REVIEW: Application of Polyphosphate Metabolism to Environmental and Biotechnological Problems
J. D. Keasling*, S. J. Van Dien, P. Trelstad, N. Renninger, and K. McMahon
Department of Chemical Engineering, University of California, Berkeley, CA 94720-1462; fax: (510) 643-1228; E-mail: keasling@socrates.berkeley.edu* To whom correspondence should be addressed.
Received November 12, 1999
The synthesis and degradation of polyphosphate (polyP) are influenced by the energy state of the cell and extracellular phosphate levels. The import of excess phosphate and its incorporation into polyP under phosphate- and energy-rich growth conditions allows organisms to survive when phosphate or energy are depleted. Under phosphate-starvation conditions, phosphate can be recovered from polyP by hydrolysis. When the organism is energy starved, energy can be recovered either by regenerating the high-energy phosphoanhydride bond donor (ATP in most cases) or by hydrolysis of polyP and subsequent secretion of orthophosphate to recharge the transmembrane proton gradient. Understanding how the energy state of the cell and environmental phosphate levels affect polyP metabolism is essential to improving such environmental processes as enhanced biological phosphorus removal, a treatment process that is widely used to remove excess phosphate from wastewater. Manipulation of the genes responsible for polyP metabolism can also be used to improve gene expression from phosphate-starvation promoters and to remove heavy metals from contaminated environments.
KEY WORDS: polyphosphates, environment, heavy metals, energy metabolism, enhanced biological phosphorus removal
Abbreviations: EBPR) enhanced biological phosphate removal; PAO) polyP accumulating organisms; PHA) polyhydroxyalkanoate; polyP) polyphosphates; PPK) polyP kinase.
The primary energy currency of living systems is the high-energy
phosphoanhydride bond. A number of organisms (including bacteria,
fungi, plants, and animals) store energy and phosphate in phosphate
polymers of three to more than a thousand residues called polyphosphate
(polyP) [1, 2]. The ubiquity of
this polymer in organisms indicates a common physiological function,
most likely energy and phosphate storage. In this paper, we describe
its important role in some environmental and biotechnological
problems.
PolyP is synthesized by polyP kinases, which reversibly transfer the phosphate residue from a high-energy donor to the polyP chain. PolyP:ADP phosphotransferase (polyP kinase, PPK), which transfers the terminal phosphate of ATP to polyP, has been purified from Escherichia coli [3], and the gene encoding that enzyme (ppk) has been cloned and overexpressed [4]. Similar genes have been cloned from a number of organisms [5-15].
Inorganic phosphate (Pi) can be released from polyP by the action of polyPase [16]. An exopolyPase (PPX), which catalyzes the removal of the terminal phosphate of polyP, was purified to homogeneity from E. coli overproducing PPK. The gene encoding PPX is located in the same operon with ppk [16]. A second polyPase in E. coli has been purified and identified as guanosine pentaphosphate phosphohydrolase (GPP), an enzyme that hydrolyzes the 5´-gamma-phosphate of pppGpp to form ppGpp [17].
PolyP, ENERGY METABOLISM, AND THE CELL MEMBRANE
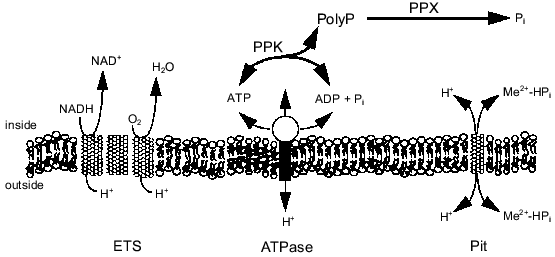
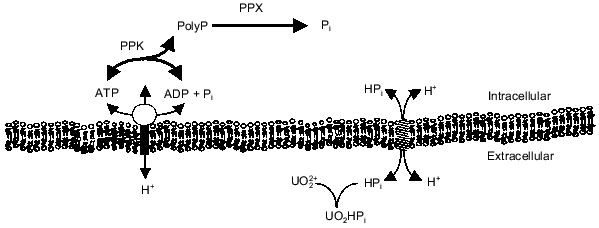
The energy state of the cell may be a key factor influencing the synthesis of polyP from ATP or regeneration of ATP from polyP. There is evidence that polyP accumulates during energy-rich growth conditions and is degraded during energy-poor conditions [18, 19]. Kornberg and coworkers have shown that a high concentration of ATP stimulates in vitro synthesis of polyP by PPK by removing the lag characteristic of polyP synthesis on low levels of ATP [3]. Furthermore, they found that ADP inhibits polyP synthesis [3, 20]. These results imply that the energy charge (([ATP] + 0.5[ADP])/([ATP] + [ADP] + [AMP])) of the cell influences polyP polymerization and depolymerization by PPK [21]. Indeed, polyP metabolism may be one way in which the cell maintains the energy charge. By maintaining a significant level of PPK in the cell, the cell could passively regulate the energy charge by regenerating ATP from ADP and polyP or by storing excess phosphoanhydride bonds (energy) as polyP. Recently, Van Veen and coworkers suggested that Pi generated from the degradation of polyP by a polyPase in Acinetobacter sp. may contribute to the transmembrane proton gradient and thereby partially conserve the energy of the phosphoanhydride bond [22]. The relationship of polyP metabolism to ATP synthesis, the transmembrane proton gradient, and Pi transport is shown in Fig. 1. The work in our laboratory has focused on the interactions of polyP metabolism and the cell membrane and how these interactions can be used for biotechnology and environmental applications.
Fig. 1. PolyP interaction with phosphate and energy metabolism. Abbreviations: ETS) electron transport; ATPase) H+-translocating ATPase; Pit) phosphate inorganic transport;PPK) polyP kinase; PPX) polyphosphatase.
PolyP AND ENHANCED BIOLOGICAL PHOSPHORUS REMOVAL
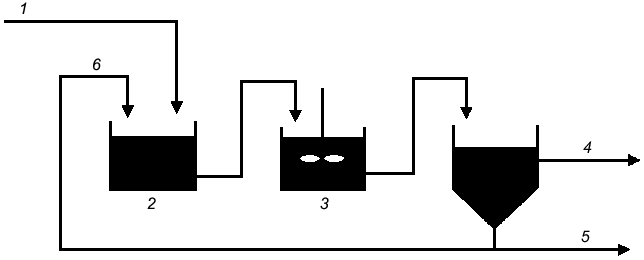
PolyP is central to enhanced biological phosphorus removal (EBPR) from municipal wastewater, a process that is accomplished through a number of surplus-starvation cycles (Fig. 2). In the anaerobic zone of the treatment system, the cells are electron-acceptor deficient and carbon rich. It has been proposed that polyP is degraded to Pi, which is secreted from the cell to increase the transmembrane proton gradient (Fig. 3). Carbon is then taken up via a proton-symport pump and stored inside the cell as polyhydroxyalkanoate (PHA). In the subsequent aerobic zone of the treatment system, the environment is electron-acceptor rich but carbon deficient. It has therefore been proposed that the PHA is degraded and polyP is synthesized from ATP generated from the metabolism of PHA. Since more Pi is taken up during the aerobic phase than is secreted during the anaerobic phase, there is a net removal of Pi from wastewater.
Fig. 2. Typical flow sheet for an EBPR reactor system: 1) phosphate-laden water; 2) anaerobic; 3) aerobic; 4) purified water; 5) settler; 6) return activated sludge.
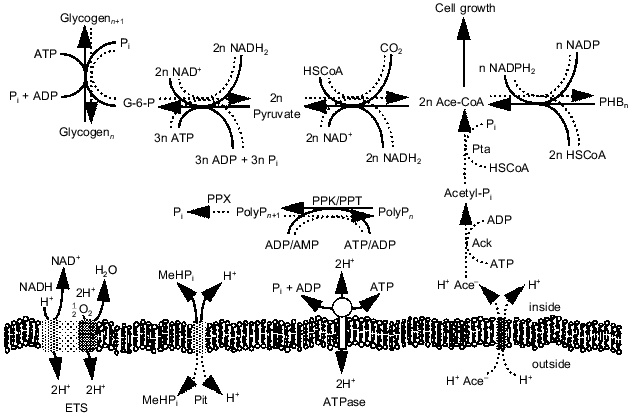
Analysis of EBPR metabolism. While the appearance and disappearance of the key biopolymers (polyP, PHA, and glycogen) in the EBPR process have been well documented, the intracellular pressures that lead to their synthesis and degradation are not well understood. To understand how carbon, energy, and redox potential are channeled through the metabolic pathways in each treatment process stage, we developed a metabolic flux model that contained a complete set of the pathways involved in biomass synthesis and energy production in bacteria [23, 24]. The model accounts for the energy requirements of macromolecule synthesis and of metabolite transport across the cell membrane. The equations for the 163 reversible and 166 irreversible reactions were solved using linear optimization. Data from a laboratory-scale sequencing batch reactor performing EBPR were used as model inputs. The model was able to make reasonable predictions of which biopolymers were consumed during each phase and at what rates, given the rates of synthesis of the pertinent biopolymers in that phase. In addition, the model yielded information on the pathways by which the energy-rich molecules ATP, NADH, and NADPH are produced and consumed during EBPR. In doing so, the model supports the hypothesis that biopolymer metabolism provides a means for organisms to balance intracellular energy supplies. Moreover, it suggests pathways at which metabolic regulation should occur. Unlike other EBPR models, the stoichiometric model provides a detailed and comprehensive account of the complete metabolism of the organisms involved.Fig. 3. Pathways proposed to be involved in EBPR. Solid lines, direction under aerobic growth conditions; dotted lines, direction under anaerobic growth conditions. Abbreviations: ETS) electron transport system; Pit) phosphate inorganic transport; ATPase) ATP synthase; Pta) phosphotransacetylase; Ack) acetyl kinase; PPK) polyP kinase; PPX) polyphosphatase.
Microbial community structure in EBPR systems. Although models of EBPR (such as that described above) describe the metabolism in a single organism, no such organism has been shown to exhibit the EBPR behavior in pure culture. The isolation of confirmed polyP accumulating organisms (PAOs) has been unsuccessful, in spite of extensive efforts for almost 25 years [25]. It appears that PAOs will not readily grow in pure culture on media routinely used to cultivate microbes. Indeed, single cells obtained directly from EBPR sludges would not grow on standard media [25]. It has been suggested that perhaps PAOs cannot grow in pure culture and that a community minimally composed of essential populations is required for successful EBPR, though no ecological data exist that support this hypothesis [25]. Further studies on the microbial ecology of EBPR sludges may suggest syntrophic interactions or specialized growth requirements that could lead to more sophisticated culturing strategies.
Acinetobacter spp. were long thought to be primarily responsible for EBPR because they were routinely isolated from EBPR sludges using classical culture-based methods. Ribosomal RNA-based studies and others have recently demonstrated that Acinetobacter, although capable of accumulating polyP under certain conditions, is not primarily responsible for EBPR in lab-scale reactors or in prototype wastewater treatment plants [25]. The rRNA-based studies led to the tentative conclusion that organisms carrying out EBPR in most systems can be phylogenetically classified as either beta-proteobacteria and/or actinobacteria [26-32]. One research group reported that actinobacteria and alpha-proteobacteria were the only PAOs in their system, while several laboratories (including our own) have determined that a Rhodocyclus-like organism was by far the dominant PAO in lab-scale sequencing batch reactors [32, 33]. Our own work has shown that Rhodocyclus spp. and close relatives dominate high phosphorus-containing cultures, and that high GC Gram-positive bacteria were not present in appreciable numbers [34]. A likely explanation for these conflicting results could be that several different kinds of organisms can carry out EBPR [25]; subtle differences between system design, operation, and/or feed characteristics could select for different kinds of PAOs. For studying PAO metabolism, any organism is acceptable as long as it exhibits all of the characteristic transformations of EBPR. However, to improve the performance and reliability of full-scale EBPR plants, we still must understand how populations of key organisms (PAOs or not) interact in these systems, and how changes in community structure affect process performance.
Biochemistry and genetics of EBPR. One of the organisms most often cited in early studies of EBPR was Acinetobacter. While this organism may not be responsible for EBPR in many wastewater treatment plants, it serves as a good model organism for developing molecular techniques to characterize metabolism and genetic control in potential EBPR organisms. A number of Acinetobacter species have been isolated, and, like many of them, Acinetobacter sp. strain ADP1 accumulates large amounts of polyP under certain conditions. Elucidating the behavior and regulation of polyP metabolism in this organism should lead to a better understanding of the metabolism of polyP in true EBPR organisms.
Recently, the ppk gene from Acinetobacter sp. strain ADP1 was cloned [35] and the PPK was purified and characterized [36]. In vivo,the PPK showed interesting behavior when the Acinetobacter cultures were subjected to a cycle of Pi starvation and surplus. Although the ppk gene was strongly induced under Pi-limited conditions, net polyP-synthesis activity declined and polyP levels became almost negligible. It was not until after the addition of excess Pi that both polyP-synthesis activities and polyP levels rose. In addition, a strong polyP-degrading activity, which appeared to be mainly due to a polyphosphatase (PPX) and not PPK working in reverse, was detected in cultures grown under low Pi conditions. This activity declined upon the addition of Pi. (Indeed, under most conditions the Acinetobacter PPK works poorly in reverse. If PPK does not work (or works only poorly) in reverse in vivo, then the cell would be forced to use PPX and the phosphate-inorganic (Pit) system to recover energy from polyP.) This work suggests that the formation of polyP-producing enzymes is linked to the formation of polyP-degrading enzymes. Thus, the same conditions (here, Pi starvation) that trigger the induction of PPX lead to the induction of PPK. When the conditions change (e.g., Pi is added) PPK is ready and available to form polyP. This type of regulatory feature could also exist for other nutritional stresses, such as the carbon starvation to surplus and anaerobic to aerobic shifts that occur in EBPR systems. In addition, this work suggests that EBPR researchers may want to consider the role of Pi levels in EBPR metabolism as well as the roles of the carbon and oxygen shifts that usually are studied.
MODULATION OF THE PHOSPHATE STARVATION RESPONSE
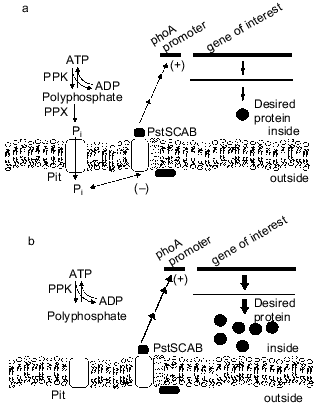
The phosphate-starvation promoter of E. coli has been widely used for the expression of heterologous genes when the desired product can be made during the non-growth stage [37]. This promoter offers the advantages of being relatively easy and inexpensive to use. Recently, we found that intracellular polyP can cause reduced yield from these constructs, as a ppk ppx mutant strain of E. coli produced more alkaline phosphatase during Pi starvation than did an isogenic control strain [38]. The basis for the reduction in expression from phosphate-starvation promoters is polyphosphate degradation by PPX, secretion of phosphate from the cell, and subsequent repression of the phosphate-starvation response [39, 40] (Fig. 4). These results have important implications for the use of the phosphate starvation promoter to express heterologous genes in industrial fermentations.
A dynamic model of the E. coli Pi starvation (Pho) regulon was extended to investigate the effect of polyP synthesis and degradation on this control system [41, 42]. In agreement with recent experimental observations, the degradation of polyP by PPX during a period of Pi limitation could repress the Pi-starvation response. This is attributed to the release of Pi from the cell into the periplasm, where it can be detected by the external Pi sensor.Fig. 4. Effect of polyP degradation on phosphate-starvation promoters. a) In the wild-type cells, polyP is degraded by PPX to Pi, which is secreted from the cells. The excess Pi in the vicinity of the cell membrane interacts with the PstSCAB system and downregulates the phosphate-starvation response. Heterologous genes under control of the phosphate-starvation promoters are downregulated, and a lower level of desired protein is produced. b) Elimination of the PPX gene prevents degradation of polyP, such that stored polyP has no effect on the phosphate-starvation response.
PolyP AND HEAVY METALS
There is strong evidence in recent literature that polyP is involved in storage of and tolerance to heavy metals. PolyP in the vacuoles of yeast, fungi, and algae, and in the granules of bacteria are often associated with positively charged, low-molecular-weight compounds (amino acids and divalent cations). The cations associated most often with polyP bodies are Ca2+, K+, and Mg2+. However, heavy metals have also been found in the polyP granules of certain bacteria [43-45], in the polyP-containing vacuoles of Saccharomyces cerevisiae [46-49], and in polyP of algae [50].
Heavy metal tolerance. It has been proposed that cells use polyP to detoxify heavy metals once they have entered the cell [43, 51-53]. It has also been suggested that surface-associated polyP may be important in chelation of cations on the surface of cells [54, 55]. However, more recent evidence suggests that polyP is degraded during growth in the presence of heavy metals [53, 56, 57]. Our own work in this area using the cyanobacterium Anacystis nidulans [58] and genetically-engineered strains of E. coli [59] supports the latter hypothesis. The polyphosphate metabolic pathways in E. coli were genetically manipulated to test the effect of polyphosphate on tolerance to cadmium. A strain mutant in the genes for polyphosphate kinase (ppk) and polyphosphatase (ppx) produced no polyphosphate, whereas the same strain carrying multiple copies of ppk on a high-copy plasmid produced significant quantities. The doubling times of both strains increased with increasing cadmium concentration. In contrast, the mutant strain carrying multiple copies of ppk and ppx produced one-tenth of the polyphosphate found in the strain carrying multiple copies of ppk only and showed no significant increase in doubling times over the same cadmium concentration range. This work [59] indicates that the ability to synthesize and degrade polyP is important for tolerance to heavy metals, in contrast to other literature that claims that only a large quantity of intracellular polyP is important.
Heavy metal removal. The apparent relationship between polyP and increased resistance to heavy metals indicates a potential application of organisms with engineered polyP pathways toward the bioremediation of heavy metal contamination in wastewater (Fig. 5). The precipitation of these metals on the cell surface, which may be coincident with depletion of polyP reserves [57], is particularly appealing. Metal phosphates are highly insoluble and will precipitate on cell surfaces [60, 61]. Recently, we found that Pseudomonas aeruginosa expressing a plasmid-encoded, inducible ppk accumulated large quantities of polyP. When the polyP-filled organism was exposed to uranyl under phosphate-limiting conditions, the polyP was degraded and the cells removed the uranyl from solution. It appears that the uranyl precipitated on the outside of the cells as a phosphate complex (Fig. 6). Cells that were not induced for ppk expression did not remove uranyl from solution.
Fig. 5. Use of stored polyP for precipitation of uranyl (UO22+) on the cell surface.
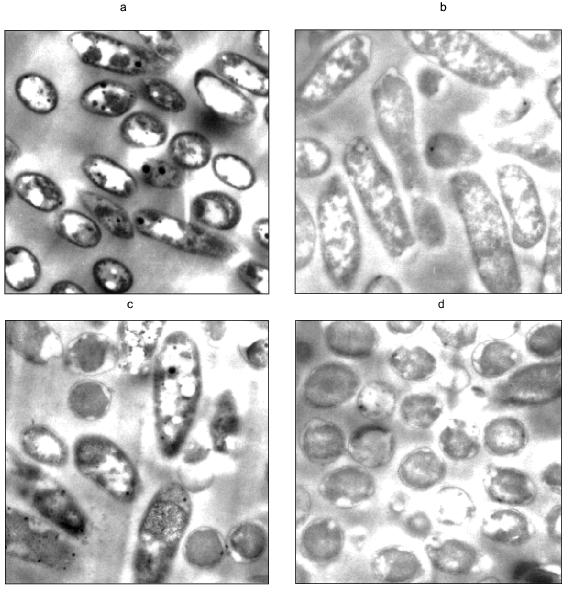
In conclusion, polyP plays a critical role in several environmental and biotechnological problems. Understanding how environmental conditions affect native polyP metabolism can lead to improvements in existing wastewater treatment processes. Manipulating polyP metabolism through genetic and metabolic engineering has resulted in improved control over heterologous gene expression from phosphate-starvation promoters and new processes to remove heavy metals contaminants.Fig. 6. Transmission electron micrographs of polyP filled cells challenged with uranyl. Pseudomonas aeruginosa was genetically engineered to overexpress ppk, such that cells induced for ppk expression accumulated large quantities of polyP. The cells were then resuspended in a solution only containing 1 mM UO2NO3. a, b) Cells resuspended in uranyl nitrate; c, d) cells resuspended in water; a, c) cells induced for ppk expression; b, d) cells not induced. Dark and white circular areas seen in cells induced for ppk expression are attributed to polyP. The darkening of the cell membranes in (a) is attributed to electron dense uranyl phosphate precipitation.
This research was funded by the U. S. National Science Foundation (BES-9612840) and the U. S. Department of Energy NABIR program.
REFERENCES
1. Kulaev, I. S. (1979) The Biochemistry of
Inorganic Polyphosphates, John Wiley & Sons, N. Y.
2. Wood, H. G., and Clark, J. E. (1988) Ann. Rev.
Biochem.,57, 235-260.
3. Ahn, K., and Kornberg, A. (1990) J. Biol.
Chem.,265, 11734-11739.
4. Akiyama, M., Crooke, E., and Kornberg, A. (1992)
J. Biol. Chem.,267, 22556-22561.
5. Clark, J. E. (1990) in Novel Biodegradable
Microbial Polymers (Dawes, E. A., ed.) Kluwer Academic, Amsterdam,
pp. 213-221.
6. Kato, J., Yamamoto, T., Yamada, K., and Ohtake, H.
(1993) Gene,137, 237-242.
7. Robinson, N. A., Goss, N. H., and Wood, H. G.
(1984) Biochem. Int.,8, 757-769.
8. Robinson, N. A., and Wood, H. G. (1986) J.
Biol. Chem.,261, 4481-4485.
9. Robinson, N. A., Clark, J. E., and Wood, H. G.
(1987) J. Biol. Chem.,262, 5216-5222.
10. Pepin, C. A., and Wood, H. G. (1987) J. Biol.
Chem.,262, 5223-5226.
11. Pepin, C. A., and Wood, H. G. (1986) J. Biol.
Chem.,261, 4476-4480.
12. Pepin, C. A., Wood, H. G., and Robinson, N. A.
(1986) Biochem. Int.,12, 111-123.
13. Skorko, R., Osipiuk, J., and Stetter, K. O.
(1989) J. Bacteriol.,171, 5162-5164.
14. Zago, A., Chugani, S., and Chakrabarty, A. M.
(1999) Appl. Environ. Microbiol.,65, 2065-2071.
15. Wood, H. G., Robinson, N. A., Pepin, C. A., and
Clark, J. E. (1986) in Phosphate Metabolism and Cellular Regulation
in Microorganisms (Torriani-Gorini, A., Rothman, F. G., Silver, S.,
Wright, A., and Yagil, E., eds.) American Society for Microbiology,
Washington, D. C., pp. 225-232.
16. Akiyama, M., Crooke, E., and Kornberg, A. (1993)
J. Biol. Chem.,268, 633-639.
17. Keasling, J. D., Bertsch, L., and Kornberg, A.
(1993) Proc. Natl. Acad. Sci. USA,90, 7029-7033.
18. Deinema, M. H., Loosdrecht, M. V., and Scholten,
A. (1985) Wat. Sci. Tech.,17, 119-125.
19. Fuhs, G. W., and Chen, M. (1975) Microb.
Ecology,2, 119-138.
20. Kornberg, A., Kornberg, S. R., and Simms, E. S.
(1956) Biochim. Biophys. Acta,20, 215-227.
21. Chapman, A. G., and Atkinson, D. E. (1977)
Adv. Microbiol. Physiol.,15, 253-306.
22. Van Veen, H. W., Abee, T., Kortstee, G. J. J.,
Periera, H., Konings, W. N., and Zehnder, A. J. B. (1994) J. Biol.
Chem.,269, 29509-29514.
23. Pramanik, J., Trelstad, P. L., and Keasling, J.
D. (1999) Wat. Sci. Tech.,37, 609-613.
24. Pramanik, J., Trelstad, P. L., Schuler, A. J.,
Jenkins, D., and Keasling, J. D. (1998) Water Res.,33,
462-476.
25. Mino, T., van Loosdrecht, M. C. M., and Heijnen,
J. J. (1998) Water Res.,32, 3193-3207.
26. Christensson, M., Blackall, L. L., and Welander,
T. (1998) Appl. Microbiol. Biotechnol.,49, 226-234.
27. Kampfer, P., Erhart, R., Beimfohr, C.,
Bohringer, J., Wagner, M., and Amann, R. (1996) Microb.
Ecology,32, 101-121.
28. Kawaharasaki, M., Tanaka, H., Kanagawa, T., and
Nakamura, K. (1999) Water Res.,33, 257-265.
29. Bond, P. L., Hugenholtz, P., Keller, J., and
Blackall, L. L. (1995) Appl. Environ. Microbiol.,61,
1910-1916.
30. Schuppler, M., Mertens, F., Schoen, G., and
Goebel, U. B. (1995) Microbiology,141, 513-521.
31. Wagner, M., Erhart, R., Manz, W., Amann, R.,
Lemmer, H., Wedi, D., and Schleifer, K.-H. (1994) Appl. Environ.
Microbiol.,60, 792-800.
32. Hesselmann, R. P. X., Werlen, C., Hahn, D., van
der Meer, J. R., and Zehnder, A. J. B. (1999) System. Appl.
Microbiol.,submitted.
33. McMahon, K. D., Dojka, M. A., Pace, N. R.,
Keasling, J. D., and Jenkins, D. (1999) 99th Gen. Meet. Amer. Soc.
Microbiol., Chicago,May 30-June 4, 1999.
34. McMahon, K. D., Pace, N. R., Jenkins, D., and
Keasling, J. D., in preparation.
35. Geißdörfer, W., Ratajczak, A., and
Hillen, W. (1998) Appl. Environ. Microbiol.,64,
896-901.
36. Trelstad, P. L., Purdhani, P., Geissdorfer, W.,
Hillen, W., and Keasling, J. D. (1999) Appl. Environ.
Microbiol.,65, 3780-3786.
37. Shin, P. K., and Seo, J.-H. (1990)
Biotechnol. Bioeng.,36, 1097-1104.
38. Sharfstein, S. T., van Dien, S. J., and
Keasling, J. D. (1996) Biotechnol. Bioeng.,51,
434-438.
39. Van Dien, S. J., Keyhani, S., Yang, C., and
Keasling, J. D. (1997) Appl. Environ. Microbiol.,63,
1689-1695.
40. Van Dien, S. J., and Keasling, J. D. (1998)
Biotechnol. Bioeng.,59, 754-761.
41. Van Dien, S. J., and Keasling, J. D. (1999)
Biotechnol. Progr.,15, 587-593.
42. Van Dien, S. J., and Keasling, J. D. (1998)
J. Theor. Biol.,190, 37-49.
43. Pettersson, A., Kunst, L., Bergman, B., and
Roomans, G. M. (1985) J. Gen. Microbiol.,131,
2545-2548.
44. Crang, R. E., and Jensen, T. E. (1975) J.
Cell Biol.,67, 80a.
45. Scott, J. A., and Palmer, S. J. (1990) Appl.
Microbiol. Biotechnol.,33, 221-225.
46. Gadd, G. M. (1986) in Microbes in Extreme
Environments (Hervert, R. A., and Codd, G. A., eds.) Academic
Press, London, pp. 83-110.
47. Klionsky, D. J., Herman, P. K., and Emr, S. D.
(1990) Microbiol. Rev.,54, 266-292.
48. Norris, P. R., and Kelly, D. P. (1977) J.
Gen. Microbiol.,99, 317-324.
49. Raguzzi, F., Lesuisse, E., and Crichton, R. R.
(1988) FEBS Lett.,231, 253-258.
50. Peverly, J. H., Adamec, H., and Parthasarathy,
M. V. (1978) Plant Physiol.,62, 120-126.
51. Rachlin, J. W., Jensen, T. E., Baxter, M., and
Jani, V. (1982) Arch. Environm. Contam. Toxicol.,11,
323-333.
52. Sicko-Goad, L., and Lazinsky, D. (1986) Arch.
Environ. Contam. Toxicol.,15, 617-627.
53. Aiking, H., Stijnman, A., van Garderen, C., van
Heerikhuizena, H., and van't Riet, J. (1984) Appl. Environ.
Microbiol.,47, 374-377.
54. Tijssen, J. P. F., and van Steveninck, J. (1984)
Biochem. Biophys. Res. Commun.,119, 447-451.
55. Rothstein, A., and Meier, R. (1951) J. Cell.
Comp. Physiol.,38, 245-270.
56. Sicko-Goad, L., and Stoermer, E. F. (1979) J.
Phycol.,15, 316-321.
57. Zhang, W., and Majidi, V. (1994) Environ.
Sci. Technol.,28, 1577-1581.
58. Keyhani, S., Lopez, J. L., Clark, D. S., and
Keasling, J. D. (1996) Microbios,88, 105-114.
59. Keasling, J. D., and Hupf, G. A. (1996) Appl.
Environ. Microbiol.,62, 743-746.
60. Macaskie, L. E., Bonthrone, K. M., and Rouch, D.
A. (1994) FEMS Microbiol. Lett.,121, 141-146.
61. Montgomery, D. M., Dean, A. C. R., Wiffen, P.,
and Macaskie, L. E. (1995) Microbiology,141,
2433-2441.