Mini-REVIEW: Eukaryotic Translation Termination Factor 1 Associates with Protein Phosphatase 2A and Targets It to Ribosomes
K. Lechward1, S. Zolnierowicz1*, and B. A. Hemmings2
1Intercollegiate Faculty of Biotechnology UG-MUG, Kladki 24, 80-822 Gdansk, Poland; fax: +48-58-301-03-81; E-mail: staszek@biotech.univ.gda.pl2Friedrich Miescher Institute, Maulbeerstrasse 66, CH-4058 Basel, Switzerland; fax: +41-61-697-39-76; E-mail: hemmings@fmi.ch
* To whom correspondence should be addressed.
Received July 10, 1999
Purification of type 2A protein phosphatase (PP2A) from rabbit skeletal muscle resulted in the isolation of a trimeric phosphatase which is composed of a catalytic (PP2Ac), a structural (PR65alpha/Aalpha), and a regulatory (PR55alpha/Balpha) subunit, together with translation termination factor 1 (eRF1) and another protein of 55 kD (EMBO J., 15, 101-112). Yeast two-hybrid system analysis demonstrated that the eRF1 interacted with PP2Acalpha but not with PR65alpha/Aalpha or PR55alpha/Balpha. The N-terminal region of PP2Acalpha, comprising 50 amino acid residues, and the C-terminal part of eRF1, corresponding to an internal region between amino acids 338-381, were found to be necessary for eRF1--PP2Acalpha interaction in yeast. Immunoprecipitations using 12CA5 antibodies and extracts from COS1 cells transiently transfected with eRF1 tagged with 9-amino acid epitope from influenza hemagglutinin (HA) demonstrated the presence of eRF1--PP2Acalpha--PR65alpha/Aalpha complex in these cells. In addition, polysomes obtained from COS1 cells overexpressing HA--eRF1 displayed several-fold higher PP2A activity than control polysomes. No effect of either PP2Ac or dimeric and trimeric PP2A holoenzymes on the rate of translation termination was detected using an in vitro reconstituted translation termination assay. In summary, eRF1 appears to represent a novel PP2A-targeting subunit that brings this phosphatase in contact with putative ribosomal substrate(s). It remains to be established whether termination of translation requires dephosphorylation of participating protein factor(s).
KEY WORDS: protein phosphatase type 2A, eukaryotic translation termination factor 1, protein purification, the yeast two-hybrid system, co-immunoprecipitation, termination of translation, polysomes, COS1 cells, transient transfection
Abbreviations: PP2As) type 2A protein phosphatases; PP2Ac) catalytic subunit of PP2A; PR65/A) structural subunit of PP2A; PR55/B, PR72/B., PP61/B´, PTPA) regulatory subunits of PP2A; PPP) phosphoprotein phosphatases; HA) hemagglutinin; eRF1) eukaryotic release factor 1; SV40) simian virus 40; PTPA) phosphotyrosine phosphatase activator; CaM) calmodulin; HSF2) heat shock transcription factor 2; FRAP) 12-rapamycin-associated FKBP protein.
In a typical eukaryotic cell approximately 30% of proteins undergo
phosphorylation that triggers changes in their conformation and
biological activity. Thus, reversible protein phosphorylation catalyzed
by protein kinases and phosphoprotein phosphatases regulates numerous
cellular processes (reviewed in [1]). Recent data
demonstrate that kinases and phosphatases are equally important in the
regulation of the phosphorylation state and activity of proteins. In
addition, the activities of both protein kinases and phosphatases are
also frequently controlled by phosphorylation, creating complicated
intracellular phosphorylation cascades and networks (reviewed in [1-4]).
Based on homology of amino acid sequences and the similarity of three-dimensional structures, phosphoprotein phosphatases (PPPs) are divided into three families designated PPP, PPM, and PTP (reviewed in [5, 6]). PPP and PPM comprise phosphoserine- and phosphothreonine-specific enzymes, whereas PTP comprises phosphotyrosine specific and dual specificity phosphatases. As suggested by the name, dual specificity phosphatases can dephosphorylate all three phosphoresidues [7]. Type 2A protein phosphatase (PP2A) together with PP1, PP4, PP6, PP2B, PP5, and PP7 are classified in the PPP family (reviewed in [8]).
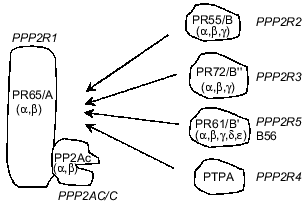
PP2As represent a large and diverse family of enzymes present in mammalian cells, mostly in the cytosol, although nucleus-, plasma membrane-, microtubule-, and microfilament-associated forms have been described [9-12]. Only two genes encoding catalytic subunits of PP2A (PP2Acalpha and PP2Acbeta) have been identified in mammalian cells, but they form multiple dimeric and/or trimeric complexes with regulatory subunits [4, 5, 13]. Dimers are formed by the association of PP2Ac with a structural PR65/A subunit which is also encoded by two genes. Therefore, four different dimers may exist. Trimers are composed from the PR65/A-PP2Ac 'core dimer' associated with a regulatory subunit belonging to one of three protein families: PR55/B, PR72/B., PR61/B´ (Fig. 1) [4, 5, 8, 12-14]. In addition, another protein designated phosphotyrosine phosphatase activator (PTPA) has been classified as a subunit of PP2A (Fig. 1). There is no amino acid sequence similarity between proteins constituting these families of PP2A regulatory subunits [15-18]. Multiplicity and diversity of PP2A holoenzymes result from at least 12 genes encoding regulatory subunits. So far, less than 10 different PP2A holoenzymes have been purified from animal tissues and their properties characterized by activity measurements against different phosphoprotein substrates [19, 20]. However, the number of genes encoding PP2A subunits predicts the formation of approximately 50 distinct holoenzymes. Adding potential subunits produced by alternative splicing of mRNA doubles this number [15, 21]. It has been shown that association of PP2Ac with different regulatory subunits changes activity, substrate specificity and the subcellular location of holoenzymes, enabling them to control numerous cellular functions [4, 12, 22, 23]. PP2As have been implicated in the regulation of metabolism, DNA replication, transcription, RNA splicing, translation, cell cycle progression, oncogenic transformation, and cell differentiation and signal transduction [4, 5, 8, 9, 13, 14].
The formation of different PP2A holoenzymes is a dynamic process occurring during development, cell differentiation, cell cycle progression, and viral infection. Some viral proteins utilize PP2As as their molecular targets and either displace constitutive subunits (simian virus 40 small t antigen) or form tetrameric/multimeric complexes with phosphatase (adenovirus protein encoded by orf4) [24-26]. Although mechanisms responsible for remodeling of PP2A holoenzymes have not been fully elucidated, data from several laboratories indicate that methylation of a C-terminal leucine of PP2Ac is important [27-29]. It remains to be established whether phosphorylation of PP2Ac, a post-translational modification reported to suppress phosphatase activity, plays any role in holoenzyme remodeling.Fig. 1. Holoenzymes of type 2A protein phosphatase (PP2A). The nomenclature recommended by a nomenclature committee formed at the FASEB Summer Research Conference in 1992 is applied (italics) as well as nomenclature used by different laboratories. The catalytic subunit, PP2Ac/C/PPP2C, associates with the subunit PR65/A/PPP2R1 and forms a 'core dimer'. The third subunit belonging to one of four classes of proteins and termed variable regulatory subunit associates to form a trimeric complex. PPP2R4, also termed phosphotyrosine phosphatase regulator (PTPA), has been designated as a subunit of PP2A, but some controversy about its relationship to the phosphatase remains. Greek letters refer to distinct genes encoding subunits of PP2A.
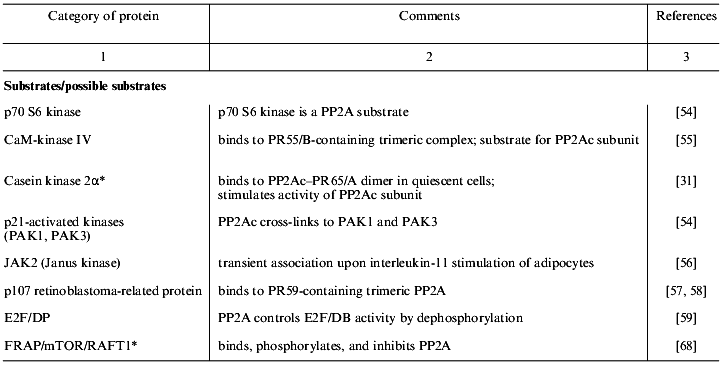
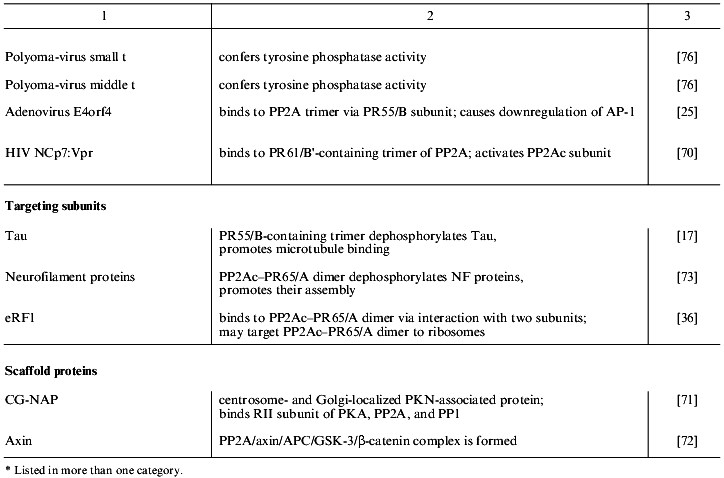
Table 1 lists proteins known to form complexes with various forms of PP2A in vivo. These are grouped into the four classes: PP2A substrates, modulators, targeting subunits, and scaffold proteins. The majority of the PP2A substrates identified so far are protein kinases, which suggests that PP2A plays an important role in the regulation of many signaling cascades [30]. Protein kinases, in addition to being substrates of PP2A, also modulate phosphatase activity. This is exemplified by casein kinase 2alpha that is dephosphorylated by PP2A and also stimulates phosphatase activity [31]. Considering the pace at which proteins that form complexes with PP2A in vivo are being discovered, many more will be discovered in the near future.
Table 1. Proteins that interact with PP2A
in vivo

PURIFICATION OF PP2A--eRF1--p55 COMPLEX FROM RABBIT SKELETAL MUSCLE
Rabbit skeletal muscle has been routinely used as a source of PP2As [15, 32-34]. Several holoenzymes made up of the PP2Ac--PR65/A core dimer (PP2A2) and trimers containing an additional regulatory subunit from one of the three unrelated protein families--PR55/B (PP2A1), PR72/B. (PP2A3), and PR61/B´ (PP2A0)--have been isolated from rabbit skeletal muscle and characterized by activity measurements and microsequencing of subunits (reviewed in [4, 8, 15, 30]). Data derived from both purification and immunoprecipitation indicate an in vivo equilibrium between dimeric and trimeric forms of PP2A [35]. Partial amino acid sequences derived from purified phosphatase subunits aided cloning of their cDNAs and revealing high complexity of PP2A subunits [14, 15, 21, 34]. In addition, PTPA was found to increase phosphatase activity against phosphotyrosine residues and was designated a PP2A subunit (Fig. 1). However, in contrast to other PP2A subunits, PTPA has not been isolated in a complex with a PR65/A--PP2Ac core dimer [18]. Viral proteins such as polyoma-virus small t and middle t antigen increase activity of PP2A against phosphotyrosine residues. Thus, in addition to being a phosphatase specific against phosphoserine and phosphothreonine residues, PP2A can also dephosphorylate phosphotyrosine residues.
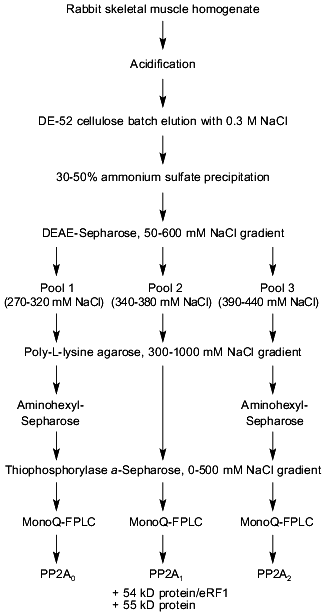
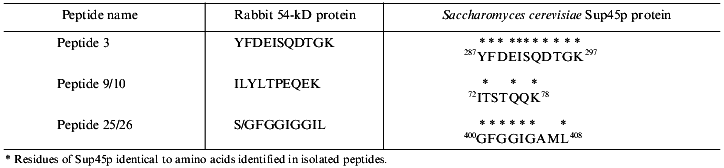
To search for novel regulatory subunits of PP2A and/or PP2A-interacting proteins, a modified protocol for PP2A purification from rabbit skeletal muscle was developed (Fig. 2) [36]. This protocol differed from those previously applied by inclusion of affinity chromatography on thiophosphorylase a-Sepharose followed by final purification on MonoQ-FPLC. Since 32P-labeled phosphorylase a in the presence of protamine (an activator of PP2A) and ammonium sulfate (an inhibitor of PP1 and synergistic activator of protamine action on PP2A) was applied as substrate, only phosphatases able to dephosphorylate phosphorylase a were detected using this procedure. A diagram of the purification steps applied is given in Fig. 2. Before anion-exchange chromatography on DEAE-Sepharose, batch DE-52 cellulose chromatography was performed. This was done to facilitate rapid separation of PP2A holoenzymes from contaminating proteases. Proteins precipitated between 30 and 50% of ammonium sulfate saturation were loaded on DEAE-Sepharose and eluted using a 50-600 mM NaCl gradient. Subsequently, PP2A obtained after the anion-exchange column was divided into three pools relating to the NaCl concentration necessary for phosphatase elution (pool 1: 270-320 mM; pool 2: 340-380 mM, and pool 3: 390-440 mM NaCl) and processed separately over polylysine-agarose, omega-aminohexyl-Sepharose (except pool 2), thiophosphorylase a-Sepharose, and MonoQ-FPLC. By comparison with previously purified PP2A holoenzymes and immunodetection with antibodies generated against established PP2A subunits, the phosphatases identified in the different pools corresponded to PP2A0 (pool 1), PP2A1 (pool 2), and PP2A2 (pool 3). Analysis of PP2A1 by Coomassie staining of SDS-polyacrylamide gels revealed the presence of two protein bands migrating very close to the PR55/B subunit. Further purification by MonoQ-FPLC resulted in the separation of a PP2A trimer (partially dissociated into core dimer and free PR55/B subunit) from two proteins of apparent molecular masses 54 and 55 kD, respectively. Neither of these protein reacted with antibodies against PR55/B and they were thus neither degradation products of this subunit nor related proteins. To identity them, partial amino acid sequences were obtained. Table 2 shows the sequences of three peptides of 30 amino acids derived from the 54-kD protein. Database searches revealed that these peptides are homologs to a yeast protein encoded by the omnipotent suppressor (SUP) 45 gene [37]. Based on amino acid sequences from the 54-kD protein, degenerate primers were designed and used to clone fragments of human cDNA. A first cDNA fragment, comprising the region between sequences encoding for peptides 3 and 9/10, was obtained by RT-PCR using poly(A+) RNA isolated from T47D human breast carcinoma cells. The isolated fragment was radiolabeled and used as a probe to screen a human fetal brain cDNA library. With this nucleotide sequence, a further search of cDNA databases was performed and the 54-kD protein was identified as a homolog of the human gene TB3-1 [38]. The mammalian homolog of Sup45 was later found to act as a eukaryotic release factor (eRF1) by Frolova and collaborators [39]. Northern blot analysis of the distribution of eRF1 mRNA in human tissues identified three widely expressed transcripts of 2, 2.5, and ~4 kb. To determine whether eRF1 affects PP2A activity, several experiments were carried out using eRF1 either purified from rabbit skeletal muscle or expressed in E. coli and purified recombinant 6xHis-eRF1. Using both 32P-labeled phosphorylase a and 32P-labeled Kempeptide as substrates, no effect on phosphatase activity was detected. Therefore, the yeast two-hybrid analysis was applied to look for interaction between eRF1 and PP2A subunits.
Table 2. Peptides derived from p54 and their homologs from Sup45pFig. 2. Schematic diagram depicting purification of protein phosphatase 2A (PP2A) holoenzymes from rabbit skeletal muscles. Two trimers, PP2A0 and PP2A1, as well as PP2A2 dimer were purified.
YEAST TWO-HYBRID ANALYSIS OF INTERACTION BETWEEN PP2Ac AND eRF1
The interaction between eRF1 and different PP2A subunits was examined using the yeast two-hybrid system as described by Fields and Song [40, 41]. By this time, another translation termination factor, termed eRF3, had been identified and found to have GTPase activity [42, 43]. Therefore, both eRF1 and eRF3 were characterized. cDNAs coding for PP2Acalpha, PR65alpha/Aalpha, and PR55alpha/Balpha were cloned into pGBT9 to produce hybrid proteins containing the Gal4 DNA-binding domain, whereas eRF1 and eRF3 were cloned into pGAD424 to produce fusions with the Gal4 transactivation domain. Yeast were transformed with appropriate constructs, grown on selective media and the interaction of PP2A subunits with eRF1 and eRF3 was detected by monitoring activity of two independent reporter genes--of beta-galactosidase encoded by LacZ in SFY526 and HIS3 in Hf7c. From these studies, it was concluded that human eRF1 specifically interacted with PP2Acalpha but not with other subunits of the PP2A1 holoenzyme. There was no interaction between eRF3 and PP2A subunits. Mapping of domains required for interaction between these proteins was performed using a series of N- and C-terminal mutants derived from both PP2Acalpha and eRF1. The N-terminal 50-amino-acid region of PP2Acalpha and the region of eRF1 between residues 338-381 were found necessary for binding to occur. Using the yeast two-hybrid assay, Merkulova and collaborators have recently demonstrated that C-terminal domains of human eRF1 and eRF3 mediate their interaction [44]. Two regions, those between residues 281-305 and 411-415 of eRF1, have been delineated as necessary for eRF3 binding. Interestingly, these regions lie outside the one (residues 338-381) mapped as an interaction site with PP2Ac. One can therefore speculate that binding between eRF1--eRF3 and eRF1--PP2Ac may be mutually exclusive due to the competition for closely situated binding sites. In agreement with this, immunoprecipitation of eRF3 overexpressed in COS-1 cells did not bring down PP2A activity (see also below). Further analysis using PR65alpha/Aalpha cloned together with the pGal4 transactivation domain and eRF1 as a fusion with a DNA-binding domain, repeatedly detected interaction between these two hybrids (Jodczyk and Zolnierowicz, unpublished data, Fig. 3). It will be interesting to identify the regions necessary for interaction between eRF1 and PR65alpha/Aalpha. Taken together, these data indicate that eRF1 interacts with both PP2Acalpha and the PR65alpha/Aalpha subunits of PP2A.
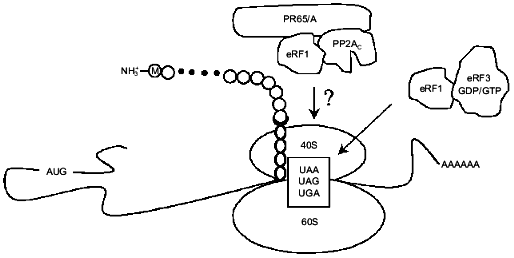
Fig. 3. eRF1 together with eRF3 terminates eukaryotic translation. Another function of eRF1 described in this review is to target a dimer of type 2A protein phosphatase (PP2A) composed of PP2Ac and PR65/A to ribosomes. The question mark indicates that substrates of PP2A among translation termination factors and/or ribosomal proteins remain to be identified.
CO-IMMUNOPRECIPITATION OF PHOSPHATASE SUBUNITS AND eRF1
To look for eRF1--PP2Acalpha complex formation, transient transfections of cultured mammalian cells were applied. For this purpose, COS-1 cells were transfected with a plasmid encoding eRF1 together with the N-terminal 9-amino-acid epitope from influenza hemagglutinin (HA) under the control of the cytomegalovirus promoter (pCMV). Cell-free extracts prepared from transfected COS-1 cells could be immunoprecipitated with 12CA5 monoclonal antibodies, demonstrating complex formation between eRF1 and PP2Acalpha. Furthermore, PR65alpha/Aalpha was detected in these immunoprecipitates. PR65alpha/Aalpha was either brought down due to association with the catalytic subunit or, as discussed above, was immunoprecipitated due to its binding to eRF1. To determine whether complex formation between eRF1 and PP2Acalpha changes the intracellular distribution of PP2A, cell fractionation experiments applying COS-1 cells overexpressing HA--eRF1 were performed. The polysomal fraction from exponentially growing COS-1 cells exhibited a significant increase in PP2A activity measured with 32P-labeled Kempeptide as substrate. Based on phosphatase activity measurements, it was calculated that only 1-2% of total cellular PP2Ac was associated with polysomes. As in the case of the in vitro experiments, no effect on the specific activity of PP2A was observed.
PP2A HAS NO DIRECT EFFECT ON THE RATE OF TRANSLATION
TERMINATION
To gain insight into the physiological role of the association between eRF1 and PP2Acalpha, an in vitro translation termination assay was performed using 80S ribosomes isolated from COS-1 cells. Termination of translation was measured as a stop codon (containing the oligonucleotide UGAAAAAA)-dependent release of formyl[35S]methionine from formyl[35S]methionyl--tRNAfMet--AUG-80S substrate [45]. Both recombinant 6xHis-eRF1 and eRF1 purified from rabbit skeletal muscle were found to activate termination of translation in this assay. In contrast, HA--eRF1 present in immunoprecipitates in the above assay yielded only approximately 5% of the activity observed for the recombinant protein. This may have been due to the presence of inhibitory protein(s) bound to immunoprecipitated eRF1 or to a steric hindrance preventing eRF1 immobilized on protein A-Sepharose beads via 12CA5 antibodies to reach its substrate. Different forms of PP2A were applied to look for their effects on the termination of translation measured as described above. It was concluded from the results that neither free catalytic subunit of this phosphatase nor different holoenzymes (PP2A1 and PP2A2) were able to affect the rate of translational termination.
ROLE OF PP2A AND OTHER PHOSPHATASES IN TRANSLATIONAL
CONTROL
Protein phosphorylation appears to play an important role in the regulation of protein translation [46]. Both ribosomal proteins and translational factors undergo reversible phosphorylation. While phosphorylation of some proteins of the translational machinery inhibits translation, phosphorylation of others stimulates this process [47, 48]. S6 protein is a constituent of the 40S ribosomal complex. Upon mitogenic stimulation of cells, S6 undergoes phosphorylation at several sites. p70 S6 kinase phosphorylates S6 and is itself activated by phosphorylation. As presented in Table 2, p70 S6 kinase forms a complex with PP2A in vivo, indicating that this phosphatase is responsible for its dephosphorylation and inactivation. FRAP (FKBP12-rapamycin-associated protein) is responsible for phosphorylation of both p70 S6 kinase and 4E-BP1, triggering initiation of translation and entry into the cell cycle. Studies carried out with an immunosuppressive macrolide rapamycin demonstrate how PP2A that dephosphorylates p70 S6 kinase is regulated. It appears that PP2A is phosphorylated and inactivated by FRAP. Both rapamycin and amino acid deprivation inactivate FRAP, which is then no longer able to inhibit PP2A.
At present, it is much more difficult to assess the role played by PP2A associated with eRF1. In Table 1, eRF1 is listed as a protein interacting with PP2A in vivo and directing it to polysomes. Data leading to this conclusion are described briefly in this review. In vitro phosphorylation studies using recombinant eRF1 and several protein kinases such as cAMP-dependent protein kinase, protein kinases C, and casein kinase 2 resulted in a very low level of phosphate incorporation into this protein. This is in contrast to the presence of consensus sites recognized by these kinases in the eRF1 primary sequence. For instance, eRF1 amino acid sequence analysis with a program designed to identify putative sites phosphorylated by protein kinases [49] identified serines 251 and 358 and threonine 133 that corresponded to the R/KXXS/T consensus recognized by cAMP-dependent protein kinase. Similarly, potential protein kinase C phosphorylation sites corresponding to the S/TXR/K consensus are present at positions 301, 311, 362, and 397 in human eRF1. Finally, casein kinase 2 sites corresponding to the S/TXXD/E consensus are serines 6 and 240 and threonines 342, 362, and 366. Since no phosphorylation of eRF1 by any of these kinases was observed, either the sites listed above are not accessible to kinases or prior phosphorylation by another kinase is required. Other potential eRF1 kinases identified by analysis of their primary sequences include myosin-I heavy chain kinase, multifunctional calmodulin-dependent kinase II, cGMP-dependent protein kinase, S6 kinase II, and casein kinase 1. Isolation of eRF1, for instance by immunoprecipitation, using extracts prepared from 32P-labeled mammalian cells, followed by autoradiography of SDS-polyacrylamide gels will be necessary to determine whether this protein undergoes phosphorylation in vivo. More work is needed to demonstrate whether eRF1 serves as a substrate for PP2A.
Similarly, no effect of PP2A on the termination of translation was detected. However, it can be argued that different preparations of eRF1 applied in the translation termination assay were either not phosphorylated or not accessible to PP2A due to the presence of interfering proteins. It is also possible that putative substrates of PP2A targeted to ribosomes by eRF1 were not present in the reconstituted system applied. Therefore, care should be taken to preserve the phosphorylation state of proteins which make up ribosomes and act as translational termination factors.
Although, at present there are no data demonstrating a role for PP2A in the process of translation termination, other protein phosphatases have been implicated in this process. One example is Saccharomyces cerevisiae allosuppressor sal3 [50]. Sal3 has been shown to correspond to cdc25, a dual-specificity protein phosphatase [51]. Recently, a phosphoprotein phosphatase termed PPQ has been implicated in the regulation of translation in S. cerevisiae [52]. cDNA encoding this phosphatase was initially identified by PCR and subsequently cloned by screening a S. cerevisiae genomic library with a 32P-labeled fragment. PPQ is composed of two distinct domains. The C-terminal domain of approximately 300 amino acids is similar to phosphoprotein phosphatases from the PPP family with highest identity to PP1 and PPZ. The N-terminal domain of PPQ comprising 236 amino acids is very rich in serine (24%) and asparagine (12%). Yeast with the PPQ gene disrupted grow slower and are hypersensitive to the protein synthesis inhibitors cycloheximide and G418. The same gene has been identified by complementation of the sal6 mutation [53]. Null mutants are viable, show an altered morphology and have a slight growth defect on several carbon sources. Furthermore, sal6 mutants show a reduced rate of protein synthesis in the exponential phase and are hypersensitive to protein synthesis inhibitors.
Disassembly of ribosomes occurs as the last step in the translation of mRNA into protein. It is possible that dephosphorylation of certain protein factors is necessary for this process to take place. PP2A may, therefore, be required for ribosome disassembly. If this is the case, then eRF1 has a double life, serving both as a translation termination factor and as a protein responsible for ribosome disassembly.
This work was supported by grants from the Howard Hughes Medical Institute (grant No. 75195-544001 to S. Z. and B. A. H.) and from the Polish State Committee for Scientific Research (grant No. 6P04A06015 to K. L. and S. Z.). We thank Patrick King for comments on the manuscript.
REFERENCES
1.Hunter, T. (1995) Cell, 80,
225-236.
2.Hubbard, M., and Cohen, P. (1993) Trends
Biochem. Sci., 18, 172-177.
3.Stone, R. L., and Dixon, J. E. (1994) J. Biol.
Chem., 269, 31323-31326.
4.Wera, S., and Hemmings, B. A. (1995) Biochem.
J., 311, 17-29.
5.Barford, D. (1996) Trends Biochem. Sci.,
21, 407-412.
6.Tonks, N. T., and Neel, B. E. (1996) Cell,
87, 365-368.
7.Keyse, S. M. (1995) Biochim. Biophys. Acta,
1265, 152-160.
8.Cohen, P. T. (1997) Trends Biochem. Sci.,
22, 245-251.
9.Walter, G., and Mumby, M. C. (1993) Biochim.
Biophys. Acta, 1155, 207-226.
10.Inagaki, A., Ito, M., Nakano, T., and Inagaki, M.
(1994) Trends Biochem. Sci., 19, 448-452.
11.Sontag, E., Nunbhakdi-Craig, V., Bloom, G. S.,
and Mumby, M. C. (1995) J. Cell. Biol., 128,
1131-1144.
12.McCright, B., Rivers, A. M., Audlin, S., and
Virshup, D. M. (1996) J. Biol. Chem., 271,
22081-22989.
13.DePaoli-Roach, A. A., Park, I.-K., Cerovsky, V.,
Csortos, C., Durbin, S. D., Kuntz, M. J., Sitikov, A., Tang, P. M.,
Verin, A., and Zolnierowicz, S. (1994) in Advances in Enzyme
Regulation (Weber, G., ed.) Vol. 34, pp. 199-224.
14.Mayer-Jaekel, R. E., and Hemmings, B. A. (1994)
Trends Cell. Biol., 4, 287-291.
15.Zolnierowicz, S., Van Hoof, C., Andjelkovic, N.,
Cron, P., Stevens, I., Merlevede, W., Goris, J., and Hemmings, B. A.
(1996) Biochem. J., 317, 187-194.
16.Mayer, R. E., Hendrix, P., Cron, P., Matthies,
R., Stone, S. R., Goris, J., Merlevede, W., Hofsteenge, J., and
Hemmings, B. A. (1991) Biochemistry, 30, 3589-3597.
17.Hendrix, P., Mayer-Jaekel, R. E., Cron, P.,
Goris, J., Hofsteenge, J., Merlevede, W., and Hemmings, B. A. (1993)
J. Biol. Chem., 268, 15267-15276.
18.Cayla, X., Van Hoof, C., Bosch, M., Waelkens, E.,
Vandekerckhove, J., Peeters, B., Merlevede, W., and Goris, J. (1994)
J. Biol. Chem., 269, 15668-15675.
19.Zolnierowicz, S., Mayer-Jaekel, R. E., and
Hemmings, B. A. (1995) in Neuroprotocols: a Companion to Methods in
Neurosciences (Nairn, A., and Shenolikar, S., eds.) Academic Press,
Inc., Vol. 6, pp. 11-19.
20.Kamibayashi, C., Estes, R., Lickteig, R. L.,
Yang, S. I., Craft, C., and Mumby, M. C. (1994) J. Biol. Chem.,
269, 20139-20148.
21.McCright, B., and Virshup, D. M. (1995) J.
Biol. Chem., 270, 26123-26128.
22.Usui, H., Inoue, R., Tanabe, O., Nishito, Y.,
Shimizu, M., Hayashi, H., Kagamiyama, H., and Takeda, M. (1998) FEBS
Lett., 430, 312-316.
23.Zhao, Y., Boguslawski, G., Zitomer, R. S., and
DePaoli-Roach, A. A. (1997) J. Biol. Chem., 272,
8256-8262.
24.Pallas, D. C., Shahrik, L. K., Martin, B. L.,
Jaspers, S., Miller, T. B., Brautigan, D. L., and Roberts, T. M. (1990)
Cell, 60, 167-176.
25.Kleinberg, T., and Shenk, T. (1993) J.
Virol., 67, 7556-7560.
26.Ruediger, R., Fields, K., and Walter, G. (1999)
J. Virol., 73, 839-842.
27.Favre, B., Turowski, P., and Hemmings, B. A.
(1997) J. Biol. Chem., 272, 13856-13863.
28.Bryant, J. C., Westphal, R. S., and Wadzinski, B.
E. (1999) Biochem. J., 339, 241-246.
29.Turowski, P., Fernandez, A., Favre, B., Lamb, N.
J., and Hemmings, B. A. (1995) J. Cell. Biol., 129,
397-410.
30.Millward, T. A., Zolnierowicz, S., and Hemmings,
B. A. (1999) Trends Biochem. Sci., 24, 186-191.
31.Heriche, J. K., Lebrin, F., Rabilloud, T., Leroy,
D., Chambaz, E. M., and Goldberg, Y. (1997) Science, 276,
952-955.
32.Cohen, P. (1989) Annu. Rev. Biochem.,
58, 453-508.
33.Shenolikar, S. (1994) Annu. Rev. Cell.
Biol., 10, 55-86.
34.Csortos, C., Zolnierowicz, S., Bako, E., Durbin,
S. D., and DePaoli-Roach, A. A. (1996) J. Biol. Chem.,
271, 2578-2588.
35.Kremmer, E., Ohst, K., Kiefer, J., Brewis, N.,
and Walter, G. (1997) Mol. Cell. Biol., 17,
1692-1701.
36.Andjelkovic, N., Zolnierowicz, S., Van Hoof, C.,
Goris J., and Hemmings, B. A. (1996) EMBO J., 15,
7156-7167.
37.Breining, P., and Piepersberg, W. (1986)
Nucleic Acids Res., 14, 5187-5197.
38.Grenett, H. E., Bournelis, P., and Fuller, G. M.
(1992) Gene, 110, 239-243.
39.Frolova, L., Le Geoff, X., Rasmunssen, H. H.,
Cheperegin, S., Drugeon, G., Kress, M., Arman, I., Haenni, A. L.,
Celis, J. E., and Philippe, M. (1994) Nature, 372,
701-703.
40.Fields, S., and Song, O. (1989) Nature,
340, 245-246.
41.Chien, C.-T., Bartel, P. L., Sternglanz, R., and
Fields, S. (1991) Proc. Natl. Acad. Sci. USA, 88,
9578-9579.
42.Zhouravleva, G., Frolova, L., Le Geoff, X., Le
Guellec, R., Inge-Vechtomov, S., Kisselev, L., and Philippe, M. (1995)
EMBO J., 14, 4065-4072.
43.Frolova, L., Le Goff, X., Zhouravleva, G.,
Davydova, E., Philippe, M., and Kisselev, L. (1996) RNA,
2, 334-341.
44.Merkulova, T. I., Frolova, L. Y., Lazar, M.,
Camonis, J., and Kisselev, L. L. (1999) FEBS Lett., 443,
41-47.
45.Tate, W. P., and Caskey, C. T. (1990) in
Ribosomes and Protein Synthesis. A Practical Approach (Spedding,
G., ed.) IRL Pressat Oxford University Press, pp. 81-100.
46.Hershey, J. W. (1991) Annu. Rev. Biochem.,
60, 717-755.
47.Riera, M., Roher, N., Miro, F., Gil, C.,
Trujillo, R., Aguilera, J., Plana, M., and Itarte, E. (1999) Mol.
Cell. Biochem., 191, 97-104.
48.Waskiewicz, A. J., Johnson, J. C., Penn, B.,
Mahalingam, M., Kimball, S. R., and Cooper, J. A. (1999) Mol. Cell.
Biol., 19, 1871-1880.
49.Wera, S. (1999) Comput. Methods Programs
Biomed., 58, 65-68.
50.Crouzet, M., and Tuite, M. F. (1987) Mol. Gen.
Genet., 210, 581-583.
51.Dunphy, W. G., and Kumagai, A. (1991)
Cell, 67, 189-196.
52.Chen, M. X., Chen, Y. H., and Cohen, P. T. (1993)
Eur. J. Biochem., 218, 689-699.
53.Vincent, A., Newnam, G., and Liebman, S. W.
(1994) Genetics, 138, 597-608.
54.Ballou, L. M., Jeno, P., and Thomas, G. (1988)
J. Biol. Chem., 263, 1189-1194.
55.Westphal, R. S., Tavalin, S. J., Lin, J. W.,
Alto, N. M., Fraser, I. D., Langeberg, L. K., Sheng, M., and Scott, J.
D. (1999) Science, 285, 93-96.
56.Fuhrer, D. K., and Yang, Y. (1996) Biochem.
Biophys. Res. Commun., 224, 289-296.
57.Voorhoeve, P. M., Hijmans, E. M., and Bernards,
R. (1999) Oncogene, 18, 515-524.
58.Voorhoeve, P. M., Watson, R. J., Fairlie, P. G.,
Bernards, R., and Lam, E. W. (1999) Oncogene, 18,
679-688.
59.Altiok, S., Xu, M., and Spiegelman, B. M. (1997)
Genes Dev., 11, 1987-1998.
60.Li, M., Makkinje, A., and Damuni, Z. (1996)
Biochemistry, 35, 6998-7002.
61.Li, M., Makkinje, A., and Damuni, Z. (1996) J.
Biol. Chem., 271, 11059-11062.
62.Adler, H. T., Nallaseth, F. S., Walter, G., and
Tkachuk, D. C. (1997) J. Biol. Chem., 272,
28407-28414.
63.Di Como, C. J., and Arndt, K. T. (1996) Genes
Dev., 10, 1904-1916.
64.Murata, K., Wu, J., and Brautigan, D. L. (1997)
Proc. Natl. Acad. Sci. USA, 94, 10624-10629.
65.Okamoto, K., Kamibayashi, C., Serrano, M.,
Prives, C., Mumby, M. C., and Beach, D. (1996) Mol. Cell. Biol.,
16, 6593-6602.
66.Kawabe, T., Muslin, A. J., and Korsmeyer, S. J.
(1997) Nature, 385, 454-458.
67.Santoro, M. F., Annand, R. R., Robertson, M. M.,
Peng, Y. W., Brady, M. J., Mankovich, J. A., Hackett, M. C., Ghayur,
T., Walter, G., Wong, W. W., and Giegel, D. A. (1998) J. Biol.
Chem., 273, 13119-13128.
68.Peterson, R. T., Desai, B. N., Hardwick, J. S.,
and Schreiber, S. L. (1999) Proc. Natl. Acad. Sci. USA,
96, 4438-4442.
69.Ogris, E., Du, X., Nelson, K. C., Mak, E. K., Yu,
X. X., Lane, W. S., and Pallas, D. C. (1999) J. Biol. Chem.,
274, 14382-14391.
70.Tung, H. Y., De Rocquigny, H., Zhao, L. J.,
Cayla, X., Roques, B. P., and Ozon, R. (1997) FEBS Lett.,
401, 197-201.
71.Takahashi, M., Shibata, H., Shimakawa, M.,
Miyamoto, M., Mukai, H., and Ono, Y. (1999) J. Biol. Chem.,
274, 17267-17274.
72.Hsu, W., Zeng, L., and Costantini, F. (1999)
J. Biol. Chem., 274, 3439-3445.
73.Saito, T., Shima, H., Osawa, Y., Nagao, M.,
Hemmings, B. A., Kishimoto, T., and Hisanaga, S. (1995)
Biochemistry, 34, 7376-7384.
74.Seeling, J. M., Miller, J. R., Gil, R., Moon, R.
T., White, R., and Virshup, D. M. (1999) Science, 283,
2089-2091.
75.Hong, Y., and Sarge, K. D. (1999) J. Biol.
Chem., 274, 12967-12970.
76.Cayla, X., Ballmer-Hofer, K., Merlevede, W., and
Goris, J. (1993) Eur. J. Biochem., 214, 281-286.
77.Kamibayashi, C., and Mumby, M. C. (1995) in
Adv. Prot. Phosphatases (Merlevede, W., ed.) Leuven University
Press, Leuven, Vol. 9, pp. 195-210.