REVIEW: Photoisomerization in Rhodopsin
H. Kandori1,2*, Y. Shichida1, and T. Yoshizawa1
1Department of Biophysics, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan; fax: +81-75-753-8502; E-mail: kandori@photo2.biophys.kyoto-u.ac.jp2Present address: Department of Applied Chemistry, Nagoya Institute of Technology, Showa-ku, Nagoya 466-8555, Japan; fax: +81-52-735-5207; E-mail: kandori@ach.nitech.ac.jp
* To whom correspondence should be addressed.
Received April 6, 2001; Revision received May 29, 2001
This article reviews the primary reaction processes in rhodopsin, a photoreceptive pigment for twilight vision. Rhodopsin has an 11-cis retinal as the chromophore, which binds covalently with a lysine residue through a protonated Schiff base linkage. Absorption of a photon by rhodopsin initiates the primary photochemical reaction in the chromophore. Picosecond time-resolved spectroscopy of 11-cis locked rhodopsin analogs revealed that the cis-trans isomerization of the chromophore is the primary reaction in rhodopsin. Then, generation of femtosecond laser pulses in the 1990s made it possible to follow the process of isomerization in real time. Formation of photorhodopsin within 200 fsec was observed by a transient absorption (pump-probe) experiment, which also revealed that the photoisomerization in rhodopsin is a vibrationally coherent process. Femtosecond fluorescence spectroscopy directly captured excited-state dynamics of rhodopsin, so that both coherent reaction process and unreacted excited state were observed. Faster photoreaction of the chromophore in rhodopsin than that in solution implies that the protein environment facilitates the efficient isomerization process. Such contributions of the protein residues have been monitored by infrared spectroscopy of rhodopsin, bathorhodopsin, and isorhodopsin (9-cis rhodopsin) at low temperatures. The crystal structure of bovine rhodopsin recently reported will lead to better understanding of the mechanism in future.
KEY WORDS: rhodopsin, photorhodopsin, bathorhodopsin, isorhodopsin, photoisomerization
Rhodopsin is the only photoreceptor protein (a visual pigment) in the outer segment of rod visual cell responsible for twilight vision. It has been extensively studied more than any other visual pigments such as cone pigments responsible for color vision because of relative ease of preparation and abundance. Rhodopsin has 11-cis retinal as its chromophore, which is embedded inside a single peptide transmembrane protein called opsin. The role of rhodopsin in the signal transduction cascade of vision is to activate transducin, a heterotrimeric G-protein, upon absorption of light (reviewed in [1-4]). Therefore, a central question in rhodopsin is how light energy is used to change protein structure through photochemical reaction of the retinal chromophore. Rhodopsin (opsin), a member of G-protein coupled receptor family, is composed of 7-transmembrane helices. The 11-cis retinal forms the Schiff base linkage with a lysine residue of the 7th helix (Lys296 in the case of bovine rhodopsin), and the Schiff base is protonated, which is stabilized by a negatively charged carboxylate (Glu113 in the case of bovine rhodopsin) [1-4]. The beta-ionone ring of the retinal is coupled with hydrophobic region of opsin through hydrophobic interactions [5]. Thus, the retinal chromophore is fixed by three kinds of chemical bonds in the retinal binding pocket of rhodopsin.
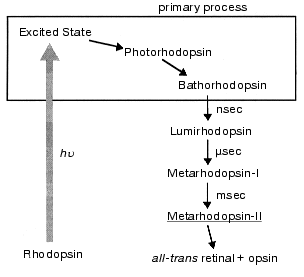
Absorption of a photon by the chromophore induces a primary photoreaction, followed by conformational changes of protein, and eventually activates transducin. This is called the “bleaching process” because rhodopsin loses its color. Several intermediate states are formed during the bleaching process, which have been identified by visible spectroscopy (Fig. 1). In such studies, both low-temperature and time-resolved spectroscopies have provided a great deal of information on the bleaching processes of rhodopsin (reviewed in [4, 6, 7]).
Among the bleaching processes of rhodopsin, we define the “primary process” as the event of formation of bathorhodopsin (Fig. 1). Bathorhodopsin is formed and stable in the picosecond time domain. It is also stable at low temperature (<-140°C) [6]. Unlike bathorhodopsin, photorhodopsin cannot be stabilized at low temperature, as well as the excited state of rhodopsin [7]. There has been a fundamental question on the nature of the primary photoprocess in rhodopsin: What is the primary reaction in vision? To address this question, the primary process of rhodopsin has been extensively studied by low-temperature and time-resolved spectroscopies.Fig. 1. Photobleaching process of bovine rhodopsin. The primary process is shown in the frame, which involves excited state of rhodopsin, photorhodopsin, and bathorhodopsin. Metarhodopsin-II that activates transducin is underlined.
In 1992, we published an article entitled “primary photochemical events in the rhodopsin molecule” [8], in which both low-temperature and time-resolved spectroscopic approaches were reviewed. Since the chromophore of rhodopsin is in 11-cis form and the final bleaching product is a mixture of all-trans retinal and opsin (Fig. 1), the cis-trans isomerization of the chromophore must occur during the bleaching process of rhodopsin. On the basis of the remarkable red-shift of rhodopsin to bathorhodopsin, Yoshizawa and Wald proposed that bathorhodopsin would have a twisted all-trans chromophore [9]. This hypothesis gave an indication that the primary reaction in vision is a cis-trans isomerization of the retinal chromophore. However, the discovery of precursors of bathorhodopsin, such as photorhodopsin and hypsorhodopsin, by low-temperature and picosecond time-resolved spectroscopies [10-12] raised a question, “what is the primary reaction in vision?” again. In addition, an application of picosecond laser photolysis led to propose another possibility, a proton translocation mechanism [13]. In the first chapter of this article, we will make a historical review of the study of the primary reaction in rhodopsin.
In the study of the primary reaction mechanism, rhodopsin analogs possessing 11-cis locked ring retinals supplied a mass of interesting and valuable information [8]. In the second chapter, we review the study of such systems, by which we are convinced that the primary reaction in vision is a cis-trans isomerization of the chromophore.
Although picosecond laser photolysis provided much information on the primary reaction in rhodopsin, it could not capture the electronically excited state of rhodopsin. Since the primary isomerization occurs in the excited state of rhodopsin, we need femtosecond pulses to examine the reaction dynamics of rhodopsin in real time. In the last decade of the 20th century, femtosecond spectroscopy was applied to rhodopsin. However, the early stage of trials was rather confusing on the mechanism of the primary processes of rhodopsin, as was for picosecond spectroscopy of rhodopsin [7, 8]. In the third chapter, recent advances in femtosecond spectroscopy of rhodopsin are described, where the situation becomes clearer now.
It is known that the cis-trans isomerization is highly efficient in rhodopsin (quantum yield: 0.67) [14], which is essential to make twilight vision highly sensitive. In fact, a human rod cell can respond to a single photon absorption. Such an efficient photoisomerization of the retinal chromophore is characteristic in the protein environment of rhodopsin, being in contrast to the rhodopsin chromophore in solution. This indicates that the protein environment facilitates the isomerization. How is it possible? To understand the mechanism, structural information is necessary, and such vibrational spectroscopies as resonance Raman and infrared spectroscopies are useful. Resonance Raman spectroscopy has experimentally revealed that light energy is stored by chromophore distortion, which was originally proposed by Yoshizawa and Wald [9]. On the other hand, infrared spectroscopy has revealed how the protein responds to the chromophore motion. In the fourth chapter, the structural study of the primary isomerization processes are mainly presented on the basis of our low-temperature infrared spectroscopy.
Very recently, bovine rhodopsin was crystallized by Okada et al. [15], and its three-dimensional structure was determined [16] as the first structure of G-protein coupled receptor family which includes over 1000 proteins. Such accomplishment promises better understanding of the primary reaction mechanism in rhodopsin in future. As a perspective, we give some comments on the rhodopsin structure in the fifth chapter.
I. HISTORY OF THE STUDIES ON THE PRIMARY REACTION OF
RHODOPSIN
To investigate the primary photoreaction processes in rhodopsin, two spectroscopic approaches have been applied; low-temperature and time-resolved spectroscopies [8]. They can detect primary photointermediate states by reducing the thermal reaction rate at low temperature (low-temperature spectroscopy) or directly probing dynamical processes at physiological temperature (time-resolved spectroscopy). Historically, the former technique was in advance of the latter, because generation of ultrashort pulses was necessary to detect primary intermediates of rhodopsin at physiological temperature, and picosecond pulses emerged in the 1970s [8].
After discovery of the chromophore (vitamin A) by George Wald in the 1930s, Yoshizawa and Kito first found a red-shifted photoproduct of bovine rhodopsin at low temperature (-186°C), which reverted to rhodopsin by illumination [17]. On warming above -140°C, this photoproduct (now called bathorhodopsin [10]) is converted to lumirhodopsin and finally decomposed to all-trans retinal and opsin through several intermediates (Fig. 1). Based on low temperature spectrophotometric experiments, Yoshizawa and Wald proposed in 1963 that bathorhodopsin has a “highly constrained and distorted” all-trans retinal as its chromophore and is on a higher potential energy level than rhodopsin and subsequent intermediates [9]. According to their prediction, the process of rhodopsin to bathorhodopsin should be a cis-trans isomerization of the chromophore.
The first challenge to the isomerization model was made in 1972 by picosecond laser photolysis [18]. Busch et al. reported the formation of bathorhodopsin within 6 psec after excitation of rhodopsin at room temperature, and interpreted that its formation would be too fast to be attributed to such a conformational change as the cis-trans isomerization of the retinal chromophore [18]. On the basis of both isotope effect and non-Arrhenius temperature dependence of the formation time of bathorhodopsin, Peters et al. proposed a model that the formation of bathorhodopsin is accompanied by proton translocation [13]. Which reaction takes place in rhodopsin, isomerization or proton translocation?
The isomerization model was favored by various low-temperature spectroscopic results [8], such as considerable angle change (26°) of the transition dipole moment between rhodopsin and bathorhodopsin [19], and no formation of bathorhodopsin for an 11-cis locked rhodopsin analog [20]. Low-temperature resonance Raman spectroscopy revealed that a chromophore isomerization occurs in the rhodopsin-bathorhodopsin transformation [21]. To determine what is the primary reaction of rhodopsin at physiological temperature, however, time-resolved spectroscopy with ultrashort pulses was necessary.
In the 1970s and 1980s, various studies were performed by picosecond time-resolved spectroscopy [8]. However, rather complicated results were obtained from picosecond time-resolved spectroscopy of rhodopsin. The results were classified into three; the primary intermediate is bathorhodopsin itself (case 1), the primary intermediate is a blue-shifted photoproduct (hypsorhodopsin [10]) that decays to bathorhodopsin (case 2), and the primary intermediate is a more red-shifted photoproduct (photorhodopsin [12]) that decays to bathorhodopsin (case 3). Such discrepancy among groups was then explained in terms of the difference in laser power for excitation of rhodopsin. Namely, little attention was paid to the energy density of the laser pulse at the early stage of investigation, and it turned out that excitation photon density is extremely important in the study of photochemistry of rhodopsin [8]. We found that photorhodopsin was formed under weak excitation conditions in bovine and invertebrate (squid and octopus) rhodopsins (case 3) [12], while strong excitation brought about different results due to multiphoton absorption. In bovine rhodopsin, no kinetic changes from photorhodopsin to bathorhodopsin were apparently observed under strong excitation conditions, which could lead to a conclusion that bathorhodopsin is formed as the primary intermediate (case 1) [22]. On the other hand, hypsorhodopsin was formed under strong excitation conditions of invertebrate (squid and octopus) rhodopsins as a two photon product of rhodopsin, decaying to bathorhodopsin (case 2) [23]. Thus, different results are most likely to originate from the difference in excitation photon densities.
Picosecond time-resolved spectroscopy of rhodopsin under physiological (i.e., weak excitation) conditions revealed that photorhodopsin is formed as a single photon product, decaying to bathorhodopsin (Fig. 1). The time constant of the transition between photorhodopsin and bathorhodopsin was determined to be 45 psec for bovine [22] and 200 psec for squid [23]. Bathorhodopsin observed at room temperature decays to lumirhodopsin, while bathorhodopsin at low temperature is converted to lumirhodopsin on warming (Fig. 1). On the other hand, photorhodopsin could not be stabilized at low temperature. This fact indicated that the properties of photorhodopsin can be examined only by time-resolved spectroscopy.
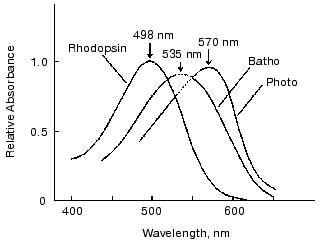
Absorption spectra of photointermediates of rhodopsin provide fundamental information. Therefore, we next attempted to determine absorption spectra of photorhodopsin and bathorhodopsin at room temperature. In determining absorption spectra of intermediates, an accurate estimation of percentage of bleaching is necessary. Although it is generally difficult in laser spectroscopy, we succeeded to estimate percentage of bleaching by picosecond laser excitation of rhodopsin molecules immobilized in polyacrylamide gel [24]. Thus, absorption spectra of bathorhodopsin and photorhodopsin were obtained at room temperature (Fig. 2). The absorption maximum of bathorhodopsin at room temperature was located at about 535 nm [23], considerably blue-shifted in comparison with that at low temperature (543 nm) [9]. In addition, the spectral half bandwidth (4660 cm-1, Fig. 2) was broader than those of rhodopsin (4300 cm-1, Fig. 2) and bathorhodopsin at low temperature (4250 cm-1 [9]), while the oscillator strength of bathorhodopsin was comparable with that of rhodopsin at room temperature.
The absorption spectrum of photorhodopsin has its maximum at 570 nm. It is also noted that both spectral half bandwidth and oscillator strength of photorhodopsin were considerably smaller than those of rhodopsin and bathorhodopsin. The small oscillator strength of photorhodopsin suggested that photorhodopsin possesses highly distorted chromophore, because the oscillator strength is sensitive to coplanarity of the conjugated double bond system [25]. Thus, the absolute absorption spectrum of photorhodopsin implied that the chromophore is distorted, possibly in an all-trans form. More plausible evidence was given by the comprehensive study of 11-cis-locked rhodopsin analogs as shown in the next chapter.Fig. 2. Absorption spectra of bovine rhodopsin, bathorhodopsin, and photorhodopsin at room temperature. Because of scattering of the excitation pulse, it was impossible to measure the spectral region from 530 to 550 nm. The spectra in the region were estimated by extrapolating the spectra in both sides of the region (dotted lines). This figure is modified from Kandori et al. [24].
II. PRIMARY REACTION IS cis-trans
PHOTOISOMERIZATION
As possible models of the primary photochemical reaction in rhodopsin, both isomerization and proton translocation were proposed as described in the previous chapter. The former was on the basis of the low-temperature spectroscopy of bathorhodopsin [9, 10, 19, 20], while the latter was on the basis of the low-temperature picosecond photolysis on bathorhodopsin [13, 18]. Since photorhodopsin is formed as the precursor of bathorhodopsin at room temperature, the primary reaction mechanism in rhodopsin had to be examined by time-resolved spectroscopy. Regarding the proton translocation model, it should be noted that the excitation photon density was extremely high in the low-temperature picosecond experiments [13, 18]. Therefore, the non-Arrhenius dependence of the formation rate of bathorhodopsin on temperature and deuterium isotope effect may be results that could be detected only under intense excitation conditions. In fact, a deuterium isotope effect was not observed in the process from photorhodopsin to bathorhodopsin under weak excitation conditions [22].
For further investigation of the primary reaction mechanism, rhodopsin analogs possessing 11-cis locked ring retinals were used. Irradiation of 11-cis locked rhodopsin analogs such as 5-membered [20] and 7-membered rhodopsins at room temperatures [26] displayed no formation of bathorhodopsin, indicating that the cis-trans isomerization of the chromophore is essential for the formation of bathorhodopsin. Then, what about photorhodopsin? In this regard, a notable study was reported in 1983. Buchert et al. described that excitation of 7-membered rhodopsin yielded three transient photoproducts, one of which was close to photorhodopsin in terms of red-shifted absorption spectrum and with a lifetime in the picosecond time domain [27]. This observation might argue against the isomerization model, even if bathorhodopsin has an all-trans chromophore.
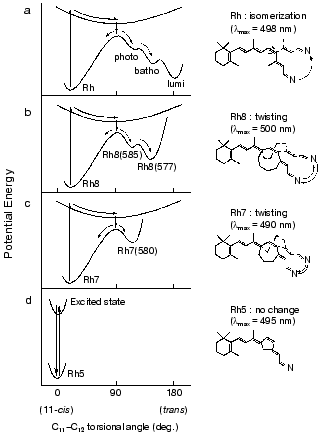
Therefore, we attempted a comprehensive study by applying picosecond spectroscopy to rhodopsin analogs possessing 11-cis-locked ring retinals as their chromophores. In these rhodopsin analogs, 11-cis-locked retinals with 5-, 7-, and 8-membered rings are incorporated into bovine opsin, so that the formed pigments become unbleachable. In spite of no photobleaching of these pigments, interestingly, picosecond time-resolved spectroscopy detected differences in photophysical and photochemical properties among them (Fig. 3).
In the case of 5-membered rhodopsin, only a long-lived excited state (tau = 85 psec) was formed without any ground-state photoproduct (Fig. 3d), giving direct evidence that the cis-trans isomerization is the primary event in vision [28]. Excitation of 7-membered rhodopsin, on the other hand, yielded a ground-state photoproduct having a spectrum similar to photorhodopsin (Fig. 3c). These different results were interpreted in terms of the rotational flexibility along C11-C12 double bond [28]. This hypothesis was further supported by the results with an 8-membered rhodopsin that possesses a more flexible ring. Upon excitation of 8-membered rhodopsin with a 21 psec pulse, two photoproducts, photorhodopsin-like and bathorhodopsin-like products were observed (Fig. 3b) [29]. Thus, the picosecond absorption studies directly elucidated the correlation between the primary processes of rhodopsin and the flexibility of C11-C12 double bond of the chromophore, and we eventually inferred the respective potential surfaces as shown in Fig. 3 [29].Fig. 3. Schematic drawing of ground- and excited-state potential surfaces along the 11-ene torsional coordinates of the chromophore of rhodopsin (a), 8-membered rhodopsin (b), 7-membered rhodopsin (c), and 5-membered rhodopsin (d). This figure is modified from Mizukami et al. [29].
III. PHOTOISOMERIZATION IN RHODOPSIN OCCURS IN
FEMTOSECONDS
As described above, the cis-trans isomerization of the chromophore of rhodopsin is the primary reaction in vision, which is likely to occur in the electronically excited state of rhodopsin. Although picosecond time-resolved spectroscopy of rhodopsin successfully detected the primary intermediates at room temperature, direct observation of the cis-trans isomerization process needs a better time resolution to capture its excited state. Doukas et al. estimated the excited-state lifetime of rhodopsin to be in a subpicosecond time domain from their observation of the fluorescence quantum yield [30]. Therefore, we had to wait for generation of femtosecond pulses to detect the isomerization process of rhodopsin in real time.
In 1991, two groups first reported femtosecond transient absorption spectroscopy of bovine rhodopsin [31, 32], whereas their conclusions were remarkably different from each other (table). One group excited bovine rhodopsin with a 35 fsec pulse and probed with a 10 fsec pulse, and concluded that product formation completed within 200 fsec [31]. An amazingly fast event! In contrast, the other group measured transient absorption of bovine rhodopsin with 300 fsec resolution, and concluded that the primary isomerized photointermediate appears in 3 psec [32]. In the following year, a different group applied femtosecond transient absorption spectroscopy to octopus rhodopsin, and reported that there are two time-constants for the formation of the primary photointermediates, 400 fsec and 2 psec [33]. Thus, the first trials provided rather confusing results on the primary processes of rhodopsin photoisomerization. Time-resolved spectroscopy of rhodopsin with femtosecond resolution so far reported are listed in the table [31-43] as well as that applied to the rhodopsin chromophore (protonated Schiff base formed from 11-cis retinal and butylamine) in solution [44].
Table 1. Femtosecond time-resolved
spectroscopy of rhodopsin
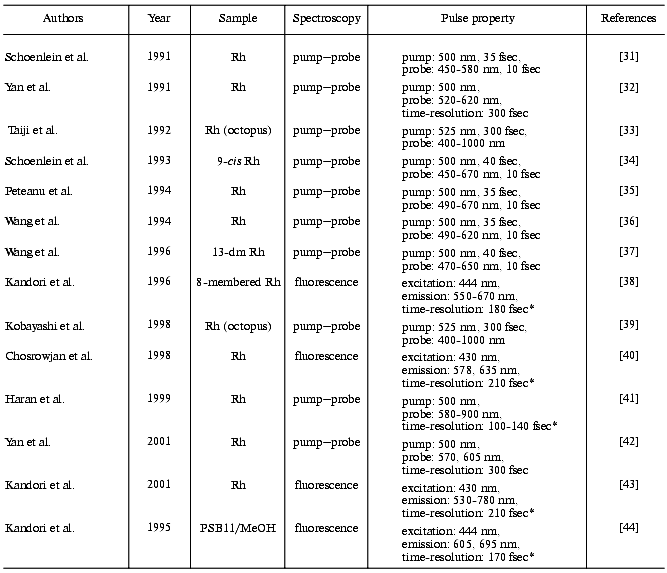
Note: In sample condition column, animals are bovine except for two reports on octopus. Rh, rhodopsin; 13-dm Rh, 13-demethylrhodopsin; PSB11/MeOH, protonated Schiff base of 11-cis retinal in methanol; *, cross correlation width.
The first group quickly reported several femtosecond pump-probe works one after another, which involved the measurement of bovine rhodopsin with wide-spectral window [35], the measurements of 9-cis rhodopsin [34], and 13-demethyl rhodopsin [37] (table). In addition, they observed oscillatory features with a period of 550 fsec (60 cm-1) on the kinetics at probe wavelengths within the photoproduct absorption band of rhodopsin, whose phase and amplitude demonstrate that they are the result of nonstationary vibrational motion in the ground state of photorhodopsin [36]. The observation of coherent vibrational motion in photorhodopsin supports the idea that the primary step in vision is a vibrationally coherent process and that the high quantum yield of the cis-trans isomerization in rhodopsin is a consequence of the extreme speed of the excited-state torsional motion.
The idea of an extremely fast isomerization also matched with the results on 8-membered rhodopsin, in which we estimated the fluorescence lifetime of 8-membered rhodopsin to be 60 fsec throughout the observed wavelengths (550-640 nm) [38]. The results were interpreted in terms of rapid deformation around the C11-C12 double bond occurring as fast as vibrational motions of the chromophore (Fig. 3). Thus, it seems that coherent isomerization becomes a general consensus in the field of ultrafast spectroscopy of rhodopsin, which was reviewed by the first group [45, 46]. Thus, the vibrationally coherent photoisomerization can lead to the high reaction quantum yield of rhodopsin. In fact, the cis-trans isomerization yield of the rhodopsin chromophore (a protonated Schiff base of 11-cis retinal) in solution (~0.15) [47, 48] is much smaller than that in protein (0.67 [14]), indicating that the isomerization of the rhodopsin chromophore is much enhanced in the protein environment of rhodopsin.
While the coherent isomerization provided a new concept on the excited-state dynamics in rhodopsin, there remained various questions on the primary cis-trans photoisomerization of rhodopsin. One of them is that the coherent isomerization could result in a quantum yield much higher than that observed in rhodopsin. Another issue is that the dynamics of rhodopsin seems far different from that of the chromophore in solution, if the photoreaction of rhodopsin is simply described by the coherent isomerization within 200 fsec. The fluorescence dynamics of the protonated Schiff base of 11-cis retinal in solution was found to possess two components, one in femtosecond (90-600 fsec, 25%) and another in the picosecond time domain (2-3 psec, 75%) [44]. These issues required further efforts for better understanding of the primary process of rhodopsin, and the mechanism of its excited-state dynamics was still an open question.
It was intriguing that the first group missed observing stimulated emission, while they observed an absorption band of the excited state at shorter wavelength side of rhodopsin [31, 35]. It might be explained assuming the product absorption rapidly appears at the same wavelength region. On the other hand, stimulated emission from octopus rhodopsin [33, 39] was observed with a time-resolution lower than the first group. The femtosecond transient absorption study by Haran et al. extended the wavelength region into the near infrared, which resulted in detection of the stimulated emission from bovine rhodopsin [41].
All these studies with femtosecond pulses on the primary photochemical processes of rhodopsin were done by means of transient absorption (pump-probe) spectroscopy. To probe the excited-state dynamics of rhodopsin, however, the absorption spectroscopy may not be advantageous, because other spectral features, such as ground-state depletion and product absorption, are possibly superimposed on the excited-state spectral features (absorption and stimulated emission) in the obtained data. Each spectral feature may even vary in the femtosecond time domain, which provides further difficulty in analyzing the data. On the other hand, fluorescence spectroscopy focuses only onto the excited-state processes, so that the excited-state dynamics can be observed more directly. Thus, we attempted to apply femtosecond up-conversion spectroscopy to detect the excited-state dynamics of rhodopsin in real time. After reports on the rhodopsin chromophore in solution [44] and 8-membered rhodopsin [38], we recently succeeded to detect fluorescence from rhodopsin in real time [40, 43] (table).
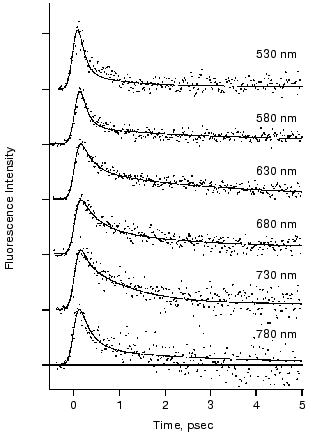
The fluorescence decay at 578 and 635 nm was clearly nonexponential and different from each other in the first report [40]. The results motivated us to measure fluorescence decay kinetics in a wider wavelength region. Figure 4 shows the recent results of the rhodopsin emission in real time that were monitored at 6 wavelengths between 530 and 780 nm [43]. The decay kinetics clearly showed nonexponential nature, where both femtosecond (125-330 fsec) and early picosecond (1.0-2.3 psec) components were predominantly obtained as the best fit. As shown in Fig. 4, no dynamic Stokes shift was detected, whereas the femtosecond component of the fluorescence at both blue (530 and 580 nm) and red (780 nm) sides of the spectrum decayed faster than that at the center. This result suggests that the conversion from the Franck-Condon state to the fluorescence state occurs within the time resolution of the apparatus (<100 fsec) owing to coupling with intra-chromophore high frequency modes and faster initial decay is due to sharpening of band shape caused by a decrease of amplitudes of high frequency modes along the reaction coordinate of twisting.
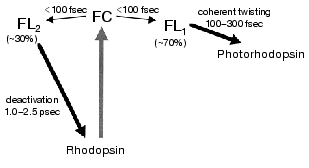
The fastest main components in the 100 fsec regime are probably correlated with the ultrafast coherent isomerization process, which leads to formation of photorhodopsin within 200 fsec [31, 45, 46]. On the other hand, the origin of the early picosecond components was not clear. We confirmed the linear relationship between the excitation laser power and the fluorescence intensity, indicating that the two components of the fluorescence decay indeed originate from the excited state of rhodopsin. The presence of two electrically excited states may be related to the observation. Interestingly, the averaged amplitude of the femtosecond components among 6 wavelengths was about 70%, being very close to the quantum yield of photoisomerization of rhodopsin (0.67). We thus concluded that the slow components (~30%) originate from the nonreactive excited state of rhodopsin, and proposed a scheme shown in Fig. 5. It is noted that such a “branch model” was first proposed for halorhodopsin, a chloride ion pump, based on the longer stimulated emission than the product rise in the pump-probe measurement [49]. In general, two kinetic components are observable which cannot be explained by a simple one-dimensional reaction pathway conventionally assumed for rhodopsin so far.Fig. 4. Fluorescence dynamics of bovine rhodopsin excited at 430 nm and monitored at 530 (133), 580 (145), 630 (204), 680 (169), 730 (106), and 780 nm (60). The numbers in parentheses show the photon counts at maximum position for each monitoring wavelength, respectively. Smooth curves are the best fits obtained by deconvolution procedure with the instrumental response function (full width of half maximum: 210 fsec). Three kinetic components are used in the fitting; femtosecond (125-330 fsec), early picosecond (1.0-2.4 psec), and >50 psec components, among which the femtosecond component is major (58-87%). This figure is modified from Kandori et al. [43].
The excited state features of rhodopsin have been mainly studied by steady-state fluorescence spectroscopy. Fluorescence quantum yields of rhodopsin have been reported to be 5*10-3 [50], 6*10-4 [51], 1.2*10-5 [30], and 9*10-6 [52], which range in almost 3 orders of magnitude. This fact suggests difficulty in estimating accurate quantum yield from weak fluorescence of rhodopsin. The value of the most recent study corresponds to 50 fsec as the excited-state lifetime [52]. The authors argued about the coincidence with the ultrafast processes of rhodopsin. However, the fluorescence lifetime measured in real time [40, 43] clearly demonstrated that the lifetime is certainly longer than 50 fsec. The excited-state dynamics of rhodopsin are still not well understood, but studies underway promise to give a better understanding.Fig. 5. Schematic diagram for the primary reaction processes of rhodopsin. FC and FL correspond to the excited Franck-Condon and fluorescence states, respectively. FC-to-FL conversion followed by the ultrafast twisting to the formation of photointermediate and the slow deactivation to the original ground state. This figure is modified from Kandori et al. [43].
Generation of femtosecond laser pulses enabled us to challenge the photochemistry of rhodopsin in real time. It was concluded that photoisomerization occurs in a coherent manner in the femtosecond time domain, leading to highly efficient formation of photorhodopsin. In addition, recent theoretical studies with the aids of deuterated retinal analogs suggested that isomerization starts at 60 fsec after the photon absorption [53]. It is in remarkable contrast to the argument 30 years ago that formation of the photoproduct within 6 psec would be too fast to be attributed to such a conformational change as cis-trans isomerization of retinal [18]. In fact, the isomerization takes place in 100 fsec regime, and the twisted all-trans state in photorhodopsin is formed within 200 fsec! Such reactions are indeed achieved in the protein environment, because a femtosecond component is rather minor for the rhodopsin chromophore (protonated Schiff base of 11-cis retinal) in solution [44]. The protein environment facilitates ultrafast motion of the retinal molecule from 11-cis to all-trans forms. In addition, femtosecond spectroscopy revealed the presence of the slower picosecond components, which could not be coupled with the isomerization. It is of interest to address the following question: when a rhodopsin molecule is photoexcited, what determines the branch? We need further efforts to understand the fate of the excited rhodopsin molecule.
IV. EFFICIENT PHOTOISOMERIZATION OCCURS IN THE SPECIFIC PROTEIN
ENVIRONMENT
It is well known that rhodopsin is an excellent molecular switch to convert light signal to the electric response of the photoreceptor cell. As mentioned in the preceding chapter, the highly efficient photoisomerization of rhodopsin (quantum yield: 0.67) is assured by the extremely fast cis-trans isomerization of the chromophore that is facilitated by the protein environment. Earlier resonance Raman spectroscopy of bathorhodopsin revealed the distorted structure of the chromophore through the analysis of hydrogen-out-of-plane (HOOP) vibrational modes [54, 55]. It could constitute one of the modes for light energy storage in bathorhodopsin (~35 kcal/mol [56]). Further secrets of the efficient isomerization and energy storage must be hidden in the protein structure and structural changes of the rhodopsin molecule.
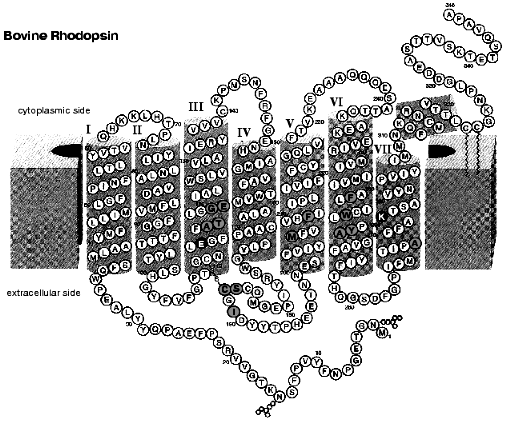
Figure 6 shows the secondary structural model of bovine rhodopsin. The retinal chromophore binds to Lys296 of the 7th helix, and Glu113 of the 3rd helix is the counterion of the protonated Schiff base [57, 58]. In rhodopsin, the chromophore is surrounded by amino acid residues, and the chromophore-protein interaction is altered by the photoisomerization. To investigate such changes of the interaction, infrared spectroscopy is a powerful tool, because it monitors conformational changes of protein as well as the chromophore structure (reviewed in [59-61]). In particular, the reliable Fourier transform infrared (FTIR) spectrophotometer provided progress in rhodopsin study. Infrared spectral changes accompanying the cis-trans isomerization of the retinal chromophore in the rhodopsin molecule were reported in the 1800-800 cm-1 region [62-64].
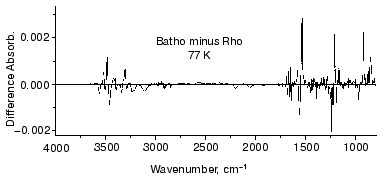
To further examine the conformational changes, we optimized the measuring system for low-temperature infrared spectroscopy of rhodopsin. As a result, we are now able to measure the difference infrared spectra among rhodopsin, bathorhodopsin, and isorhodopsin (9-cis rhodopsin) at liquid nitrogen temperature (77 K) in the whole mid-infrared region (4000-800 cm-1) [65]. In the measurement, the improved equipment could limit the fluctuation within an order of 10-4 absorbance unit in the whole region. Furthermore, several repeated recordings allowed to detect the absorbance change as small as 10-5, which is sufficient to detect a single vibration in rhodopsin. Figure 7 shows the bathorhodopsin minus rhodopsin spectrum. Large spectral changes in the 1600-800 cm-1 originate from vibrational bands of the retinal chromophore such as C=C stretch (1600-1500 cm-1), C-C stretch (1300-1100 cm-1) and hydrogen-out-of-plane vibrations (1000-800 cm-1). In contrast, spectral changes in the higher frequency region (>1600 cm-1) are mainly due to protein bands. In particular, O-H and N-H stretching vibrations appearing in the 3700-2700 cm-1 region are good probes of hydrogen bonds. Thus, accurate spectral detection provided a new tool for experimental study of the protein changes in response to the chromophore motion.Fig. 6. Secondary structure of bovine rhodopsin. Including Lys296 forming a Schiff base linkage with 11-cis retinal, and Glu113 as the protonated Schiff base counterion, 16 highlighted amino acid residues are located within 4.0 Å from the retinal chromophore according to the recent crystallographic structure of rhodopsin [16]. This figure is a gift from Dr. T. Okada.
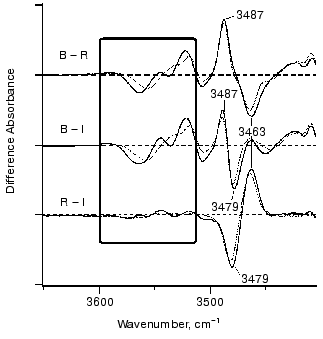
Figure 8 shows infrared spectral changes of bovine rhodopsin in the 3650-3410 cm-1 region. Vibrational bands in the frame originate from water molecules because they exhibit isotope shift in H218O (dotted lines). Infrared spectroscopy is indeed a powerful technique to monitor water structural changes during the functional processes in rhodopsins [61, 66]. It is noted that the spectral feature is similar between bathorhodopsin minus rhodopsin (top) and bathorhodopsin minus isorhodopsin (middle) spectra, indicating that water structures are identical in rhodopsin and isorhodopsin. In fact, no clear spectral feature is visible for the rhodopsin minus isorhodopsin spectrum (bottom). Since only the Schiff base region is polar in the retinal chromophore (Fig. 9), we inferred that these water molecules are in the Schiff base region [65]. Subsequently, location of two water molecules has been assigned by the use of mutant proteins; one is close to Glu113 [67], and the other is in the Asp83-Gly120 region [68]. The former water molecule is indeed located in the Schiff base region, and the latter water molecule is in the region, which is probably connected by hydrogen-bonding network to the Schiff base.Fig. 7. Bathorhodopsin minus rhodopsin infrared spectrum of bovine rhodopsin measured at 77 K. The data originate from Kandori and Maeda [65].
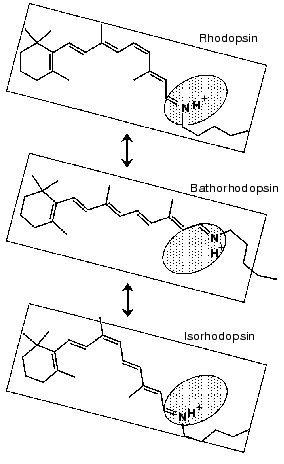
The identical water structures between rhodopsin and isorhodopsin provide an insight into the isomerization mechanism in rhodopsin. Namely, there has been a long-standing question; “which side of the retinal chromophore moves upon isomerization, the beta-ionone ring or the Schiff base?”. Motion of the Schiff base side was previously suggested by low-temperature visible spectroscopy of fluorinated rhodopsin analogs [69]. The observation of the water structural changes provided additional experimental suggestion that the Schiff base side actually moves (rotates), so that water structures are identical between rhodopsin and isorhodopsin (Fig. 9). The initial movement of the Schiff base region is consistent with the cross-linking studies using a retinal analog having a photo-affinity substituent in its beta-ionone ring [70]. That is, the cross-linking group is attached to Trp265 in the 6th helix in rhodopsin, as the crystal structure presumes [16], and is still attached in bathorhodopsin after photoisomerization. However, the beta-ionone ring flips over to the position near Ala169 in the 4th helix during the transition from bathorhodopsin to lumirhodopsin.Fig. 8. Bathorhodopsin minus rhodopsin (top), bathorhodopsin minus isorhodopsin (middle) and rhodopsin minus isorhodopsin (bottom) infrared spectra of bovine rhodopsin in the 3650-3410 cm-1 region. The sample was hydrated with H2O (solid lines) or H218O (dotted lines). One division of Y-axis corresponds to 0.0015 absorbance unit. Solid frame indicates O-H stretching vibrations of water molecules. This figure is modified from Kandori and Maeda [65].
These facts suggest the rotation of the Schiff base side at the stage of bathorhodopsin. It is however noted that the strong hydrogen bond of the Schiff base remains unchanged between rhodopsin and bathorhodopsin [71]. To explain this observation, the new hydrogen-bonding acceptor of the Schiff base proton is necessarily taken into account after the Schiff base motion. Another possibility is that the hydrogen-bonding acceptor moves together with the Schiff base. We suggested a water molecule playing such a role [67]. Presence of a water molecule between the Schiff base and Glu113 was supported by recent NMR studies [72, 73]. Nevertheless, X-ray crystallographic structure [16] does not show such water molecules in the Schiff base region as described below, which requires further clarification on the Schiff base structure.Fig. 9. Schematic drawing of the structure of the chromophore in rhodopsin, bathorhodopsin, and isorhodopsin. The Schiff base region shown by shaded oval is the only polar region in the retinal chromophore. This figure is modified from Kandori and Maeda [65].
Figure 8 also shows an interesting protein band. Like water bands, protein bands were generally similar between rhodopsin and isorhodopsin. This sounds reasonable, because they are stable at room temperature, while bathorhodopsin decays to subsequent intermediates. One exceptional band is the X-H stretching vibration in the 3500-3450 cm-1 region. The isomer-specific protein band appears at 3463, 3487, and 3479 cm-1 for rhodopsin, bathorhodopsin, and isorhodopsin, respectively (Fig. 8) [65]. The band, being due to either O-H or N-H stretch, is not exchangeable for D2O, indicating that the group is embedded in the hydrophobic region. Identification of this band in the future will provide better understanding of protein structural changes accompanying retinal isomerization.
V. PERSPECTIVE: AFTER THE EMERGENCE OF THE CRYSTAL
STRUCTURE
Recently, bovine rhodopsin was crystallized by Okada et al. [15], and its three-dimensional structure was determined with 2.8 Å resolution [16]. These accomplishments promise better understanding of the primary reaction mechanism in rhodopsin, and existing data by various techniques have to be reexamined on the basis of the structural background. Because of the limited space, we would like to make brief comments in the following.
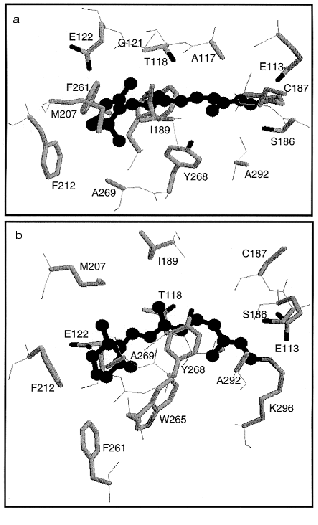
Figure 10 shows the retinal molecule and surrounding amino acids according to the crystallographic structure of bovine rhodopsin [16]. All 16 amino acid residues within 4.0 Å from the retinal are shown including Lys296 (Figs. 6 and 10). Two bulky side chains, Ile189 and Trp265 sandwich the retinal vertically in Fig. 10b, while Thr118 and Tyr268 are located at both sides of the polyene chain (Fig. 10, a and b). Regarding water molecules, a water is confirmed in the vicinity of Asp83-Gly120 as predicted by Nagata et al. [68], whereas no water molecules were shown in the Schiff base region involving Glu113. The latter is of particular interest, because the Schiff base structure is closely related to the mechanisms of spectral tuning, high pKa of the Schiff base, and primary photoisomerization. Further experimental efforts will provide information on the Schiff base structure.
Extensive studies by means of ultrafast spectroscopy of rhodopsin have provided an answer to the question, “What is the primary reaction in vision?” We now know that it is isomerization from 11-cis to all-trans. Femtosecond spectroscopy of rhodopsin eventually captured the excited state of rhodopsin, and as the consequence, we know that unique photochemistry takes place in our eyes. Namely, the cis-trans isomerization in vision is a coherent reaction through the barrierless potential surface, in which formation of photorhodopsin occurs in 200 fsec. Such amazingly fast reaction is facilitated in the protein environment, and vibrational analysis of primary intermediates, such as resonance Raman and infrared spectroscopies, have provided insight into the isomerization mechanism from structural background. The atomic structure of bovine rhodopsin has further encouraged detailed understanding of the mechanism.Fig. 10. Crystallographic structure of bovine rhodopsin (PDB: 1F88) [16]. The retinal molecule is viewed from 9-methyl group and I189 (a), or viewed from Y268 and A292 (b). Totally 16 amino acids are shown that have atoms within 4.0 Å from the retinal chromophore (drawn by ball and stick in black).
When we published the last review article on primary photochemical events of rhodopsin in 1992 [8], we did not make a conclusion concerning the nature of the primary reaction in rhodopsin. We had no femtosecond pulses and we did not know the crystallographic structure of rhodopsin. Remarkable progress has taken place during the past 10 years, and this progress is shown in this article. What will be the progress in the next 10 years? Regarding the dynamics, we still do not know the origin of the nonreactive pathways. On the other hand, we have started to study the role of the protein environment during the retinal isomerization from the structural background. Further efforts by means of spectroscopy, diffraction, and theory will provide us with new insights into the structure and dynamics of photoisomerization in rhodopsin.
We would like to thank Drs. K. Nakanishi of Columbia University (USA), M. Ito of Kobe Pharmaceutical University (Japan), N. Mataga of Institute for Laser Technology (Japan), A. Maeda of University of Illinois (USA), and A. Terakita of Kyoto University (Japan) for their contributions to the results shown in this review article. We thank Dr. T. Okada for a kind gift of the figure of the secondary structure of bovine rhodopsin (Fig. 6). We also thank many collaborators in the references. Some of the research described herein was supported by the Human Frontier Science Program to H. K.
REFERENCES
1.Khorana, H. G. (1992) J. Biol. Chem.,
267, 1-4.
2.Hofmann, K.-P., and Helmreich, E. J. M. (1996)
Biochim. Biophys. Acta, 1286, 285-322.
3.Sakmar, T. P. (1998) Prog. Nucleic Acid Res.
Mol. Biol., 59, 1-33.
4.Shichida, Y., and Imai, H. (1998) CMLS, Cell.
Mol. Life Sci., 54, 1299-1315.
5.Matsumoto, H., and Yoshizawa, T. (1975)
Nature, 258, 523-526.
6.Yoshizawa, T., and Shichida, Y. (1982) Meth.
Enzymol., 81, 333-354.
7.Shichida, Y. (1986) Photobiochem.
Photobiophys., 13, 287-307.
8.Yoshizawa, T., and Kandori, H. (1992) in
Progress in Retinal Research (Osborne, N., and Chader, G., eds.)
Pergamon Press, Oxford, pp. 33-55.
9.Yoshizawa, T., and Wald, G. (1963) Nature,
197, 1279-1286.
10.Yoshizawa, T. (1972) in Handbook of Sensory
Physiology (Dartnall, H. J. A., ed.) Springer-Verlag, Berlin, pp.
69-81.
11.Shichida, Y., Kobayashi, T., Ohtani, H.,
Yoshizawa, T., and Nagakura, S. (1978) Photochem. Photobiol.,
27, 335-341.
12.Shichida, Y., Matuoka, S., and Yoshizawa, T.
(1984) Photobiochem. Photobiophys., 7, 221-228.
13.Peters, K., Applebury, M. L., and Rentzepis, P.
M. (1977) Proc. Natl. Acad. Sci. USA, 74, 3119-3123.
14.Dartnall, H. J. A. (1967) Vision Res.,
8, 339-358.
15.Okada, T., Trong, I. L., Fox, B. A., Behnke, C.
A., Stenkamp, R. E., and Palczewski, K. (2000) J. Struct. Biol.,
130, 73-80.
16.Palczewski, K., Kumasaka, T., Hori, T., Behnke,
C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada,
T., Stenkamp, R. E., Yamamoto, M., and Miyano, M. (2000)
Science, 289, 739-745.
17.Yoshizawa, T., and Kito, Y. (1958) Nature,
182, 1604-1605.
18.Busch, G. E., Applebury, M. L., Lamola, A. A.,
and Rentzepis, P. M. (1972) Proc. Natl. Acad. Sci. USA,
69, 2802-2806.
19.Kawamura, S., Tokunaga, F., Yoshizawa, T., Sarai,
A., and Kakitani, T. (1979) Vision Res., 19, 879-884.
20.Fukada, Y., Shichida, Y., Yoshizawa, T., Ito, M.,
Kodama, A., and Tsukida, K. (1984) Biochemistry, 23,
5826-5832.
21.Callender, R. (1982) Meth. Enzymol.,
88, 625-633.
22.Kandori, H., Matuoka, S., Shichida, Y., and
Yoshizawa, T. (1989) Photochem. Photobiol., 49,
181-184.
23.Matuoka, S., Shichida, Y., and Yoshizawa, T.
(1984) Biochim. Biophys. Acta, 765, 38-42.
24.Kandori, H., Shichida, Y., and Yoshizawa, T.
(1989) Biophys. J., 56, 453-457.
25.Sperling, W. (1973) in Biochemistry and
Physiology of Visual Pigments (Langer, H., ed.) Springer-Verlag,
Heidelberg, pp. 19-28.
26.Mao, B., Tsuda, M., Ebrey, T. G., Akita, H.,
Balogh-Nair, V., and Nakanishi, K. (1981) Biophys. J.,
35, 543-546.
27.Buchert, J., Stefancic, V., Doukas, A. G.,
Alfano, R. R., Callender, R. H., Pande, J., Akita, H., Balogh-Nair, V.,
and Nakanishi, K. (1983) Biophys. J., 43, 279-283.
28.Kandori, H., Matuoka, S., Shichida, Y.,
Yoshizawa, T., Ito, M., Tsukida, K., Balogh-Nair, V., and Nakanishi, K.
(1989) Biochemistry, 28, 6460-6467.
29.Mizukami, T., Kandori, H., Shichida, Y., Chen,
A.-H., Derguini, F., Caldwell, C. G., Bigge, C., Nakanishi, K., and
Yoshizawa, T. (1993) Proc. Natl. Acad. Sci. USA, 90,
4072-4076.
30.Doukas, A. G., Junnarker, M. R., Alfano, R. R.,
Callender, R. H., Kakitani, T., and Honig, B. (1984) Proc. Natl.
Acad. Sci. USA, 81, 4790-4794.
31.Schoenlein, R. W., Peteanu, L. A., Mathies, R.
A., and Shank, C. V. (1991) Science, 254, 412-415.
32.Yan, M., Manor, D., Weng, G., Chao, H., Rothberg,
L., Jedju, T. M., Alfano, R. R., and Callender, C. H. (1991) Proc.
Natl. Acad. Sci. USA, 88, 9809-9812.
33.Taiji, M., Bryl, K., Nakagawa, M., Tsuda, M., and
Kobayashi, T. (1992) Photochem. Photobiol., 56,
1003-1011.
34.Schoenlein, R. W., Peteanu, L. A., Wang, Q.,
Mathies, R. A., and Shank, C. V. (1993) J. Phys. Chem.,
97, 12087-12092.
35.Peteanu, L. A., Schoenlein, R. W., Wang, Q.,
Mathies, R. A., and Shank, C. V. (1993) Proc. Natl. Acad. Sci.
USA, 90, 11762-11766.
36.Wang, Q., Schoenlein, R. W., Peteanu, L. A.,
Mathies, R. A., and Shank, C. V. (1994) Science, 266,
422-424.
37.Wang, Q., Kochendoerfer, G. G., Schoenlein, R.
W., Verdegem, P. J. E., Lugtenburg, J., Mathies, R. A., and Shank, C.
V. (1996) J. Phys. Chem., 100, 17388-17394.
38.Kandori, H., Sasabe, H., Nakanishi, K.,
Yoshizawa, T., Mizukami, T., and Shichida, Y. (1996) J. Am. Chem.
Soc., 118, 1002-1005.
39.Kobayashi, T., Kim, M., Taiji, M., Iwasa, T.,
Nakagawa, M., and Tsuda, M. (1998) J. Phys. Chem. B, 102,
272-280.
40.Chosrowjan, H., Mataga, N., Shibata, Y.,
Tachibanaki, S., Kandori, H., Shichida, Y., Okada, T., and Kouyama, T.
(1998) J. Am. Chem. Soc., 120, 9706-9707.
41.Haran, G., Morlino, E. A., Mathes, J., Callender,
R. H., and Hochstrasser, R. M. (1999) J. Phys. Chem. A,
103, 2202-2207.
42.Yan, M., Rothberg, L., and Callender, R. H.
(2001) J. Phys. Chem. B, 105, 856-859.
43.Kandori, H., Furutani, Y., Nishimura, S.,
Shichida, Y., Chosrowjan, H., Shibata, Y., and Mataga, N. (2001)
Chem. Phys. Lett., 334, 271-276.
44.Kandori, H., Katsuta, Y., Ito, M., and Sasabe, H.
(1995) J. Am. Chem. Soc., 117, 2669-2670.
45.Kochendoerfer, G. G., and Mathies, R. A. (1995)
Isr. J. Chem., 35, 211-226.
46.Mathies, R. A. (1999) in Rhodopsins and
Phototransduction (Novartis Foundation Symposium)
(Yoshizawa, T., ed.) John Wiley & Sons, Chichester, pp. 70-84.
47.Becker, R. S., and Freedman, K. (1985) J. Am.
Chem. Soc., 107, 1477-1485.
48.Koyama, Y., Kubo, K., Komori, M., Yasuda, H., and
Mukai, Y. (1991) Photochem. Photobiol., 54, 433-443.
49.Kandori, H., Yoshihara, K., Tomioka, H., and
Sasabe, H. (1992) J. Phys. Chem., 96, 6066-6071.
50.Guzzo, A. V., and Pool, G. L. (1967)
Science, 159, 312-314.
51.Sineshchekov, V. A., Balashov, S. P., and Litvin,
F. F. (1983) Dokl. AN SSSR, 270, 1231-1235.
52.Kochendoerfer, G. G., and Mathies, R. A. (1996)
J. Phys. Chem., 100, 14526-14532.
53.Kakitani, T., Akiyama, R., Hatano, Y., Imamoto,
Y., Shichida, Y., Verdegem, P., and Lugtenburg, J. (1998) J. Phys.
Chem. B, 102, 1334-1339.
54.Eyring, G., Curry, B., Broek, A., Lugtenburg, J.,
and Mathies, R. A. (1982) Biochemistry, 21, 384-393.
55.Palings, I., van den Berg, E. M. M., Lugtenburg,
J., and Mathies, R. A. (1989) Biochemistry, 28,
1498-1507.
56.Cooper, A. (1979) Nature, 282,
531-533.
57.Zhukovsky, E. A., and Oprian, D. D. (1989)
Science, 246, 928-930.
58.Sakmar, T. P., Franke, R. R., and Khorana, H. G.
(1989) Proc. Natl. Acad. Sci. USA, 86, 8309-8313.
59.Rothschild, K. J. (1992) J. Bioenerg.
Biomembr., 24, 147-167.
60.Siebert, F. (1995) Isr. J. Chem.,
35, 309-323.
61.Maeda, A., Kandori, H., Yamazaki, Y., Nishimura,
S., Hatanaka, M., Chon, Y.-S., Sasaki, J., Needleman, R., and Lanyi, J.
K. (1997) J. Biochem., 121, 399-406.
62.Siebert, F., Mäntele, W., and Gerwert, K.
(1983) Eur. J. Biochem., 136, 119-127.
63.Bagley, K. A., Balogh-Nair, V., Croteau, A. A.,
Dollinger, G., Ebrey, T. G., Eisenstein, L., Hong, M. K., Nakanishi,
K., and Vittitow, J. (1985) Biochemistry, 24,
6055-6071.
64.DeGrip, W. J., Gray, D., Gillespie, J., Bovee, P.
H. M., van den Berg, E. M. M., Lugtenburg, J., and Rothschild, K. J.
(1988) Photochem. Photobiol., 48, 497-504.
65.Kandori, H., and Maeda, A. (1995)
Biochemistry, 34, 14220-14229.
66.Kandori, H. (2000) Biochim. Biophys. Acta,
1460, 177-191.
67.Nagata, T., Terakita, A., Kandori, H., Kojima,
D., Shichida, Y., and Maeda, A. (1997) Biochemistry, 36,
6164-6170.
68.Nagata, T., Terakita, A., Kandori, H., Shichida,
Y., and Maeda, A. (1998) Biochemistry, 37,
17216-17222.
69.Shichida, Y., Ono, T., Yoshizawa, T., Matsumoto,
H., Asato, A. E., Zingoni, J. P., and Liu, R. S. H. (1987)
Biochemistry, 26, 4422-4428.
70.Borhan, B., Souto, M. L., Imai, H., Shichida, Y.,
and Nakanishi, K. (2000) Science, 288, 2209-2212.
71.Siebert, F. (1995) Israel J. Chem.,
35, 309-323.
72.Eilers, M., Reeves, P. J., Ying, W., Khorana, H.
G., and Smith, S. O. (1999) Proc. Natl. Acad. Sci. USA,
96, 487-492.
73.Verhoeven, M. A., Creemers, A. F. L.,
Bovee-Geurts, P. H. M., de Grip, W. J., Lugtenburg, J., and de Groot,
H. J. M. (2001) Biochemistry, 40, 3282-3288.