A Specific DNA-Dependent DNA Polymerase Is Associated with Saccharomyces cerevisiae Telomerase
A. V. Petrov1, P. V. Dmitriev1, K. A. Sokolov2, and O. A. Dontsova1*
1School of Chemistry, Lomonosov Moscow State University, Moscow, 119899 Russia; fax: (095) 939-3181; E-mail: dontsova@genebee.msu.su2Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, ul. Vavilova 32, Moscow, 117312 Russia; fax: (095) 135-1405
* To whom correspondence should be addressed.
Received August 22, 2001
The telomere DNA of most eucaryotes consists of tandem DNA repeats and a number of associated proteins. The synthesis of the G-rich DNA strand is performed by the telomerase complex. The complementary C-strand is synthesized by DNA-dependent DNA polymerases. Using telomerase reverse transcriptase tagging followed by immunoprecipitation coupled with two-step purification, we have found a specific DNA-dependent DNA polymerase activity associated with telomerase of Saccharomyces cerevisiae. Investigation of the biochemical properties of this DNA polymerase activity confirms the hypothesis of tight interaction of DNA polymerase delta with telomerase.
KEY WORDS: yeast telomerase, DNA-polymerase delta, affinity tagging, telomerase reverse transcriptase
The regulation of telomere length is an important process that is essential for the majority of eucaryotic cells [1, 2]. Telomeric DNA is elongated by a telomerase-ribonucleoprotein complex that consists of telomerase RNA, telomerase reverse transcriptase, and a number of other subunits necessary for regulation of telomerase activity in vivo [3, 4].
The components of Saccharomyces cerevisiae telomerase are now supposed to include TLC1 (telomerase RNA), Est2p (telomerase reverse transcriptase), and Est1p (accessory protein containing RRM motifs and responsible for binding of single-stranded nucleic acids) [4-6]. Est3p is another possible component of yeast telomerase complex, but the function of this protein is not still defined [7].
Telomerase activity is influenced by many telomere-associated proteins: Cdc13p [8], Rap1p [9], Rif1p, Rif2p [10], Tel2p [11], Stn1p [12], Pif1p [13], and Ku70/Ku80 [14]. None of them are actually telomerase components as these proteins could not be coimmunoprecipitated with telomerase.
It was recently shown that telomerase-mediated telomere synthesis in yeast depends on DNA polymerases alpha and delta [15]. However, it remained unknown whether they interact with telomerase complex directly or indirectly and how strong such interaction might be.
Our previous in vitro studies of the functional properties of yeast telomerase [16] revealed DNA-dependent DNA polymerase activity in a partially purified fraction of telomerase complex that might be evidence of a tighter interaction of one of the DNA polymerases with telomerase than was supposed before. To prove this, the procedure of telomerase purification used before was modified and an additional affinity purification step was added. Using an affinity tagging method, we have obtained a number of yeast mutants synthesizing Est2p with c-myc epitopes at the carboxyl terminus under control of native promoter. Precipitation of the telomerase by Myc antibodies showed that telomerase complex indeed contains associated DNA-dependent DNA polymerase activity. Investigation of the biochemical properties of this activity allowed us for the first time to suggest association of DNA polymerase delta with telomerase in vitro.
MATERIALS AND METHODS
Est2p affinity tagging with c-myc epitopes. To introduce 10 c-myc epitopes into the carboxyl terminus of Est2p we used a strategy based on a double crossingover event between DNA fragments flanked by two homologous regions [17]. For this purpose total DNA of JD833 (MATa pep4::HIS3 Nuc1::LEU2 trpD1 ura3-52 leu2 his3) was used as a template for PCR amplification with primers E2S7 (5´-CCGTAAGCTCCCAATCAG-3´) and E2D500 (5´-CGCCATGGGTAAGGCATTAGAAACTTTCG-3´). The PCR fragment was cloned into SmaI-digested M13mp18. The resulting construct MEP1 was used for Kunkel site-directed mutagenesis [18] for introduction of a SmaI site before the EST2 stop codon. This site was used for cloning of a DNA fragment containing 10 tandem repeats of c-myc epitopes and a TRP marker. Such fragment was PmeI/SmaI cut from pFA6a-10myc-TRP [19] that was kindly presented by N. Faergemann (South Denmark University, Odense). Subsequent site-directed mutagenesis resulted in insertion of a TEV-protease cleavage site between the Est2p carboxyl terminus and c-myc epitopes (E2-mut3 5´-GCATATATATATATATACATATCCGAGAATCTTTATTTTCAGGAGAACGGTGAACAAAAGC-3´).
The resulting construct MEM4 was used to obtain corresponding linear fragment by PCR amplification using primers E2S7 and E2D500. Strain JD833 of Saccharomyces cerevisiae was transformed by this PCR-fragment [20]. Transformants were analyzed by PCR-screening and immunoblotting of total cell lysates using Myc antibodies (9E10). Thus, strain JD833M4 was constructed.
Isolation and purification of telomerase. Cell-free extract preparation, centrifugation in glycerol gradient, and DEAE chromatography were performed as described elsewhere [16, 21, 22]. The telomerase complex was immunoprecipitated by Sepharose-immobilized Myc antibodies at 4°C. The telomerase-containing fraction after DEAE-chromatography was loaded onto the equilibrated column at the rate of 0.5 ml/min, washed with TMG150-5 buffer (50 mM Tris-HCl, pH 7.5, 150 mM sodium glutamate, 1.2 mM MgCl2, 0.1 mM EDTA, 0.1 mM EGTA, 5% glycerol, v/v). TEV-protease was diluted with 200 µl TMG150-5 buffer to the final concentration 0.25 unit/µl, loaded onto the column, and incubated for 10 h.
Telomerase activity assay. A sample of extract (2-25 µl) was incubated with 1 µM of the corresponding primer in buffer containing 10 mM Tris-HCl, pH 7.8, 1 mM MgCl2, 1 mM DTT, 50 µM dTTP, and 50 µM alpha-[32P]dGTP (3000 Ci/mmol) for 30 min at 30°C. RNAse A solution (1 µl, 10 mg/ml) was added to the mixture which was then incubated for another 30 min at 37°C. Primer elongation products were separated by electrophoresis in 12% denaturing polyacrylamide gel.
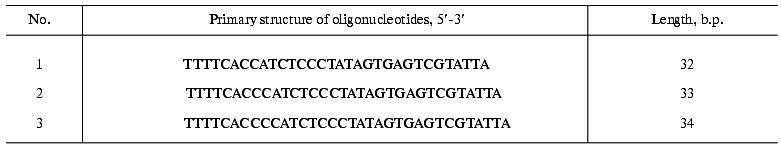
DNA-dependent DNA polymerase activity assay. DNA-dependent DNA polymerase activity was assayed in the presence of the oligonucleotides listed in the table under the conditions described above for telomerase assay besides the presence of 50 µM dATP. The products of elongation were separated in 12% polyacrylamide gel and visualized on a Phosphor Imager SI apparatus (Molecular Dynamics). The level of the activity was calculated using ImageQuant 5.0 software (Molecular Dynamics).
Oligonucleotides elongated in
RNA-independent manner
To test biochemical properties of the telomerase-associated DNA polymerase activity, a mixture of 50 µM oligonucleotides T7 (5´-ATTATGGACTCACTATAG-3´) and 3837 (5´-GGCCATACAACTCCTTCCCTATAGTGAGTCGTATTA-3´) were annealed at 70°C in buffer containing 10 mM Tris-HCl, pH 7.8, 1 mM MgCl2, 40 mM NaCl, 1 mM DTT. Then 4 µl of the mixture was incubated in the presence of 830 µM dTTP, dCTP, dATP, 1 µl alpha-[32P]dGTP (3000 Ci/mmol), and 10 µl extract for 30 min at 37°C. Primer elongation products were separated in 15% denaturing polyacrylamide gel and visualized on the Phosphor Imager SI. Specific inhibition reaction of the DNA-dependent DNA polymerase activity was performed at the presence of 333 mM NaCl, or 80 mM MgCl2, or 1 µl 150 µM ddTTP.
Deletion of the telomerase RNA gene. To delete the telomerase RNA gene we used a strategy [20] similar to that described above. The PCR-amplified DNA fragment (TR500 5´-GGTGGCATCTATAAGAGC-3´ and TR28 5´-CAGTAAATATTAAGAGGCATACC-3´) was cloned into AvaI-digested pUC19 and blunt-ended by Klenow fragment of DNA-polymerase I. The resulting vector was cut by AflII and used for cloning of the DNA fragment containing the TRP marker of K. lactis from PvuII and SmaI digested pGBT9. Thus obtained DNA (pYTR + TRP) was used to synthesize a linear PCR fragment for yeast transformation. Deletion of TLC1 was checked both with PCR and Northern blot analysis.
RESULTS AND DISCUSSION
Est2p affinity tagging. Earlier we applied the strategy of two-step purification of the yeast telomerase complex by glycerol gradient ultracentrifugation of the cell-free extract followed by anion-exchange chromatography [16]. The telomerase purification degree of this method was about 5*103, which was insufficient for investigation of telomerase-associated proteins. Thus, it was necessary to introduce an additional step for considerable enhancement of the purification degree.
In the last several years introduction the affinity epitopes to the protein of interest (affinity tagging) has been widely applied as a method for investigation of large protein and RNA containing complexes [23]. Affinity epitopes represent the protein domains (for instance, IgG-binding domain of Staphylococcus aureus protein A[24, 25], glutathione-S-transferase of Schistosoma japonicum [26], chitin- or maltose-binding domain [27-29]), or short peptides (7-20 amino acids): human c-myc(Myc) epitope [29], influenza virus hemagglutinin (HA) epitope [30]. The presence of such epitopes allows mild and rapid purification of both tagged protein and associated factors (other proteins or nucleic acids).
We applied this strategy to yeast telomerase complex. Ten tandem Myc epitopes were introduced at the carboxyl terminus of the Est2p [29]. As a result, we obtained the strain JD833M4 that expressed modified EST2 under control of the native promoter (Fig. 1). The affinity epitopes were separated from the protein of interest by TEV protease cleavage site. Despite low level of EST2 gene expression, this approach allowed visualization of Est2p directly in cell-free lysates by the immunoblotting method, thus greatly simplifying the procedure of isolation and purification of the telomerase complex.
Isolation of telomerase complex. We performed total cell-free extract preparation using JD833M4 strain as described [16, 22]. Then S100 extract was fractionated subsequently using glycerol gradient ultracentrifugation and DEAE chromatography. Active telomerase fractions were identified by telomerase assay and Northern blot analysis to reveal the presence of telomerase RNA.Fig. 1. Est2p affinity tagging. a) Schematic representation of the wild-type (strain JD833) and fusion protein (strain JD833M4); b) western blot of the cell free lysates from the strains JD833 and JD833M4 using antibodies to Myc. Arrows indicate the position of the molecular weight markers.
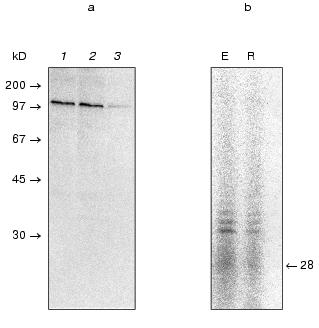
Then we carried out affinity chromatography of partially purified complex using Sepharose-immobilized antibodies to Myc (9E10). Affinity epitope was separated from the carboxyl terminus of Est2p by the Tobacco Etch Virus (TEV) protease recognition site that allowed elution of the complex under native conditions [31]. TEV protease itself carries seven histidine residues at the C-terminus, so it was removed from the eluate by NiNTA chromatography (Fig. 2). The degree of telomerase complex purification calculated as ratio of the total protein quantity before the first stage to protein quantity after the last stage was about 5*106.
DNA polymerase activity is associated with telomerase in vitro. Telomeric DNA replication results from telomerase elongation of the 3´ terminus of the G-strand and C-strand synthesis carried out by DNA-dependent DNA polymerases involved in lagging strand synthesis [32]. In fact, participation of DNA polymerases alpha and delta in telomerase-dependent telomere homeostasis was demonstrated in vivo in a clever manner [15]. Taking into account the fact that synthesis of leading and lagging strands during genome replication should be strictly coordinated, one could suppose that DNA polymerase carrying out C-strand synthesis is associated with the telomerase complex.Fig. 2. Affinity chromatography of the telomerase complex on Sepharose-immobilized Myc antibodies. a) Western blot of the different fractions: 1) crude S100 extract; 2) fraction bound to the affinity resin; 3) fraction remaining bound to the affinity matrix after TEV-protease cleavage. Arrows point the position of the molecular weight markers; b)telomerase activity in fractions after immunoprecipitation. E, activity eluted by TEV protease treatment; R,the activity remaining bound to the affinity resin after TEV-cleavage. The arrow corresponds to the position of the oligonucleotide used in the telomerase assay.
Indirect evidence supporting this hypothesis was based on telomerase-dependent addition of telomeric repeats to non-telomeric substrates by partially purified telomerase fraction [16]. It was shown that telomeric and some non-telomeric oligonucleotides were substrates for the yeast enzyme, and their elongation occurred in an RNA-dependent manner. But further, we found that some non-telomeric substrates are effectively elongated even after RNAse A digestion of the telomerase active fraction (Fig. 3a).
The elongation spectrum of such oligonucleotides depended on dATP (besides commonly used in telomerase assay dTTP, dGTP). We carried out computer analysis of these oligonucleotides and sequenced the products of elongation by the Maxam-Gilbert method [33]. The combination of these two methods showed that these oligonucleotides were able to form partial duplexes due to self-complement interactions (Fig. 3b). Two different structures with 5´ overhangs can be formed and are substrates for DNA-dependent DNA synthesis.Fig. 3. a) RNA-independent elongation of non-telomeric oligonucleotides. The numbers of the lanes correspond to the numbers of the oligonucleotides listed in the table; + and - mean preliminary RNAse A digestion or the absence of it, accordingly; b)proposed duplex structures. Arrows indicate the direction of the DNA synthesis. The unbolded bases are supposed to be eliminated before the synthesis occurs.
It is known that deletion of each yeast telomerase component leads to telomerase inactivation in vivo and in the case of TLC1 and Est2p in vitro [34, 35]. Such event is possibly caused by the dissociation of the factors associated with the wild-type complex. That is why, to check the hypothesis of the association between DNA-polymerase and telomerase, we constructed the strain JD833 Deltatlc1 with deleted telomerase RNA gene (TLC1) and comparatively analyzed glycerol gradient mobility and DEAE-chromatography elution conditions of activity while investigating the mutant and wild-type strains.
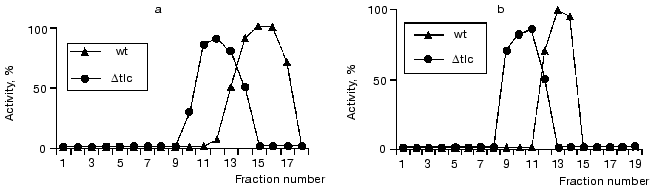
As expected, telomerase activity was not detected in the extract prepared from mutant cells; nevertheless, RNA-independent elongation of non-telomeric oligonucleotides (table) was still detectable after partial purification in both cases. However, the position of the activity peak was changed comparing the mutant and wild-type strains (Fig. 4, a and b). Thus, it was shown that DNA-dependent DNA polymerase activity was dependent on telomerase composition and did not simply co-purify with it.
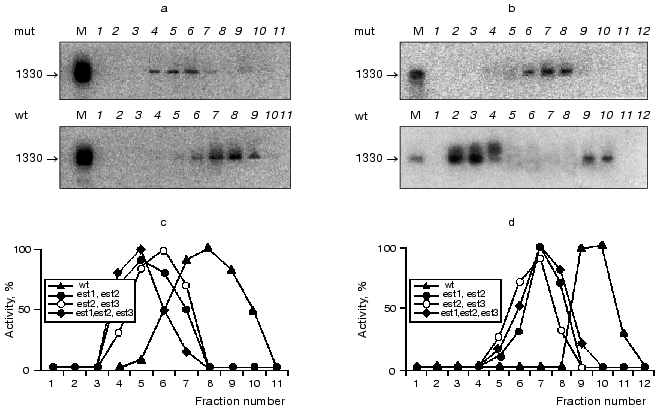
It is known that reverse transcriptases can use a DNA strand as a template, demonstrating properties of DNA-dependent DNA polymerase [36]. One could suggest that telomerase reverse transcriptase Est2p can act as a DNA-dependent DNA polymerase elongating non-telomeric substrates in RNA-independent manner. To check this hypothesis, we used Saccharomyces cerevisiae strains with deleted genes of telomerase components est1, est2, and est3 that were kindly given by P. Donini (University di Roma La Sapienza, Italy). These strains were used for extract preparation followed by ion-exchange chromatography and glycerol gradient centrifugation consecutively. All isolated fractions were subjected to the Northern blot analysis to detect the presence of telomerase RNA and assayed for DNA-dependent DNA polymerase activity as described before (Fig. 5).Fig. 4. Telomerase is associated with DNA-dependent DNA polymerase activity. a) DNA-dependent DNA polymerase activity profile of the glycerol gradient fractionating of lysates prepared from wild-type (wt) and Deltatlc1 cells. Fraction numbers correspond to increasing glycerol concentration from 15 to 40%; b) DNA-dependent DNA polymerase activity profile after DEAE-chromatography of the lysates prepared from wild-type (wt) and Deltatlc1 cells. Fraction numbers correspond to increasing salt concentration from 50 to 1000 mM. The ordinate axis represents relative activity calculated by ImageQuant 5.0 software.
Deletion of each of telomerase RNP protein components shifts the TLC1 peak [22]. However, as in the case of TLC1 deletion, DNA-dependent DNA polymerase activity was not abolished but the corresponding fraction did not coincide with the telomerase RNA fraction. Thus, detected DNA-dependent DNA polymerase activity seemed to be due to the enzymatic activity of a single protein (or protein complex) which dissociates from telomerase in case of deletion of telomerase RNP components.Fig. 5. Distribution of telomerase RNA and specific telomerase-associated DNA-dependent DNA polymerase activity after fractionating lysates prepared from wild-type cells (wt) and mutant cells (mut) with deleted genes encoding for telomerase protein components: est1, est2, est3. a, b) Northern blot analysis of TLC1 presence in the different fractions. M, T7-transcript of TLC1 gene. Numbers represent fraction numbers and increase according to the concentration of the glycerol (a) or sodium acetate (b); c, d) distribution of the specific telomerase associated DNA-dependent DNA polymerase activity during the fractionating of S100 extracts isolated from wild-type or double (triple) mutants in glycerol gradient (c) or DEAE chromatography (d). The abscissa axis represents the number of fractions; the ordinate axis, relative activity calculated by the ImageQuant 5.0 program.
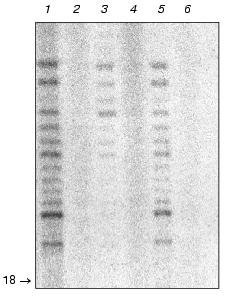
DNA polymerase delta is possibly associated with telomerase complex. Results presented above raise the question of which of the known DNA polymerases might be associated with telomerase. This task can be solved using antibodies to different yeast DNA polymerases or utilizing the corresponding thermo sensitive mutant strains. There is another approach that is reliable enough based on the known differences in the enzymatic properties among the now characterized yeast DNA polymerases [37]. We carried out ordinary DNA polymerase assay using telomerase isolated by the three-step purification scheme described above (Fig. 6).
It turned out that this kind of DNA-dependent DNA polymerase activity is unable to initiate DNA synthesis de novo (lane 2), poorly inhibited by increasing ionic strength (lane 3), fully eliminated by increased Mg2+ concentration (lane 4), and not inhibited by 2´,3´-dideoxythymidine-5´-triphosphate (lane 5). Among the reasonably well-characterized DNA polymerases, only DNA polymerase delta possesses all these properties [37].Fig. 6. Biochemical properties of the telomerase associated DNA-dependent DNA polymerase activity: 1) control reaction of primer elongation; 2) no input primer; 3) reaction under increased ionic strength conditions (200 mM NaCl); 4) reaction in the presence of 50 mM MgCl2; 5) reaction in the presence of 100 µM 2´.3´-dideoxythymidine-5´-triphosphate; 6) no input template. Arrow indicates the position of the primer.
However, we cannot exclude the possibility that telomerase is associated with a special DNA polymerase. This enzyme could be functionally similar to the DNA polymerase delta and is supposed to be responsible exclusively for telomeric DNA replication (C-strand synthesis). It would not be extremely surprising because the number of known specialized DNA polymerases has grown rapidly during recent years [38-40].
This kind of an association of a DNA polymerase activity with telomerase complex in vitro was shown here for the first time. Perhaps there is a much tighter connection among different proteins and RNPs participating in telomere homeostasis than was thought before. The diversity of the functional activities of the telomerase suggests the complexity of the structural organization of the complex that still remains to be investigated.
This work was supported by the grants from the Howard Hughes Medical Institute #55000303, NATO “Investigation of protein composition of S. cerevisiae telomerase complex”, and Russian Foundation for Basic Research: 96-04-55009, 00-15-97950, and 98004-48177.
REFERENCES
1.Shay, J. W., Zou, Y., Hiyama, E., and Wright, W. E.
(2001) Hum. Molec. Genet., 10, 677-685.
2.Evans, S. K., and Lundblad, V. (2000) J. Cell
Sci., 113, 3357-3364.
3.Nugent, C. I., and Lundblad, V. (1998) Genes
Dev., 12, 1073-1085.
4.O'Reilly, M., Teichmann, S. A., and Rhodes, D.
(1999) Curr. Op. Struc. Biol., 9, 56-65.
5.Lingner, J., Cech, T. R., Hughes, T. R., and
Lundblad, V. (1997) Proc. Natl. Acad. Sci. USA, 94,
11190-11195.
6.Hughes, T. R., Evans, S. K., Weilbaecher, R. G.,
and Lundblad, V. (2000) Curr. Biol., 10, 809-812.
7.Zhou, J., Hidaka, K., and Futcher, B. (2000)
Mol. Cell. Biol., 20, 1947-1955.
8.Nugent, C. I., Hughes, T. R., Lue, N. F., and
Lundblad, V. (1996) Science, 274, 249-252.
9.Marcand, S., Wotton, D., Gilson, E., and Shore, D.
(1997) Ciba Found. Symp., 211, 76-103.
10.Wotton, D., and Shore, D. (1997) Gen.
Dev., 11, 748-760.
11.Runge, K. W., and Zakian, V. A. (1996) Mol.
Cell. Biol., 16, 3094-3105.
12.Wang, M. J., Lin, Y. C., Pang, T. L., Lee, J. M.,
Chou, C. C., and Lin, J. J. (2000) Nucleic Acids Res.,
28, 4733-4741.
13.Zhou, J.-Q., Monson, E. K., Teng, S.-C., Schulz,
V. P., and Zakian, V. A. (2000) Science, 289,
771-774.
14.Peterson, S. E., Stellwagen, A. E., Diede, S. J.,
Singer, M. S., Haimberger, Z. W., Johnson, C. O., Tzoneva, M., and
Gottschling, D. E. (2001) Nature Genetics, 27, 64-67.
15.Diede, S., and Gottschling, D. E. (1999)
Cell, 99, 723-733.
16.Petrov, A. V., Dokudovskaya, S. S., Sokolov, K.
A., Lavrik, O. I., Favre, A., Dontsova, O. A., and Bogdanov, A. A.
(1998) FEBS Lett., 436, 35-40.
17.Guthrie, C., and Fink, G. R. (1991) Meth.
Enzymol., 194, 281-302.
18.Kunkel, T. A. (1985) Proc. Natl. Acad. Sci.
USA, 82, 488-492.
19.Longtine, M. S., McKenzie, A., 3rd, Demarini, D.
J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.
R. (1998) Yeast, 14, 953-961.
20.Guthrie, C., and Fink, G. R. (1991) Meth.
Enzymol., 194, 281-302.
21.Cohn, M., and Blackburn, E. H. (1995)
Science,269,404-409.
22.Lingner, J., Hughes, T. R., Shevchenko, A., Mann,
M., Lundblad, V., and Cech, T. R. (1997) Science, 276,
561-567.
23.Deutscher, M. P. (1990) Meth. Enzymol.,
182, 318-331.
24.Knop, M., and Schiebel, E. (1997) EMBO J.,
16, 6985-6995.
25.Siegers, K., Waldmann, T., Leroux, M. R., Grein,
K., Shevchenko, A., Schiebel, E., and Hartl, F. U. (1999) EMBO
J., 18, 75-84.
26.Smith, D. B., and Johnson, K. S. (1988)
Gene, 67, 31-40.
27.Chong, S., Mersha, F. B., Comb, D. G., Scott, M.
E., Landry, D., Vence, L. M., Perler, F. B., Benner, J., Kucera, R. B.,
Hirvonen, C. A., Pelletier, J. J., Paulus, H., and Xu, M. Q. (1997)
Gene, 192, 271-281.
28.Kellerman, O. K., and Ferenci, T. (1982) Meth.
Enzymol., 90, 459.463.
29.Evan, G. I., Lewis, G. K., Ramsay, G., and
Bishop, J. M. (1985) Mol. Cell. Biol., 5, 3610-3616.
30.Field, J., Nikawa, J., Broek, D., MacDonald, B.,
Rodgers, L., Wilson, I. A., Lerner, R. A., and Wigler, M. (1988)
Mol. Cell. Biol., 8, 2159-2165.
31.Dougherty, W. G., and Semler, D. L. (1993)
Microbiol. Rev., 57, 781-822.
32.Price, C. M. (1997) Biochemistry (Moscow),
62, 1216-1223.
33.Maxam, A. M., and Gilbert, W. (1977) Proc.
Natl. Acad. Sci. USA, 74, 560-564.
34.Singer, M. S., and Gottschling, D. E. (1994)
Science, 266, 404-409.
35.Lendvay, T. S., Morris, D. K., Sah, J.,
Balasubramanian, B., and Lundblad, V. (1996) Genetics,
144, 1399-1412.
36.Perach, M., and Hizi, A. (1999)
Virology, 259, 176-189.
37.Burgers, M. J. (1995) Meth. Enzymol.,
262, 49-62.
38.Ruiz, J. F., Dominguez, O., Lain de Lera, T.,
Garcia-Diaz, M., Bernad, A., and Blanco, L. (2001) Philosophical
Transactions: Biological Sciences, 356, 99-109.
39.Kajiwara, K., O-Wang, J., Sakurai, T., Yamashita,
S., Tanaka, M., Sato, M., Tagawa, M., Sugaya, E., Nakamura, K., Nakao,
K., Katsuki, M., and Kimura, M. (2001) Genes Cells, 6,
99-106.
40.Wang, Z., Castano, I. B., De Las Penas, A.,
Adams, C., and Christman, M. F. (2000) Science, 289,
774-779.