696-1704.
17.Mottagui-Tabar, S., Björnsson, A., and
Isaksson, L. A. (1994) EMBO J., 13, 249-257.
18.Nakamura, Y., Ito, K., Matsumura, K., Kawazu, Y.,
and Ebihara, K. (1995) in Frontiers in Translation (Matheson, A.
T., Davies, J. E., Dennis, P. P., and Hill, W. E., eds.) University of
Toronto Press, Canada, pp. 1113-1122.
19.Keiler, K. C., Waller, P. R., and Sauer, R. T.
(1996) Science, 271, 990-993.
20.Williams, K. P. (1999) Nucleic Acids Res.,
27, 165-166.
21.Beremand, M. N., and Blumenthal, T. (1979)
Cell, 18, 257-266.
22.Atkins, J. F., Gesteland, R. F., Reid, B. R., and
Anderson, C. W. (1979) Cell, 18, 1119-1131. 23.Craigen,
W. J., and Caskey, T. (1986) Nature, 322, 273-275.
24.Donly, B. C., Edgar, C. D., Adamski, F. M., and
Tate, W. P. (1990) Nucleic Acids Res.,18, 6517-6522.
25.Geller, A. I., and Rich, A. (1980) Nature,
283, 41-46.
26.Weiss, R. B., Dunn, D. M., Dahlberg, A. E.,
Atkins, J. F., and Gesteland, R. F. (1988) EMBO J., 7,
1503-1507.
27.Persson, B. C., and Atkins, J. F. (1998) J.
Bacteriol., 180, 3462-3466.
28.Nakamura, Y., and Ito, K. (1998) Genes
Cells, 3, 265-278.
29.Hayashi, S., Murakami, Y., and Matsufuji, S.
(1996) Trends Biochem. Sci., 21, 27-30.
30.Matsufuji, S., Matsufuji, T., Miyazaki, Y.,
Murakami, Y., Atkins, J. F., Gesteland, R. F., and Hayashi, S. I.
(1995) Cell, 80, 51-60.
31.Ivanov, I. P., Gesteland, R. F., Matsufuji, S.,
and Atkins, J. F. (1998) RNA, 4, 1230-1238.
32.Ivanov, I. P., Simin, K., Letsou, A., Atkins, J.
F., and Gesteland, R. F. (1998) Mol. Cell. Biol., 18,
1553-1561.
33.Irvine, J. H. (1996) B.Med.Sci. Thesis,
University of Otago, New Zealand.
34.Böck, A., Forchhammer, K., Heider, J., and
Baron, C. (1991) Trends Biochem. Sci.,16, 463-467.
35.Zinoni, F., Birkmann, A., Stadtman, T. C., and
Böck, A. (1986) Proc. Natl. Acad. Sci. USA,83,
4650-4654.
36.Chambers, I., Frampton, J., Goldfarb, P., Affara,
N., McBain, W., and Harrison, P. R. (1986) EMBO J., 5,
1221-1227.
37.Heider, J., and Böck, A. (1993) Adv.
Microb. Physiol.,35, 71-109.
38.Hüttenhofer, A., Westhof, E., and Böck,
A. (1996) RNA,2, 354-366.
39.Liu, Z., Reches, M., Groisman, I., and
Engelberg-Kulka, H. (1998) Nucleic Acids Res.,26,
896-902.
40.Tormay, P., Sawers, A., and Böck, A. (1996)
Mol. Microbiol.,21, 1253-1259.
41.Mansell, J. B. (1999) Ph.D. Thesis, University of
Otago, New Zealand.
42.Suppmann, S., Persson, B. C., and Böck, A.
(1999) EMBO J., 18, 2284-2293.
43.Adamski, F. M., McCaughan, K. K., Jorgensen, F.,
Kurland, C. G., and Tate, W. P. (1994) J. Mol. Biol.,
238, 302-308.
44.Berry, M. J., Banu, L., Harney, J. W., and
Larsen, P. R. (1993) EMBO J., 12, 3315-3322.
45.Hill, K. E., Lloyd, R. S., Yang, J. G., Read, R.,
and Burk, R. F. (1991) J. Biol. Chem., 266,
10050-10053.
46.Hill, K. E., and Burk, R. F. (1997) Biomed.
Environ. Sci., 10, 198-208.
47.Knight, R. D., Freeland, S. J., and Landweber, L.
F. (1999) Trends Biochem. Sci., 24, 241-247.
48.Osawa, S., and Jukes, T. H. (1988) Trends
Genet., 4, 191-198.