REVIEW: The Development of A. N. Belozersky's Ideas in Polyphosphate Biochemistry
I. S. Kulaev*, V. M. Vagabov, T. V. Kulakovskaya, L. P. Lichko, N. A. Andreeva, and L. V. Trilisenko
Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, Pushchino, Moscow Region, 142292 Russia; fax: (095) 923-3602; E-mail: alla@ibpm.serpukhov.su* To whom correspondence should be addressed.
Received December 20, 1999
This review covers some trends and approaches to the study of inorganic polyphosphates that originated from the fruitful ideas and pioneering works of A. N. Belozersky. This is, first of all, the elucidation of a close relationship between these biopolymers and nucleic acids in organisms at different evolutionary stages; second, the study of “fossil” reactions in polyphosphate metabolism that permit an understanding of their role in the evolution of phosphorus turnover and cell bioenergetics; third, the possible use of the conservative enzymes of polyphosphate metabolism, e.g., exopolyphosphatases, as molecular chronometers for obtaining additional data concerning the theory of the endosymbiotic origin of eukaryotic cells from prokaryotes.
KEY WORDS: polyphosphates, enzymes, metabolism, bioenergetics, evolution, endosymbiosis, yeast, polyphosphatase
Abbreviations: Dol-P-P-mannose) dolichyl-pyrophosphate mannose; Dol-P) dolichol phosphate; polyP) inorganic polyphosphates.
The attention of researchers to high-molecular-weight polyphosphates
(polyP) emerged as early as the start of molecular and cell biology.
These polymers are widespread in microorganisms and found in the cells
of animals and plants. The pioneers in the field of polyP investigation
were Wiame in Belgium, Ebel in France, Lohman in Germany, Kornberg and
Harold in the USA, and Szymona in Poland.
A. N. Belozersky, the outstanding researcher of nucleic acids and one of the founders of molecular biology in Russia, was also a pioneer in the field of inorganic polyP biochemistry. His interest in their functions arose in connection with studies of nuclear acids. Based on the fact that polyP were coextracted with RNA from yeast and fungal cells, he suggested the possible existence of special RNA-polyP complexes as an active form of polyP participation in cell metabolism. Being inspired with the ideas of the two coming Nobel Prize laureates, S. Spiegelman and M. D. Kamen [1], that these complexes may function in cells as special energetic donors for protein synthesis, Belozersky started to study the nature of the association between RNA and polyP and their localization in yeast and fungal cells.
A. N. Belozersky [2] and his coworker, V. B. Korchagin [3] studied the composition of volutin in diphtheria bacteria and yeasts. They showed the presence of both polyP and RNA in volutin granules of these organisms [3]. Later it was established, in collaboration with I. S. Kulaev, that the polyP-RNA complexes isolated from microorganisms had no covalent bonds, and the interaction between them was effected by means of divalent metal cations [4]. This suggested that the RNA-polyP complexes isolated by many researchers were artificial forms. However, the idea of interrelation of polyP and nucleic acids in the cell is being developed further.
The localization of polyP granules in the vicinity of bacterial nucleoid suggests their possible involvement in the regulation of gene activity [5, 6]. It has been shown that polyP are involved both in the regulation of RNA synthesis and hydrolysis [7, 8]. The most brilliant are the following examples. RNA polymerase isolated from the stationary-phase cells of Escherichia coli is found to be closely associated with polyP. These polyP may play an important role in the promoter selectivity control of RNA polymerase in E. coli both at the stationary growth phase and under high osmolarity [9, 10]. PolyP inhibit RNA processing and degradation in degradosomes, special multienzyme complexes of bacteria [7].
The study of the interrelation between polyP metabolism and the regulation of gene activity in prokaryotes is nowadays one of the new lines in the biochemistry of these compounds [11-13]. PolyP were shown to be readily involved in the regulation of RNA polymerase activity in bacteria [8-10], in the realization of bacterial response to nitrogen starvation [14], as well as their adaptation to the stationary growth phase and stresses [15]. All these polyP functions associated with the turning on and shutdown of large gene groups are covered in detail in a number of reviews [11-13] including those presented in this issue of Biochemistry (Moscow).
The totality of genetic and biochemical data obtained in the last few years brilliantly supports the idea of a close interrelation of nucleic acids and high-molecular-weight polyP. Belozersky's disciples used also another way of polyP study in their work. For example, I. S. Kulaev and E. Bukhovich investigated polyP synthesis and degradation in fungi and yeasts [16]. Further advancement of these works led to the development of two new ways of polyP biosynthesis and investigation of a number of enzymes involved in polyP synthesis and degradation both in prokaryotes and eukaryotes.
In 1957 at the Moscow International Symposium on Life's Origin, Belozersky was the first to suggest that polyP might precede ATP in evolution as a compound coupling energy [17]. He took the floor at this Symposium after the report by the famous German scientist, G. Schramm, who presented some experiments in favor of using polyP and their esters in abiogenic processes, which could lead to non-enzyme synthesis of nucleic acids and other biopolymers on the primary Earth. Nowadays, many noted researchers working in the polyP field are taking an active interest in this idea [11, 18, 19].
This review focuses our attention first of all on the development of some of Belozersky's ideas concerning the physiological significance of polyP at different stages of evolution. Among these ideas we will emphasize the suggestion of polyP involvement in bioenergetic processes as a possible precursor or a substitute for ATP. Furthermore, we will try to cover our recent studies on some enzymes of polyP metabolism in connection with the hypothesis of the endosymbiotic origin of the eukaryotic cell.
POLYPHOSPHATES AS POSSIBLE PRECURSORS OF ATP IN BIOCHEMICAL
EVOLUTION
PolyP are simpler molecules as compared with ATP. According to the data of a number of authors, they might have abiogenic origin [5, 6, 20, 21]. Therefore, the hypothesis about their preceding ATP in evolution as molecules converting energy in the cell is quite reasonable.
There is a point in favor of this concept: in the modern prokaryotes, the role of polyP in bioenergetics is still quite substantial. In these organisms there is a close relationship between polyP metabolism and the ATP-ADP-AMP system. To realize the interaction between polyP and adenyl nucleotide pools, the prokaryotes have a number of enzymes, which are unavailable in the eukaryotes. They are polyphosphate: AMP phosphotransferase [22], polyphosphate: NADP phosphotransferase [23], and polyphosphate glucokinase [24-26].
While the first two enzymes have been revealed only in some bacteria [22, 23], the polyphosphate glucokinase is more widespread and actively studied [19, 25, 26]. This enzyme, first revealed by Szymona, was purified from Propionibacterium shermanii [25] and Mycobacterium tuberculosis [26]. It was shown that the bacteria possessing this activity carried out the reaction of glucose phosphorylation due to polyP and ATP using therewith one and the same enzyme with two different active centers. In the most phylogenetically ancient microorganisms, polyphosphate glucokinase activity is several times higher than ATP-glucokinase activity, whereas the reverse is true for much younger systematic groups. No polyP-glucokinase activity has been detected in eukaryotes [19].
It was suggested that the enzyme carrying out the glucose phosphorylation due to polyP emerged at early evolutionary stages, when polyP was more significant for bioenergetics than ATP [19]. As the function of ATP in cell metabolism increased, a new active center for ATP evolved in this enzyme until, in the course of further mutations, the polyP-binding center began to lose its activity and became totally extinct in eukaryotes [19].
Scientists classify the polyP-glucokinase reaction with the so-called “fossil reactions” which are conserved in a number of modern microorganisms. One more reaction of this kind is the formation of polyP instead of ATP from 1,3-diphosphoglycerate, the most important intermediate of glycolysis. At our laboratory, 3-phosphoglyceroylphosphate polyphosphate phosphotransferase, which acts like the well-known enzyme of glycolytic phosphorylation, phosphoglycerate kinase, was revealed in some prokaryotes [5, 6].
This enzyme was also found in eukaryotic microorganisms but only under special conditions, when ATP synthesis cannot be detected. It was found in a mutant of Neurospora crassa deficient in adenine, with markedly decreased concentrations of ATP and other adenyl nucleotides [27].
An important function of polyP in the bioenergetics of prokaryotes is supported by the marked activity of a polyphosphate kinase, the key enzyme of polyP metabolism in bacteria [28]. The polyphosphate kinase, catalyzing the reactions of reverse transfer of phosphate residues from ATP to polyP, thus links two energy-rich pools. Depending on the cultivation conditions and growth stage, this enzyme can perform the function of both polyP synthesis and ATP regeneration. Moreover, this enzyme has been shown recently to perform a polyP-dependent synthesis of some other nucleoside triphosphates, thus regulating their level in the cell [29].
The question of the existence of polyphosphate kinase in lower eukaryotes is still controversial. For example, this activity was first revealed in isolated yeast vacuoles [30], where the reaction of ATP synthesis with polyP usage proceeded at a higher rate than the reverse process. Along with this, polyphosphate kinase purified earlier from yeast homogenate by Felter and Stahl [31] turned out to be a diadenosine-5´,5´´´-P1,P4-tetraphosphate-alpha,beta-phosphorylase [32]. It seems that the function of this enzyme in the yeast cell is associated not with bioenergetics but with the regulation of the levels of adenosine polyphosphates, important secondary messengers.
It is not improbable, however, that the polyphosphate kinase reaction in the membranes of eukaryotic cells may be significant in relation to the functioning of the polyP-polyhydroxybutyrate channels, which play an important role in Ca2+ transport and some other processes according to recent data [18, 33].
Nevertheless, the role of polyP in the storage and conversion of energy in cells of eukaryotic microorganisms is retained, though it is considerably modified as compared with bacteria. The involvement of polyP in the energy supply of some processes in eukaryotes often has no direct relation to ATP. For example, the study of changes in the polyP content in Saccharomyces cerevisiae at different growth stages suggests that at certain stages polyP behave as a contributor of energy to keep high growth rate rather than as a phosphate source [34]. The biomass and the total polyP content increase in parallel until the glucose level in the medium is exhausted. However, under glucose depletion but still sufficient phosphate content in the medium, the polyP content sharply decreases and then increases again. It was suggested that at this growth stage, when glycolysis cannot support the necessary ATP level in the cell, the energy of polyP was employed. Later on, the growth rate decreases and polyP can be synthesized again [34]. An active polyP synthesis observed at the logarithmic growth stage of S. cerevisiae in phosphate- and glucose-rich medium and followed by a substantial decrease in their chain length suggests that the energy released under polyP hydrolysis is critical to support the high rate of culture growth [34].
The K+ gradient is one of the energetic reserves in the yeast cell. The transport of some metal cations into a yeast cell is performed using its energy. A substantial synthesis of high-molecular-weight polyP was shown as K+ exits from cells. Some part of the energy of the K+ gradient is suggested to be stored in this way [35].
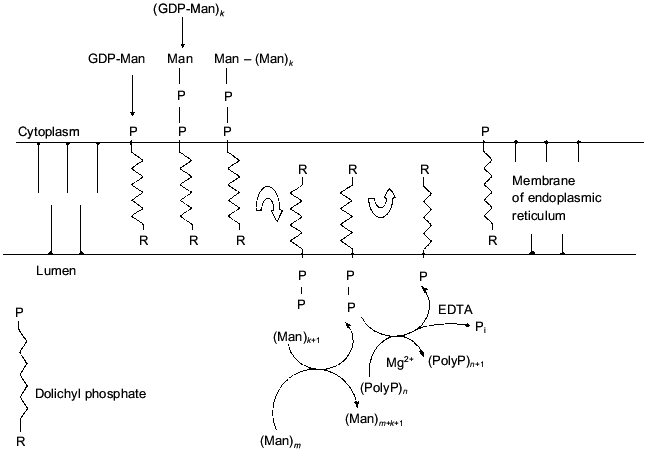
A process combining bioenergetic, structural, and transport functions of polyP in the yeast cell wall was revealed at our laboratory [36-38]. The polyP localized on the yeast cell surface are synthesized by means of a special metabolic pathway closely connected with mannoprotein synthesis in the cell wall. This process begins in the endoplasmic reticulum (ER). The formation of Dol-P-P-mannose from GDP-mannose and Dol-P occurs at the cytoplasmic side of the ER membrane. Dol-P-P-mannose is transported across the ER membrane, and the mannosyltransferase and Dol-P-P-polyP phosphotransferase reactions proceed on the lumen side of the ER membrane. The released Dol-P crosses the membrane in the opposite direction. Produced mannoproteins and polyP are transferred to the cell wall by special transport vesicles (Fig. 1). Dolichyl:polyP phosphotransferase synthesizing polyP with beta-phosphate groups of dolichyl pyrophosphate was solubilized from preparations of ER membranes using Triton X-100. A specific activity in the preparation of solubilized enzyme was 20-fold higher than that in protoplast homogenate. The enzyme was metal-dependent, exhibited the highest activities in the presence of Mg2+ or Ca2+, and was completely inhibited by EDTA. The availability of this enzyme enables the cell to store some part of the energy consumed under mannoprotein synthesis in the form of polyP.
Thus, the role of polyP in cell bioenergetics under evolutionary development from prokaryotes to eukaryotes even if somewhat decreased took some new features and peculiarities in various compartments. However, the role of direct interaction between polyP and adenyl nucleotides in eukaryotes is likely to be modest. It appears to show itself only in mitochondria [39] and very likely in vacuoles [30].Fig. 1. Correlation between mannoprotein synthesis in the cell wall and polyphosphates. (Man)n is mannan, where n is the number of mannose residues; GDP-Man is GDP-mannose.
DIVERSITY OF EXOPOLYPHOSPHATASES IN SEPARATE COMPARTMENTS OF THE
YEAST CELL IS AN ARGUMENT IN FAVOR OF THE ENDOSYMBIOTIC ORIGIN OF
EUKARYOTIC CELLS
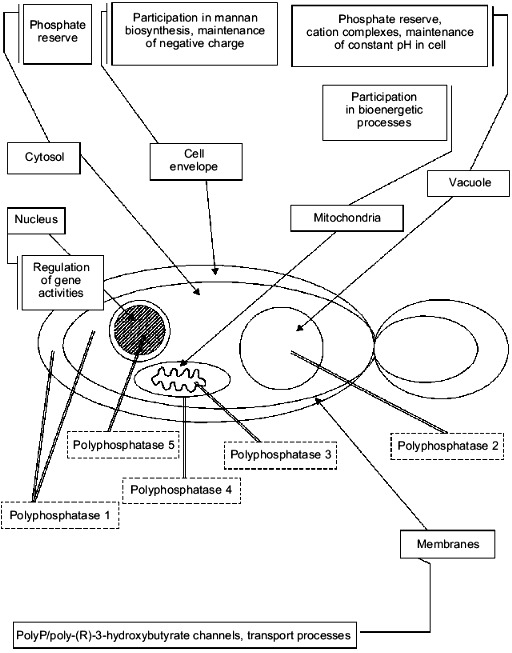
The central enzyme of polyP metabolism in the yeast is an exopolyphosphatase--an enzyme splitting Pi from the polyP chain end. Its activity is about two orders higher in the yeast cell homogenate than in the cells of E. coli. The cytosol fraction contains no less than 60% of the polyphosphatase activity of S. cerevisiae, and the remaining activity is associated with other compartments [40]. We revealed that polyphosphatase activities are characteristic not only of the cytosol of S. cerevisiae but of cell envelopes [41], nuclei [42], mitochondria [43], and vacuoles [44] as well. It is not improbable that similar activities can be revealed in some other compartments, such as various cell membranes. We managed to purify some of the yeast polyphosphatases, namely, the cell envelope [41], cytosol [40], and vacuolar polyphosphatases [45]. Isolation and purification of a soluble polyphosphatase of mitochondria are presented in this issue of Biochemistry (Moscow). Polyphosphatase activities of isolated nuclei [42] and preparations of mitochondrial membranes [46] have been characterized as well. All the activities under study exhibit some similar properties, i.e., they are able to hydrolyze polyP with different chain lengths, have optimal activity at neutral pH, and do not hydrolyze pyrophosphate, p-nitrophenylphosphate, and nucleoside triphosphates.
Further investigation of their properties (table) allowed us to conclude that only polyphosphatases of the cell envelope and cytosol (polyphosphatase 1, see also Fig. 2) are similar. Both these enzymes have the same molecular masses (~40 kD), the same sensitivity to antibodies against the purified cell-envelope polyphosphatase, and differ slightly in their substrate specificity, requirement for divalent cations, and the effect of EDTA. The comparison of their properties with those of the known polyphosphatase from the yeast homogenate PPX [47] suggests that these enzymes seem to be products of the same PPX gene with possible posttranscriptional modifications.
Properties of exopolyphosphatases from S. cerevisiae yeast
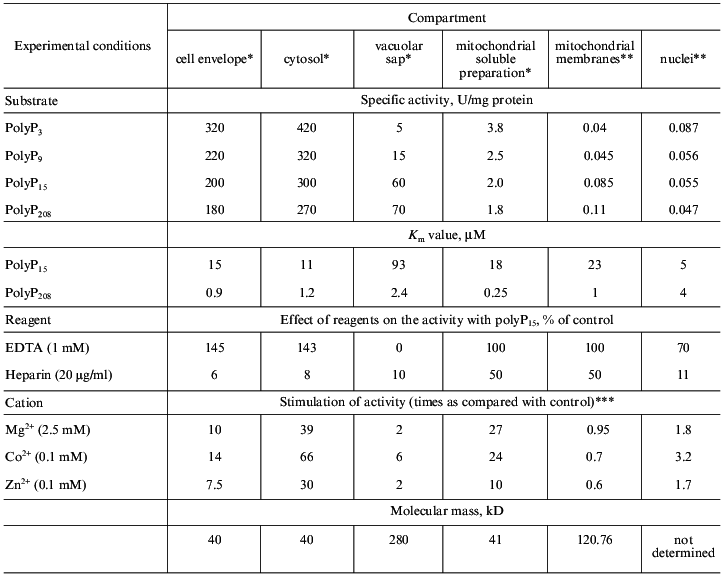
*Purified enzymes.
**Isolated subcellular fractions.
***In the absence of cations, the specific activities with polyP15
as a substrate were 20, 7.5, 10, 0.08, 0.09, and 0.028 U/mg
protein in cell envelope, cytosol, vacuolar sap, mitochondrial soluble
fraction, mitochondrial membranes, and nuclei, respectively.
The distinctions in the extent of stimulation of polyphosphatases by divalent cations are probably due to different methods of purification of these enzymes. The enzymes could retain different quantities of bound cations present in exopolyphosphatase molecules. Under the optimal conditions, the specific activities of cytosol and cell envelope polyphosphatase preparations that we obtained were nearly the same.Fig. 2. Localization, polyP functions, and diversity of exopolyphosphatases in different compartments of the yeast cell.
It should be particularly emphasized that under immunoblotting, the presence of a protein with molecular mass of ~40 kD was observed in none of the subcellular fractions under study, and none of the protein reliably reacted with antibodies. This was an indication of the absence of polyphosphatase 1 in isolated organelles.
All the other polyphosphatases under study were distinguished from the above two by their molecular masses, substrate specificities, and requirements for divalent cations (table). It is unlikely that with such distinctions they could be products of the same PPX gene [47]. The study of the N-terminal sequence of mitochondrial polyphosphatase demonstrated that the PPX gene does not encode it.
The most interesting question is, of course, the functional significance of the occurrence of different polyphosphatases in separate compartments of the yeast cell. So far we can put forward only some speculations on this subject. First, there are a number of essential differences between some of them in substrate specificity. For example, cytosol polyphosphatase splitting tripolyphosphate with the highest activity can also hydrolyze adenosine tetra- and guanosine tetraphosphates [48], which are precursors of important secondary signal molecules involved in the survival of the culture under stresses and during the stationary stage. Therefore, the cytosol polyphosphatase could participate in the regulation of the cytoplasmic content of these compounds. The ability of the cytosol and cell-envelope polyphosphatases to hydrolyze polyP to pyrophosphate suggests as well their participation in polyP usage as a phosphate reserve. It is significant that some of their properties, in particular, requirement for the detergent Triton X-100 to retain their activity under purification, point to their possible ability to be bound to a membrane under certain conditions [40, 41]. We suggest that these enzymes could be involved in the polyP synthesis under certain conditions, such as the absence of cations and association with the membrane.
The vacuolar polyphosphatase is almost inactive with tripolyphosphate and does not hydrolyze tetraphosphate nucleoside derivatives. In our opinion, its function may involve the maintenance of acid pH in vacuoles under unfavorable conditions (in particular, under alkaline stress) by means of a partial polyP hydrolysis in these organelles. The retained tripolyphosphate under changing conditions could serve as a primer for the synthesis of new polyP chains. It should be mentioned that there is a polyphosphatase activity in the vacuolar membrane [44], which seems to be involved in phosphate transport into these organelles. This transport process does not depend on ATP and is not likely to be associated directly with the electrochemical gradient of hydrogen ions across the vacuolar membrane [49], in contrast to a number of other transport processes across the tonoplast. It is not improbable that phosphate transport into a vacuole is connected with the polyP metabolism just as it is in lysosomes.
The unique properties of mitochondrial membrane polyphosphatase, i.e., its strong association with the membrane and negative sensitivity to divalent cations [46], suggest that its functional significance may be due to the functioning of polyP/poly-(R)-3-hydroxybutyrate channels revealed recently by Reusch [50]. Their functioning is directly related both to the elongation of the polyP chain from one membrane side and hydrolysis of their terminal residues from the other side.
Certainly, we are only approaching an understanding of polyphosphate functions in yeast cell compartments, but even a simple analysis of their properties leaves no doubt that all the distinctions we have found are related to the functions of these enzymes (Fig. 2).
One more argument for the existence of all these forms seems to be the evolutionary origin of eukaryotic cells. According to the modern concept, organelles originated from various ancient prokaryotes by means of their incorporation in the primary host cell [51, 52]. The properties of some enzymes, say, ATPases, are well suited to this concept. For example, ATPases of mitochondria and eubacteria relate to the F1F0 type and those of vacuoles and some archae to the V1V0 type. The polyphosphatases do not fit the above scheme. None of the yeast mitochondrial polyphosphatases that we revealed shows any similarity with that of E. coli [53, 54]. This implies that either such an enzyme was not conserved in eukaryotes during evolution or the mitochondrial precursor differed greatly in its polyP metabolism and enzymes from that of E. coli. The archae polyphosphatases have not yet been studied, and we have no data for comparison. Further investigations, including the sequencing of yeast polyphosphatases and comparison of the genes that encode them with those of bacteria, would elucidate the question of the origin of eukaryotic microbial polyphosphatases.
Our view of the functions of polyP in microorganisms has been expanded substantially in the last decade. The essential regulatory functions on a well-grounded basis are now assigned to polyP in addition to quite an evident function of phosphagens. The polyP have assumed quite a new significance for modern biotechnology. Their study is accepted throughout the world to be promising for the solution of the problem of wastewater clean-up from phosphate [55, 56], creation of biodegradable fibers for medicine and technical needs [57], and development of new approaches to combating pathogenic microorganisms and viruses [12, 13, 58]. We should be impressed with the sagaciousness of the pioneers in the field of molecular biology, who drew attention to these compounds and put forward a number of ideas that turned out to be very fruitful in further development of polyP biochemistry.
The experimental part of this work was supported by the Russian Foundation for Basic Research (grant No. 99-04-48246) and INCO-Copernicus PL 971185.
REFERENCES
1.Spiegelman, S., and Kamen, M. D. (1947) Cold
Spring Harbor Symp. Quant. Biol., 12, 211.
2.Belozersky, A. N. (1945) Mikrobiologiya,
14,29-31.
3.Belozersky, A. N., Korchagin, V. B., and Smirnova,
T. I. (1950) Dokl. Akad. Nauk SSSR, 71,89-91.
4.Kulaev, I. S., and Belozersky, A. N. (1957)
Biokhimiya, 22, 587-596.
5.Kulaev, I. S. (1979) Biochemistry of Inorganic
Polyphosphates, John Wiley and Sons, Chichester, N. Y.
6.Kulaev, I. S., and Vagabov, V. M. (1983) Adv.
Microbiol. Physiol., 24, 83-171.
7.Blum, E., Py, B., Carpousis, A. J., and Higgins, C.
F. (1997) Mol. Microbiol., 26, 387-398.
8.Kim, H. Y., Schlictman, D., Shankar, S., Xie, Z.
D., Chakrabarty, A. M., and Kornberg, A. (1998) Mol. Microbiol.,
27, 717-725.
9.Kusano, S., and Ishihama, A. (1997) Genes
Cells, 2, 433-441.
10.Shiba, T., Tsutsumi, K., Yano, H., Ihara, Y.,
Kameda, A., Tanaka, K., Takahashi, H., Munekata, M., Rao, N. N., and
Kornberg, A. (1997) Proc. Natl. Acad. Sci. USA, 94,
11210-11215.
11.Kornberg, A. (1995) J. Bacteriol.,
177, 491-496.
12.Kornberg A. (1999) in Inorganic
Polyphosphates. Biochemistry, Biology, Biotechnology (Schroder, H.
C., and Muller, W. G. E., eds.) Springer-Verlag,Berlin-Heidelberg-N.
Y., pp. 1-19.
13.Kornberg, A., Rao, N. N., and Ault-Rich, D.
(1999) Ann. Rev. Biochem., 68, 89-125.
14.Rao, N. N., Liu, S., and Kornberg, A. (1998)
J. Bacteriol., 180, 2186-2193.
15.Rao, N. N., and Kornberg, A. (1996) J.
Bacteriol., 178, 1394-1400.
16.Buhovich, E., and Belozersky, A. N. (1958)
Biokhimiya, 23, 254-259.
17.Belozersky, A. N. (1959) Speech in Discussion
at Symposium “Origin of Life on the Earth” [in Russian],
Izd-vo AN SSSR, Moscow, p. 370.
18.Reusch, R. N. (1999) in Inorganic
Polyphosphates. Biochemistry, Biology, Biotechnology (Schroder, H.
C., and Muller, W. G. E., eds.) Springer-Verlag,Berlin-Heidelberg-N.
Y., pp. 151-183.
19.Phillips, N. F. B., Hsien, P. C., and Kowalczyk,
T. H. (1999) in Inorganic Polyphosphates. Biochemistry, Biology,
Biotechnology (Schroder, H. C., and Muller, W. G. E., eds.)
Springer-Verlag,Berlin-Heidelberg-N. Y., pp. 101-127.
20.Yamagata, Y., Watanabe, H., Saitoh, M., and
Namba, T. (1991) Nature, 352, 516-519.
21.Kulaev, I. S. (1994) J. Biol. Phys.,
24, 255-273.
22.Bonting, C. F., Korstee, G. J., and Zehnder, A.
J. (1991) J. Bacteriol., 173, 6484-6488.
23.Murata, K., Kato, J., and Chibata, I. (1979)
Biotechnol. Bioeng., 21, 887-895.
24.Szymona, M. (1957) Bull. Acad. Pol. Sci. Ser.
Sci. Biol., 5, 379-382.
25.Phillips, N. F., Horn, P. J., and Wood, H. G.
(1993) Arch. Biochem. Biophys., 300, 309-319.
26.Hsieh, P. C., Shenoy, B. C., Jentoft, J. E., and
Phillips, N. F. B. (1993) Protein. Exp. Purif., 4,
76-84.
27.Kulaev, I. S., and Bobyk, M. A. (1971)
Biokhimiya, 36, 426-429.
28.Ahn, K., and Kornberg, A. (1990) J. Biol.
Chem., 265, 11734-11739.
29.Kuroda, A., and Kornberg, A. (1997) Proc.
Natl. Acad. Sci. USA, 94, 439-442.
30.Shabalin, Yu. A., Vagabov, V. M., Tsiomenko, A.
B., Zemlyanuhina, O. A., and Kulaev, I. S. (1977) Biokhimiya,
42, 1642-1648.
31.Felter, S., and Stahl, A. J. C. (1973)
Biochimie, 55, 245-251.
32.Booth, J. W., and Guidotti, G. (1995) J. Biol.
Chem., 270, 19377-19382.
33.Reusch, R. N., Huang, R. P., and Koskkosicka, D.
(1997) FEBS Lett., 412, 592-596.
34.Vagabov, V. M., Trilisenko, L. V., Shchipanova,
I. N., Sibeldina, L. A., and Kulaev, I. S. (1998)
Mikrobiologiya, 67,188-193.
35.Okorokov, L. A., Lichko, L. P., and Andreeva, N.
A. (1983) Biochem. Int., 6, 481-488.
36.Shabalin, Yu. A., Vagabov, V. M., and Kulaev, I.
S. (1979) Dokl. Akad. Nauk SSSR, 249, 243-246.
37.Shabalin, Yu. A., Vagabov, V. M., and Kulaev, I.
S. (1985) Dokl. Akad. Nauk SSSR, 283, 720-723.
38.Shabalin, Yu. A., and Kulaev, I. S. (1989)
Biokhimiya, 54, 68-75.
39.Beauvoit, B., Rigonlet, M., Guerin, B., and
Canioni, P. (1989) FEBS Lett., 252, 17-22.
40.Andreeva, N. A., Kulakovskaya, T. V., and Kulaev,
I. S. (1996) Biochemistry (Moscow), 61, 1213-1220.
41.Andreeva, N. A., and Okorokov, L. A. (1993)
Yeast, 9, 127-139.
42.Lichko, L. P., Kulakovskaya, T. V., and Kulaev,
I. S. (1996) Biochemistry (Moscow), 61, 361-366.
43.Lichko, L. P., Kulakovskaya, T. V., and Kulaev,
I. S. (1996) Biochemistry (Moscow), 61, 1176-1181.
44.Andreeva, N. A., Lichko L. P., Kulakovskaya T.
V., and Okorokov, L. A. (1993) Biochemistry (Moscow), 58,
737-744.
45.Andreeva, N. A., Kulakovskaya, T. V., and Kulaev,
I. S. (1998) FEBS Lett., 429, 194-196.
46.Lichko, L. P., Kulakovskaya, T. V., and Kulaev,
I. S. (1998) Biochim. Biophys. Acta, 1372, 153-162.
47.Wurst, H., Shiba, T., and Kornberg, A. (1995)
J. Bacteriol., 177, 898-906.
48.Kulakovskaya, T. V., Andreeva, N. A., and Kulaev,
I. S. (1997) Biochemistry (Moscow), 62, 1051-1052.
49.Kulakovskaya, T. V., and Kulaev, I. S. (1997)
Microbiologia SEM, 13, 71-74.
50.Reusch, R. N., and Sadoff, H. L. (1988) Proc.
Natl. Acad. Sci. USA, 85, 4176-4180.
51.Margulis, L. (1993) Symbiosis in Cell
Evolution, Freeman, San Francisco.
52.Nelson, N. (1992) Biochim. Biophys. Acta,
1100, 109-124.
53.Akiyama, M., Crooke, E., and Kornberg, A. (1992)
J. Biol. Chem., 267, 22556-22561.
54.Keasling, J. D., Bortish Le Roy, and Kornberg, A.
(1993) Proc. Natl. Acad. Sci. USA, 90, 7029-7033.
55.Mino, T., van Loosdrecht, M. C. M., and Heijnen,
J. J. (1998) Wat. Res., 32, 3193-3207.
56.Kortstee, G. J. J., and van Veen, H. W. (1999) in
Inorganic Polyphosphates. Biochemistry, Biology, Biotechnology
(Schroder, H. C., and Muller, W. G. E., eds.)
Springer-Verlag,Berlin-Heidelberg-N. Y., pp. 275-299.
57.Griffith, E. J. (1992) Chemtech.,
22, 220-226.
58.Lorenz, B., Leuck, J., Kohl, D., Muller, W. E.
G., and Schroeder, H. C. (1997) J. Acquired Immun. Defic. Syndr.
Hum. Retrovir., 14, 110-118.