REVIEW: Recent Developments in the Biochemistry and Ecology of Enhanced Biological Phosphorus Removal
G. J. J. Kortstee1*, K. J. Appeldoorn2, C. F. C. Bonting1, E. W. J. van Niel1, and H. W. van Veen3
1Laboratory of Microbiology, Department of Biomolecular Sciences, Wageningen University, Hesselink van Suchtelenweg 4, 6703 CT, Wageningen, The Netherlands; fax: +31-317-483829; E-mail: Gerard.Kortstee@Algemeen.MICR.WAU.NL2BKH Consulting Engineers, Poortweg 10, 6212 PA, Delft, The Netherlands
3Department of Microbiology, University of Groningen, Kerklaan 30, 9751 NN, Haren, The Netherlands
* To whom correspondence should be addressed.
Received December 27, 1999
Most of the genes encoding the enzymes involved in polyP synthesis and degradation and in phosphate transport have been studied in various Gram-negative bacteria. Progress has also been made in studying the biochemical mechanisms underlying the process of enhanced biological phosphorus removal (EBPR), in particular in lab-scale systems fed with acetate or acetate plus glucose as the sole carbon and energy sources. By applying 13C-NMR, previous models concerning anaerobic carbon metabolism have been advanced and the role of glycogen in providing reducing equivalents in EBPR is definitely demonstrated. The role of the citric acid cycle in supplying reducing equivalents for the conversion of acetyl-CoA into poly-beta-hydroxybutyrate and poly-beta-hydroxyvalerate has been discussed. An incomplete citric acid cycle has been proposed to provide a small part of the reducing equivalents. Polyphosphate:AMP phosphotransferase and polyphosphatase were readily detectable in EBPR sludge fed with acetate plus glucose, but polyphosphate kinase remained undetected. In a lab-scale EBPR system, fed for several months with only acetate as carbon source, a Rhodocyclus-like bacterium (R6) was highly enriched and is therefore probably responsible for EBPR in systems fed with acetate only. This R6-type bacterium was however also present in other EBPR sludges (but to a lesser extent), and may therefore play an important role in EBPR in general. This organism accumulates polyhydroxyalkanoates anaerobically and polyP under aerobic conditions. Unlike members of the genus Rhodocyclus, bacterium R6 cannot grow phototrophically. Therefore a provisional new genus Candidatus and species Accumulibacter phosphatis was proposed.
KEY WORDS: inorganic polyphosphates, enzymes, enhanced biological phosphorus removal, bacteria, ecology
Abbreviations: EBPR) enhanced biological phosphorus removal; F&D) fill-and-draw system; Pit) phosphate inorganic transport; PHA) polyhydroxyalkanoates; PHB) poly-beta-hydroxybutyrate; PHV) poly-beta-hydroxyvalerate; polyP) polyphosphates; PPK) polyphosphate kinase.
Inorganic polyphosphate (polyP) is a linear polymer of many tens or
hundreds of inorganic phosphate (Pi) residues linked by
high-energy phosphoanhydride bonds and usually consists of mixtures of
different molecular sizes. Thermodynamically the standard free energy
of hydrolysis of the anhydride linkage yields about 38 kJ per phosphate
bond at pH 5. The energy-storage function of polyP depends on the
ability of the bond cleavage reaction to effect phosphorylation and
thereby conserve the energy associated with the hydrolytic action [1].
The presence of considerable amounts of Pi in wastewaters due to runoff of fertilizers and discharges of industrial and household activities represents a major problem since destructive algal blooms can develop in lakes and other waterways where under normal conditions Pi is most often the limiting factor for algae growth [2]. Between 1965 and 1975 it became clear that conventional wastewater treatment systems could be induced to accumulate considerably more phosphate than required for normal bacterial growth. This process is called enhanced biological phosphorus removal (referred to as EBPR) [3]. The process of EBPR is now an accepted and lower-cost strategy in controlling eutrophication. The excess of Pi taken up is stored in the form of polyP granules. During the past ten years reviews on the biochemistry [4], biology [2], and molecular biology [5] of polyP metabolism in pure cultures have been published. In the present review we will therefore attempt to analyze the recent progress made in understanding the mechanism of EBPR. This analysis is preceded by a brief description of the polyP metabolism of Acinetobacter johnsonii (strain 210A) since bacteria belonging to the genus Acinetobacter were originally thought to be responsible for EBPR. For more detailed reviews on the enzymes involved in polyP metabolism and on phosphate transport in bacteria the reader is referred to Kortstee et al. [6] and Kortstee and van Veen [7], respectively.
POLYPHOSPHATE METABOLISM IN Acinetobacter johnsonii
210A
Biosynthesis of polyP. The only bacterial pathway that has been established so far in polyP formation is the pathway that involves polyphosphate kinase (EC 2.4.7.1; PPK), catalyzing the synthesis of long chain polyP with about 1,000 Pi residues (reviewed by Kornberg [5]): ATP + polyPn <--> ADP + polyPn+1. We have never been able to spectrophotometrically detect PPK activities in cell-free extracts of the polyP-accumulating A. johnsonii 210A [8]. Indirect evidence was produced for a PPK-mediated synthesis of polyP in this organism by using phosphate-starved cells and the H+-ATPase inhibitor N,N´-dicyclohexylcarbodiimide, and measuring orthophosphate and polyP with 31P-NMR [9]. These results are in line with the detection of the gene ppk in Acinetobacter strain ADP1 and an increased amount of ppk transcript in cells of this organism grown under phosphate-limited conditions [10]. Based on 31P-NMR experiments, a specific activity of 60-120 nmol/min per mg protein for PPK was calculated in phosphate-deficient cells of A. johnsonii 210A.
Mutants of E. coli devoid of ppk synthesize small amounts of low-molecular-mass polyP of 60-70 phosphate residues, indicating that this polyP is formed by a pathway distinct from the PPK one [11]. Attempts to detect low-molecular-mass polyP in A. johnsonii 210A and in its cytoplasmic membrane have been unsuccessful so far.
Biodegradation of polyP. When high phosphate grown cells of the strictly aerobic A. johnsonii 210A are incubated anaerobically, their polyP is degraded and orthophosphate is excreted. The organism contains two enzymes catalyzing the degradation of polyP, polyphosphate:AMP phosphotransferase (see below) and polyphosphatase (PPX) catalyzing the reaction: polyPn + H2O --> polyPn-1 + Pi . Both enzymes have been purified and their properties examined [12, 13]. Polyphosphate:AMP phosphotransferase along with the ubiquitous adenylate kinase are responsible for the direct formation of ATP from polyP:
polyPn + AMP --> polyPn-1 + ADP,
2 ADP <--> ATP + AMP.
The regeneration of AMP by adenylate kinase can keep polyP degradation going for a long time [12].
Uptake and efflux of phosphate. Orthophosphate (Pi) is taken up against a concentration gradient by energy-dependent, carrier-mediated processes [14]. The organism contains two uptake systems for Pi: an inducible, high-affinity, unidirectional, ATP-driven and binding protein-dependent and a constitutive, bidirectional, low-affinity system that is driven by the proton motive force [15]. Their Km values are 0.7 ± 0.2 µM and 9 ± 1 M, respectively. The primary ATP-driven transport system is only involved in uptake and not in efflux and mediates the translocation of HPO42- and H2PO4- but not that of a metal-phosphate complex (MeHPO4). It is 6-10-fold stimulated by transfer from excess Pi to Pi-free medium [16, 17]. The constitutive system mediates the uptake and efflux of MeHPO4, but not that of the two above phosphate species [18]. When acting in concert, both transport systems enable A. johnsonii 210A to efficiently acquire Pi from its environment through uptake of the predominant Pi species if divalent cations are in excess of Pi. The efflux of MeHPO4 generates a proton motive force [19]. The uptake and efflux of MeHPO4 seems to be a general feature of Gram-negative bacteria [20].
Energy source. It has been pointed out previously that in vivo PPK is probably not directly involved in ATP formation from polyP in spite of the fact that it catalyzes the reversible formation of long chain polyP from ATP [6]. In A. johnsonii 210A polyP serves as an energy source during anaerobiosis by: 1) the direct synthesis of ATP via the polyphosphate:AMP phosphotransferase/adenylate kinase pathway, and 2) the generation of a proton motive force by the coupled excretion of MeHPO4 and H+. PPX may enhance the latter energy recycling mechanism by providing the efflux process with a continuous supply of Pi and divalent metal ions [19].
ENHANCED BIOLOGICAL PHOSPHORUS REMOVAL (EBPR)
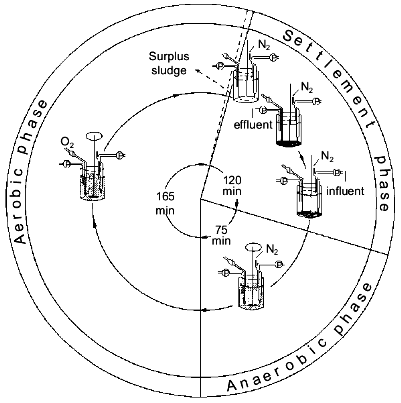
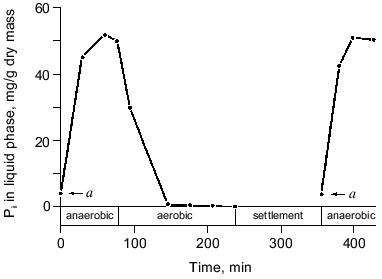
The process. Conventional activated sludge systems have been used for many decades to purify domestic wastewaters with molecular O2 as oxidant. The essential feature of these systems is that degradable organic carbon compounds are partially converted into microbial biomass and partially oxidized to carbon dioxide and that a major part of the microbes, except the waste sludge, is retained in the system as an activated sludge [21]. The observation that wastewater treatment plants removed much more phosphate than could be expected [22] and the discovery that full-scale treatment plants designed for nitrogen removal were able to eliminate up to 97% of the phosphate as well [23], lead to the discovery that the presence of an anaerobic zone in the treatment plant was compulsory in order to obtain significant biological phosphorus removal [24]. Thus the simplest design was a reactor with an anaerobic zone at the beginning of the plant where influent and return sludge come together, and an aerobic zone at the end of the plant. An anaerobic zone is considered to be a zone in which dissolved oxygen is absent. However, in full-scale wastewater treatment systems omitting aeration, allowing anaerobiosis to occur, approximates this. EBPR from domestic wastewaters in full-scale activated sludge plants is currently perceived to hinge on the provision of alternate stages in which the activated sludge is subjected to anaerobic (non-aerated) conditions and aerobic (aerated) conditions, respectively [3]. The characteristic feature of such plants is that orthophosphate (Pi), after being released from the biomass in the non-aerated stage, is reincorporated together with the influent Pi during the aerated stage. In a simple, one-vessel system Appeldoorn et al. [25] exposed activated sludge for 6-8 weeks to cycles with three consecutive periods: first an anaerobic period where the sludge was mixed with fresh mineral medium with acetate and glucose as carbon and energy sources, then an aerobic period and finally a settlement period in which the surplus sludge was removed and the medium was partly refreshed (Fig. 1). In this so-called fill-and-draw system (F&D-system), Pi was released during the anaerobic period and taken up during the aerobic period (Fig. 2), similar to the situation in full-scale treatment systems [3].
Fig. 1. Operational cycle of the fill-and-draw system [25].
Most of the treatment plants with EBPR also incorporate zones for biological removal of nitrogen by nitrification in an aerobic zone and by denitrification in a second anaerobic (non-aerated) zone. Although separate reactor combinations in series can be used for carbon removal, nitrification, and denitrification, the current practice is to have single-sludge systems in which the biomass is present as a fully mixed community of microorganisms catalyzing all desired processes, including EBPR. A well-known system for the removal of carbon, nitrogen, and phosphorus is the UCT (University of Cape Town) process in which an anoxic basin (an anaerobic, non-aerated basin with nitrate present) is positioned between the anaerobic, non-aerated zone and the aerated zone for the removal of nitrate. The reviews by Meganck and Faup [26] and Toerien et al. [3] summarize the variety of treatment systems used in practice in the combined removal of carbon, nitrogen, and phosphorus from wastewaters. The maximum phosphorus contents of sludges of a number of phosphate removal systems have been summarized by Appeldoorn [27] and varied from 35 to 180 mg of phosphorus per gram of sludge (dry weight).Fig. 2. Phosphate release and uptake in the fill-and-draw system (a, amount of phosphate present in refreshed medium [25]).
Nitrate as electron acceptor in EBPR. A prerequisite for obtaining EBPR in a full-scale treatment system is the absence of nitrate in the anaerobic, non-aerated zone where influent and sludge are mixed (Fig. 1). Feeding influents with relatively high concentrations of nitrate to a treatment plant results within a few weeks in a complete absence of EBPR. Normal denitrifying bacteria, which do not accumulate polyP, are enriched and the polyP-accumulating bacteria are out-competed [28].
Yet solid evidence has been provided for the occurrence of polyP-accumulating bacteria in activated sludge that are able to denitrify as well. In some instances enrichment of this type of organism does not need nitrate plus an anaerobic, non-aerated period [28-31]. Apparently sufficient nitrate is formed during nitrification to induce the enzymes involved in denitrification. In other instances the introduction of an anaerobic period plus nitrate (but without extracellular readily degradable carbon compounds) after the primary anaerobic period is required to obtain a bacterial population that accumulates Pi not only in the presence of oxygen but also in the presence of nitrate as terminal electron acceptor [32-34], thus showing that nitrate can be used as electron acceptor in EBPR during uptake of Pi. In conventional treatment systems denitrification and phosphorus removal proceed separately. When nitrate is used as electron acceptor in the uptake of Pi, a more efficient use is made of the available organic carbon, which is often rate-limiting in conventional treatment systems for nitrogen and phosphorus removal [34]. The presence of two bacterial populations capable of accumulating polyP, one with oxygen as terminal electron acceptor and one, which can also use nitrate as electron acceptor, can elegantly explain why in some sludges phosphorus release is reduced under anaerobic conditions by nitrate [35].
The suggestion that denitrifying bacteria may have a potential for EBPR [32, 33] has created some confusion. It was stated [36] that this suggestion contradicts the present view that nitrate is inhibitory to phosphate removal. However, only if nitrate is introduced into the anaerobic, non-aerated phase along with the influent containing readily degradable carbon compounds, rapid growth of normal denitrifying bacteria occurs. These organisms use their substrates more efficiently and out-compete the polyP bacteria, which may be able to denitrify as well [33]. Nitrate introduced into an anaerobic, non-aerated phase without readily degradable extracellular carbon compounds favors the growth of denitrifying bacteria that possess intracellular carbon substrates like the polyP bacteria. Recently evidence was produced that one of the denitrification intermediates, nitric oxide (NO), inhibits phosphate release in sludge that did not take up phosphate in an enhanced way in the presence of nitrate [27]. Further research revealed that adenylate kinase, an enzyme indirectly involved in the degradation of polyP, was severely inhibited by NO [37]. At an initial concentration of about 0.3 mM NO in the liquid phase, phosphate release by the sludge was almost completely inhibited; about 0.15 mM NO was required for complete inhibition of adenylate kinase in the crude cell-free extracts of the sludge. These levels are significantly higher than the NO levels found in wastewater treatment plants [38]. However, NO can also be formed by chemo-denitrification, a spontaneous reduction of nitrite to NO with electrons from reduced metal ions like ferrous iron [39, 40]. In conclusion, the reduction in anaerobic phosphate release in treatment systems with EBPR by nitrate may be explained as follows: 1) competition for the same substrate between normal denitrifying bacteria and polyP bacteria; 2) uptake of Pi and accumulation of polyP by polyP bacteria which are able to denitrify as well, and 3) through its conversion to NO, nitrate inhibits polyP degradation at the level of adenylate kinase.
A BIOCHEMICAL MODEL OF EBPR
Carbon metabolism under anaerobic conditions. Until about 1985 several, rather primitive, biochemical models of EBPR were put forward [41-44]. These models were all based on the feature that alternating anaerobic (non-aerated), and aerobic (aerated) conditions favor the growth of bacteria that accumulated phosphate in the form of intracellular polyP granules. Under non-aerated conditions organic substrates from the sewage were broken down to acetate and this compound was taken up and stored as carbon reserve in the form of poly-beta-hydroxybutyrate (PHB). In some cases poly-beta-hydroxyvalerate (PHV) (3-hydroxyvalerate units synthesized from acetyl-CoA and propionyl-CoA) was also found (see [45]). The energy required for these processes was thought to come from the enzymatic hydrolysis of polyP. During the subsequent aerobic phase (see Fig. 1) bacterial growth and polyP and glycogen replenishment occur at the expense of the stored polyhydroxyalkanoates (PHA). The models were well accepted since the above-mentioned set of observations was consistent and the models explained many of these observations. Around 1986 the models of Comeau et al. [28] and Wentzel et al. [46] became much more advanced (reviewed by Kortstee et al. [2]). Different hypotheses were put forward to explain the supply of reducing equivalents that are required for conversion of acetyl-CoA into PHB and of acetyl-CoA plus propionyl-CoA into PHV. In the models of Comeau et al. and Wentzel et al. reducing power is supplied by the oxidation of acetyl-CoA in the citric acid cycle, operating under anaerobic conditions. Mino et al. [47] suggested that reducing power is produced by conversion of glycogen to acetyl-CoA via pyruvate, and not by oxidation of acetyl-CoA via the citric acid cycle. This suggestion is in agreement with the earlier observation of Bordacs and Chiesa [48] that almost no 14CO2 is produced from [14C]acetate during the anaerobic period.
This model of carbon metabolism during the anaerobic period has recently being confirmed by Smolders et al. [49]. By using phosphorus removing sludge from a well-controlled sequencing batch reactor, it was shown that under really anaerobic conditions: 1) extracellular acetate was completely converted to PHA; 2) the molar hydroxybutyrate/ acetate ratio was 1.2-1.3; 3) cellular glycogen was consumed during the conversion of acetate into PHB, and 4) all cells contained glycogen and polyP at the start of the experiment (see also [50]). The data show also that glycogen serves not only as a source of reducing equivalents but also as an energy source in addition to polyP. In an elegant in vivo 13C-NMR study using [13C-2]acetate the involvement of glycogen in EBPR was recently definitely established [45] using a lab-scale EBPR system with acetate as the sole carbon and energy source. A second observation made by these authors was that the propionyl moiety of PHV is not derived from the extracellular acetate during anaerobiosis, but exclusively from the intracellular glycogen. This implicates, assuming normal biochemistry, that propionyl-CoA is formed via the acrylate pathway (pyruvate --> lactate --> acrylate --> propionate) or via the succinate-propionate pathway (pyruvate + CO2 --> oxaloacetate --> malate --> fumarate --> succinate --> succinyl-CoA --> methylmalonyl-CoA --> propionyl-CoA + CO2). The enzymatic evidence for either of these pathways is weak, but according to Mino et al. [51] the latter pathway is favored since intermediates of this pathway were easily metabolized by sludge enriched with polyP-accumulating organisms (summarized by Mino et al. [51]; see also Pereira et al. [45]). A third important suggestion of Pereira et al. [45] was that oxidation of acetyl-CoA via the citric acid cycle is a source of reducing power in addition to the oxidation of glycogen to acetyl-CoA: about 30% of the reducing equivalents required for PHB and PHV formation were calculated to be generated by the citric acid cycle and 70% by oxidation of glycogen to acetyl-CoA. For more definite conclusions as to the role of the citric acid cycle in the anaerobic period, the following problems have to be solved. 1) The failure to detect significant 14CO2 formation from [14C]acetate in a previous investigation [48]. 2) Formation of succinyl-CoA from [13C-2]acetate and oxaloacetate via citrate, isocitrate, and 2-oxoglutarate leads to the formation of succinyl-CoA labeled at the C-3 position (COOH-13CH2-CH2-COCoA) [52] and succinyl-CoA is the precursor of propionyl-CoA [45]. The failure to detect labeled propionate (in PHV) upon incubation with [13C-2]acetate during anaerobiosis is therefore difficult to understand. 3) Reoxidation of FADH2 produced by succinate dehydrogenase has to occur to keep the citric acid cycle going, and this is still an open question under anaerobic conditions [51, 53]. 4) In the citric acid cycle succinyl-CoA is oxidized to oxaloacetate via succinate, fumarate, and malate. In the metabolic model proposed by Pereira et al. [45], however, oxaloacetate is simultaneously reduced to succinyl-CoA via malate, fumarate, and succinate.
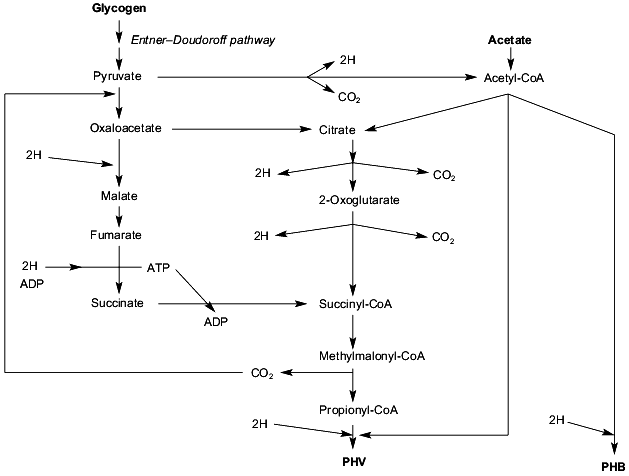
Based on the work of Mino et al. [47], Smolders et al. [49], Pereira et al. [45], and the work by many other research groups reviewed by them, we propose the tentative metabolic model for conversion of acetate and glycogen into PHB and PHV under anaerobic conditions as depictured in Fig. 3. The five characteristic features of this tentative scheme are: 1) conversion of glycogen into pyruvate via the the Entner-Doudoroff pathway [54]; 2) conversion of extracellular acetate into PHB via the well-known pathway and the conversion of acetyl-CoA plus propionyl-CoA into PHV; 3) conversion of pyruvate (derived from glycogen) into propionyl-CoA might occur via transcarboxylation to oxaloacetate and subsequent reduction to methylmalonyl-CoA or via the first part of the citric acid cycle; 4) the citric acid cycle is incomplete since succinyl-CoA is converted into propionyl-CoA; 5) formation of propionyl-CoA from pyruvate via malate and fumarate requires reducing equivalents whereas formation of propionyl-CoA from pyruvate via the first part of the citric acid cycle is accompanied by the formation of reducing equivalents. Thus dependent on the availability of reducing equivalents, propionyl-CoA is formed by one of the two pathways. This proposed model must be considered as a working hypothesis.
Carbon metabolism under aerobic conditions. When the sludge is entering the aerobic phase, in all properly functioning lab-scale phosphorus removing treatment systems no extracellular, easily degradable carbon compounds are left (see, for instance, [27, 50, 55]). This means that only intracellular compounds such as PHB and PHV serve as substrates for growth of the strictly aerobic polyP-accumulating bacteria. Growth under these conditions is a rather slow process, the maximum growth rate of the biological phosphorus-removing organisms being in the range of 0.04 h-1 [50]. During growth of these organisms Pi is taken up in considerable amounts, up to a maximum phosphorus content of 180 mg P per g dry weight [56], and largely stored as polyP and glycogen is resynthesized (see, for instance [50]). Why these organisms spend significant parts of the intracellular PHA to accumulate polyP and glycogen, and not merely to cell growth is presently unknown and is, as far as we know, a unique and fascinating characteristic of polyP-accumulating bacteria. Apparently the biosynthesis of certain cellular components is slow compared to the conservation of metabolic energy and the excess of energy is spent on transport of Pi, conversion of Pi into polyP, and accumulation of glycogen from PHA. Alternatively, the genes encoding these reactions are for some unknown reasons overexpressed and as a result, less energy is left for growth. Very recently, the phosphate uptake kinetics in relation to PHB were studied [56]. The uptake of Pi increased the rate of PHB degradation significantly and at low cellular PHB concentrations hardly any Pi was taken up. As much as 30% of the PHB was estimated to be used for uptake of Pi and conversion of Pi to polyP.Fig. 3. Anaerobic metabolism of extracellular acetate and intracellular glycogen in activated sludge showing enhanced biological phosphorus removal (EBPR). The sludge was highly enriched for EBPR by feeding acetate as sole carbon source for several months. For further details see text. PHB, poly-beta-hydroxybutyrate; PHV, poly-beta-hydroxyvalerate.
Polyphosphates are strongly negatively charged molecules and are complexed in bacteria almost exclusively with K+ and Mg2+ [19, 57] unless unusually high concentrations of other metals in the growth medium are provided. PolyP metabolism in activated sludge is also accompanied by the accumulation/release of both ions. Sometimes polyP is complexed also slightly with Ca2+ in activated sludge. From observations with a variety of phosphorus-removal sludges it appears that K+ and Mg2+, and sometimes Ca2+ to a minor extent, are co-transported with Pi molecules in a total molar ionic charge ratio of about one, irrespective of the origin of the sludge and the direction of transport (summarized by Comeau et al. [28], see also Rickard and McClintock [58]). In enhanced phosphorus removing sludges obtained by enrichment using the mineral salts medium with acetate as the sole carbon compound [50] a metabolically rather homogenous group of bacteria might be expected. In this sludge a total charge ratio between cations and Pi of about unity was observed as well and the molar Mg2+/Pi and K+/Pi ratios were 0.42 and 0.34, respectively [33]. In the polyP-accumulating A. johnsonii 210A, the molar Mg2+/Pi ratio was however close to unity during Pi efflux (Clijssen and Kortstee, unpublished observations) as could be expected from the work of van Veen et al. [15] which shows that Pi is excreted as a neutral metal phosphate (MgHPO4) by the reversible phosphate inorganic transport (Pit) system. Thus in spite of the fact that K+ and Mg2+ are the counter-ions of polyP in this organism, the molar Mg2+/Pi ratio during Pi efflux is about one.
Enzymes involved in EBPR. As far as polyP synthesis is concerned, no significant PPK activities have been demonstrated in extracts of activated sludges showing EBPR derived from full-scale treatment systems. In spite of many attempts, PPK was also not detectable in sludge from the F&D system. However, significant specific activities of polyphosphate:AMP phosphotransferase and adenylate kinase were present in this sludge, along with a reasonable polyphosphatase and readily detectable polyphosphate glucokinase activities [59]. The failure to detect PPK in F&D sludge may be due to the absence of the enzyme that catalyzes the synthesis of only long chain polyP up to 1000 Pi residues. F&D sludge contains mainly short chain polyP [60]. Mutants of E. coli devoid of ppk also contain significant amounts of short chain polyP [11]. The alternatives for PPK to catalyze polyP formation have been reviewed recently [7]. During the aerobic period the EBPR bacteria in the sludge use intracellular PHB and PHV as carbon sources. The labeling pattern of glycogen is in line with synthesis from acetyl-CoA via the glyoxylic acid cycle. The presence of isocitrate lyase and malate synthase has not been examined in EBPR sludge. Their specific activities may be low because of the low growth rate [50]. In addition, during PHV degradation propionyl-CoA may be formed, and this compound may be further metabolized via succinyl-CoA. This would make replenishment of the citric acid cycle by the glyoxylate bypass redundant or partially redundant.
THE BACTERIA INVOLVED IN EBPR
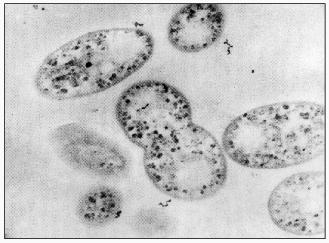
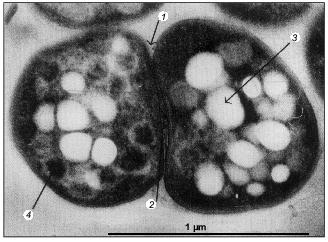
Activated sludge showing EBPR, in addition to carbon and nitrogen removal, as is the case in of the well-known UCT-process, contains a wide variety of bacteria: acidogenic bacteria catalyzing the degradation of complex substrates to acetate, nitrifying organisms catalyzing the oxidation of ammonium to nitrite and further to nitrate, denitrifying organisms catalyzing the conversion of nitrate to nitrogen, and strictly aerobic bacteria [3]. The bacteria involved in nitrification and denitrification have been isolated and shown to perform their function in pure culture as well as in activated sludge [61], although the occurrence of polyP bacteria able to denitrify as well was not anticipated at that time. The identity of the bacteria involved in EBPR is far less clear. Fuhs and Chen [41] were the first to focus attention on Acinetobacter spp. as being important in this process. In spite of the impressive number of treatment systems showing EBPR, from which polyP-accumulating Acinetobacter strains were isolated, based on the oxidation of glucose to gluconic acid [62], ubiquinone profiles [63], and polyamine profiles [64], it had to be concluded that Acinetobacter spp. are probably not involved in EBPR in full-scale treatment systems and certainly not involved in EBPR in the lab-scale systems of Appeldoorn et al. [62] and Smolders et al. [49] where acetate + glucose and acetate, respectively, were applied as carbon sources. All stable isolated strains, including the best studied A. johnsonii 210A, failed to accumulate PHA from extracellular acetate. In addition, strain 210A contains an adenylate kinase not sensitive to NO, unlike the situation in EBPR sludge grown in the F&D system [59]. This strain is however able to accumulate polyP in the absence of extracellular energy sources [9]. It has been stated that the process of EBPR probably depends critically on obscure interactions among different organisms present in the sludge [45]. However, the system used by these authors was basically similar to that of Appeldoorn et al. [25] and Smolders et al. [49]. In these EBPR systems, all cells or almost all cells in the sludge contain polyP and glycogen (Fig. 4) [49] and all cells containing the dark polyP granules also contain PHA granules with a high refractivity (Fig. 5) [65]. Thus, each single cell of the sludges grown in the systems of Appeldoorn and Smolders contained all three polymers. Thus, as far as the three polymers are concerned, there is no need for specific interactions.
Fig. 4. Electron micrograph of EBPR sludge at the end of the aerobic period [49]. The dark spots are stained glycogen granules. Polyphosphate had disappeared from the cells by the fixation and staining procedure.
Studies on the community structures of municipal wastewater treatment plants using in situ hybridization with rRNA-targeted probes suggested that the EBPR bacteria belong to the Gram-positive bacteria with a high G+C DNA content [66, 67]. Christensson et al. [68] confirmed this conclusion by analyzing a 16S rDNA clone library that was retrieved from a lab-scale EBPR system. This conclusion seemed at first sight to be confirmed by the isolation of two Gram-positive bacteria with a high G+C DNA content, of which one was identified as Microlunatis phosphovorus [69] and the other remained unidentified [70] but resembles the organism isolated by Nakamura. In spite of the fact that this organism displays the correct phosphate characteristics, it is unable to accumulate acetate and propionate in the form of PHA [71]. Instead the organism anaerobically sequesters glucose. This indicates that the organism is different from the much more common polyP-accumulating organism(s) in EBPR plants that is able to accumulate PHA from acetate and propionate. But Microlunatis phosphovorus or closely resembling bacteria may well be involved in EBPR in plants receiving glucose-rich wastewater. In contrast to the above three research groups, Bond et al. [72] concluded that not Gram-positive bacteria with a high G+C DNA content were involved in EBPR, but rather bacteria related to the genus Rhodocyclus. In a more recent study, Hesselmann et al. [73] confirmed this conclusion: acetate-grown EBPR sludge was dominated by bacteria phylogenetically related to the Rhodocyclus group within the beta-Proteobacteria (accounting for 89% of the cells), the EBPR sludge grown on complex medium was also predominated by this group of bacteria but to a lesser extent (accounting for 34% of the cells). The so-called R6-type bacterium represented the dominant bacteria. Staining of intracellular polyP and PHA confirmed that this bacterium accumulates these polymers in the same way as activated sludge showing EBPR. Although the R6-type bacterium is closely related to the genus Rhodocyclus, it did not grow phototrophically. Therefore the provisional new genus Candidatus and species Accumulibacter phosphatis was proposed. Once isolated, its polyP metabolism may be studied along the lines followed in A. johnsonii 210A [7] and E. coli [5]. The isolated R6-type organism also provides the opportunity to verify the biochemical model proposed for the anaerobic metabolism of extracellular acetate (and other substrates) and intracellular glycogen, the enzymes involved in polyP biosynthesis, and degradation and the transport systems of Pi, K+, and Mg2+.Fig. 5. Transmission electron micrograph of EBPR sludge [65]. Arrows indicate septa (1), exopolymeric substance (2), poly-beta-hydroxyalkanoate granule (3), and dark polyphosphate granule (4).
The study of Hesselmann et al. [73] also nicely showed that to a certain extent the number of bacterial groups in EBPR sludges was proportional to the number of different carbon sources. Besides, in spite of being not required for EBPR, Acinetobacter spp. may still have a significant impact on phosphorus removal.
REFERENCES
1.Dawes, E. A. (1990) in Novel Biodegradable
Microbial Polymers,Kluwer Academic Publishers, Dordrecht, pp.
3-16.
2.Kortstee, G. J. J., Appeldoorn, K. J., Bonting, C.
F. C., van Niel, E. W. J., and van Veen, H. W. (1994) FEMS
Microbiol. Rev.,15, 137-153.
3.Toerien, D. F., Gerber, A., Lotter, L. H., and
Cloete, T. E. (1990) Adv. Microb. Ecol.,11,173-230.
4.Wood, H. G., and Clark, J. E. (1988) Ann. Rev.
Biochem.,57, 235-260.
5.Kornberg, A. (1995) J.
Bacteriol.,177, 491-496.
6.Kortstee, G. J. J., Appeldoorn, K. J., Bonting, C.
F. C., van Niel, E. W. J., and van Veen, H. W. (2000) Adv. Microb.
Ecol., 16, in press.
7.Kortstee, G. J. J., and van Veen, H. W. (1999) in
Progr. Mol. Subcel. Biol. (Schroder, H. C., and Muller, W. E.
G., eds.) Vol. 23,pp. 275-297.
8.Bonting, C. F. C. (1993) Polyphosphate
Metabolism in Acinetobacter johnsonii 210A, Ph. D. Thesis,
Agricultural University, Wageningen, The Netherlands.
9.Van Niel, E. W. J., de Best, J. H., Kets, E. P. W.,
Bonting, C. F. C., and Kortstee, G. J. J. (1999) Appl. Microbiol.
Biotechnol.,51, 639-646.
10.Geißdörfer, W., Ratajczak, A., and
Hillen, W. (1998) Appl. Environ. Microbiol., 64,
896-901.
11.Castuma, C. E., Huang, R., Kornberg, A., and
Reusch, R. N. (1995) J. Biol. Chem., 270,
12980-12983.
12.Bonting, C. F. C., Kortstee, G. J. J., and
Zehnder, A. J. B. (1991) J. Bacteriol.,173,
6484-6488.
13.Bonting, C. F. C., Kortstee, G. J. J., and
Zehnder, A. J. B. (1993) Antonie van Leeuwenhoek,64,
75-81.
14.Van Veen, H. W. (1994) Energetics and
Mechanisms of Phosphate Transport in Acinetobacter
johnsonii. Ph. D. Thesis, Agricultural University, Wageningen, The
Netherlands.
15.Van Veen, H. W., Abee, T., Kortstee, G. J. J.,
Konings, W. N., and Zehnder, A. J. B. (1993) J.
Bacteriol.,175, 200-206.
16.Bonting, C. F. C., van Veen, H. W., Taverne, A.,
Kortstee, G. J. J., and Zehnder, A. J. B. (1992) Arch.
Microbiol.,158, 139-144.
17.Van Veen, H. W., Abee, T., Kortstee, G. J. J.,
Konings, W. N., and Zehnder, A. J. B. (1994) J. Biol.
Chem.,269, 16212-16216.
18.Van Veen, H. W., Abee, T., Kortstee, G. J. J.,
Konings, W. N., and Zehnder, A. J. B. (1993) J. Biol. Chem.,
268, 19377-19383.
19.Van Veen, H. W., Abee, T., Kortstee, G. J. J.,
Pereira, H., Konings, W. N., and Zehnder, A. J. B. (1994) J. Biol.
Chem.,269, 29509-29514.
20.Van Veen, H. W., Abee, T., Kortstee, G. J. J.,
Konings, W. N., and Zehnder, A. J. B. (1994) in Phosphate in
Microorganisms: Cellular and Molecular Biology (Torriani-Gorini, A.
M., Silver, S., and Yagil, Y., eds.) American Society for Microbiology,
Washington, DC, pp. 43-49.
21.Ardern, E., and Lockett, W. T. (1914) J. Soc.
Chem. In.,33, 523-539.
22.Srinath, E. G., Sastry, C. A., and Pillai, S. C.
(1959) Wat. Waste Treat.,11, 410-415.
23.Barnard, J. L. (1976) Wat. Sth.
Afr.,2, 136-144.
24.Davelaar, D., Davies, T. R., and Wiechers, S. G.
(1978) Wat. Sth. Afr., 4, 54-59.
25.Appeldoorn, K. J., Kortstee, G. J. J., and
Zehnder, A. J. B. (1992) Wat. Res., 26, 453-460.
26.Meganck, M. T. J., and Faup, G. M. (1988)
Biotreatment Syst.,3, 11-204.
27.Appeldoorn, K. J. (1993) Ecological Aspects of
the Biological Phosphate Removal from Waste Waters. Ph. D. Thesis,
Agricultural University, Wageningen, The Netherlands.
28.Comeau, Y., Hall, K. J., Hancock, R. E. W., and
Oldham, W. K. (1986) Wat. Res.,20, 1511-1521.
29.Shin, H. S., Jun, H. B., and Park, H. S. (1992)
Biodegradation,3, 105-111.
30.Kerrn-Jespersen, J. P., and Henze, M. (1993)
Wat. Res.,27, 617-624.
31.Ostgaard, K., Christensson, M., Lie, E., Jonsson,
K., and Welander, T. (1997) Wat. Res.,31, 2719-2726.
32.Kuba, T., Smolders, G., van Loosdrecht, M. C. M.,
and Heijnen, J. J. (1993) Wat. Sci. Technol.,27,
241-252.
33.Kuba, T., van Loosdrecht, M. C. M., and Heijnen,
J. J. (1995) in Toekomstige Generatie Riool Water
Zuiveringsinrichtingen RWZI 2000, RIZA/STOWA, Lelystad/Utrecht, pp.
1-83.
34.Kuba, T., van Loosdrecht, M. C. M., and Heijnen,
J. J. (1996) Wat. Res.,30, 1702-1710.
35.Schon, G., Geywitz, S., and Mertens, F. (1993)
Wat. Res.,27, 349-354.
36.Egli, T., and Zehnder, A. J. B. (1994) Curr.
Opin. Biotechnol.,5, 275-284.
37.Van Niel, E. W. J., Appeldoorn, K. J., Zehnder,
A. J. B., and Kortstee, G. J. J. (1998) Appl. Environ.
Microbiol.,64, 2925-2930.
38.Von Schulthess, R., Wild, D., and Gujer, W.
(1940) Wat. Sci. Tech.,30, 123-132.
39.Brons, H. J., Hagen, W. R., and Zehnder, A. J. B.
(1991) Arch. Microbiol.,155, 341-348.
40.Van Cleemput, O., and Baert, L. (1984) Plant
Soil, 86, 233-241.
41.Fuhs, G. W., and Chen, M. (1975) Microbiol.
Ecol.,2, 119-138.
42.Nicholls, H. A., and Osborn, D. W. (1979) J.
Wat. Pollut. Control Fed.,51, 557-569.
43.Rensink, J. H., Donker, H. G. J. W., and de
Vries, H. P. (1981) Biological P-Removal in Domestic Wastewater by
the Activated Sludge Process, Proc. 5th Envir. Sewage Symp.,
Munchen, pp. 487-502.
44.Marais, G. v. R., Loewenthal R. E., and Siebritz,
I. P. (1983) Wat. Sci. Technol.,17, 15-41.
45.Pereira, H., Lemos, P. C., Reis, M. A. M.,
Crespo, P. S. G., Carrondo, M. J. T., and Santos, H. (1996) Wat.
Res.,30, 2128-2138.
46.Wentzel, M. C., Lotter, L. H., Loewenthal, R. E.,
and Marais, G. v. R. (1986) Wat. Sth. Afr.,12,
209-224.
47.Mino, T., Arun, V., Tsuzuki, Y., and Matsuo, T.
(1987) in Advances Water Pollution Control: Biological
Removal of Phosphate from Wastewater (Ramadori, R., ed.) Oxford,
UK, Pergamon Press, pp. 27-38.
48.Bordacs, K., and Chiesa, S. C. (1989) Wat.
Sci. Technol.,21, 387-396.
49.Smolders, G. J. F., van der Mey, J., van
Loosdrecht, M. C. M., and Heijnen, J. J. (1994) Biotechnol.
Bioeng.,43, 461-470.
50.Smolders, G. J. F. (1995) A Metabolic Model of
the Biological Phosphorus Removal - Stoichiometry, Kinetics and
Dynamic Behaviour. Ph. D. Thesis, Delft University of Technology,
Delft, The Netherlands.
51.Mino, T., van Loosdrecht, M. C. L., and Heijnen,
J. J. (1998) Wat. Res.,32, 3193-3207.
52.Schlegel, H. G. (1986) Gen.
Microbiol.,Cambridge University Press, Cambridge, p. 234.
53.Thauer, R. K. (1988) Eur. J.
Biochem.,176, 497-508.
54.Maurer, M., Gujer, W., Hany, R., and Bachmann, S.
(1997) Wat. Res.,31, 907-917.
55.De Vries, H. P., and Rensink, J. H. (1985) in
Proc. Int. Conf. Management Strategies Phosphorus Environment,
Seeper Ltd., London, pp. 54-65.
56.Petersen, B., Temmink, H., Henze, M., and Isaacs,
S. (1998) Wat. Res.,32, 91-100.
57.Bonting, C. F. C., Kortstee, G. J. J.,
Boekestein, A., and Zehnder, A. J. B. (1993) Arch. Microbiol.,
159, 428-434.
58.Rickard, L. F., and McClintock, S. A. (1992)
Wat. Res.,26,2203-2206.
59.Van Niel, E. W. J., Appeldoorn, K. J., Zehnder,
A. J. B., and Kortstee, G. J. J. (1998) Appl. Environ.
Microbiol.,64, 2925-2930.
60.Appeldoorn, K. J., Boom, A. J., Kortstee, G. J.
J., and Zehnder, A. J. B. (1992) Wat. Res.,26,
937-943.
61.Jenkins, D., and Tandoi, V. (1991) Wat.
Res.,25, 1471-1478.
62.Bonting, C. F. C., Willemsen, B. M. F.,
Akkermans-van Vliet, W., Bouvet, P. J. M., Kortstee, G. J. J., and
Zehnder, A. J. B. (1992) FEMS Microbiol. Ecol.,102,
57-64.
63.Hiraishi, A., Masamune, K., and Kitamura, H.
(1989) Appl. Microbiol.,55, 897-901.
64.Auling, G., Pilz, F., Busse, H.-J., Karrasch, S.,
Streichan, M., and Schon, G. (1991) Appl. Environ.
Microbiol.,57, 3585-3592.
65.Christensson, M. (1997) Enhanced Biological
Phosphorus Removal. Carbon Sources, Nitrate as Electron
Acceptor, and Characterization of the Sludge Community. Ph.
D. Thesis, Department of Biotechnology, Lund University, Sweden.
66.Wagner, M., Erhart, R., Manz, W., Amann, R.,
Lemmer, H., Wedi, D., and Schleiffer, K.-L. (1994) Appl. Environ.
Microbiol.,56,1919-1925.
67.Kampfer, P., Erhart, R., Beimfohr, C., Bohringer,
J., Wagner, M., and Amann, R. (1996) Microb. Ecol.,32,
101-121.
68.Christensson, M., Blackall, L. L., and Welander,
T. (1998) Appl. Microbiol. Biotechnol.,49, 226-234.
69.Nakamura, K., Hiraishi, A., Yoshimi, Y.,
Kawaharasaki, M., Masuda, K., and Kamagata, Y. (1995) Int. J. Syst.
Bacteriol.,45, 17-24.
70.Ubukata, Y., and Takii, S. (1994) Wat.
Res.,28, 247-249.
71.Nakamura, K., Ishikawa, S., and Kawaharasaki, M.
(1995) J. Ferment. Bioeng.,80, 377-382.
72.Bond, P. L., Hugenholtz, P., Keller, J., and
Blackall, L. L. (1995) Appl. Environ. Microbiol.,61,
1910-1916.
73.Hesselmann, R. P. X., Werlen, Ch., Hahn, D., van
der Meer, J. R., and Zehnder, A. J. B. (1999) System. Appl.
Microbiol., 22, 454-465.