Novel Class of Polytopic Proteins with Domains Associated with Putative Protease Activity
A. P. Grigorenko1,2, Y. K. Moliaka1, G. I. Korovaitseva1, and E. I. Rogaev1*
1Research Center of Mental Health, Russian Academy of Medical Sciences, Zagorodnoe Shosse 2/2, Moscow, 113152 Russia; fax: (095) 952-8940; E-mail: rogaev@dol.ru or Evgeny.Rogaev@umassmed.edu2Institute of Molecular Genetics, Russian Academy of Sciences, pl. Kurchatova 46, Moscow, 123182 Russia; fax: (095) 196-0221
* To whom correspondence should be addressed.
Received April 11, 2002; Revision received May 6, 2002
A significant proportion of early onset Alzheimer's disease (AD) is caused by mutations in human genes for amyloid precursor protein (APP), presenilins 1 and 2 (PSEN1,2). AD associated mutations in PSEN1,2 genes alter the gamma-secretase cleavage activity of APP resulting in increased production of amyloidogenic Abeta42. PSEN dependent intramembrane proteolysis was described as an important step required for cleavage of Notch receptors, Notch-dependent signal transduction, and processing of other proteins. It is still unclear whether presenilins are unusual intramembrane proteases or they are necessary cofactors of gamma-secretase cleavage of APP and Notch. Identification of other proteins similar to presenilins may resolve this dilemma. We describe here the identification of novel families of genes encoding polytopic transmembrane proteins of Eukaryotes (IMPASes) and Arachaea(membrases). These proteins have a predicted structure similar to presenilins. The amino acid similarity is significant in domains carrying invariant amino acid residues, which are critical in specific presenilin-regulated endoproteolysis. Many members of the IMPAS family have protease associated domains (PA) typical of proteases. We identified and cloned five human IMPAS genes. Expression analysis of the hIMP1 gene (located on chromosome 20) was performed in human cell tissues and transfected cell cultures. The data demonstrate that a conservative class of putative protease-related polytopic proteins related to presenilins exists in multicellular eukaryotes and microorganisms.
KEY WORDS: Alzheimer's disease, presenilins, APP, Notch, intramembrane proteolysis, gamma-secretase, aspartate, transmembrane domain, PA domain
Abbreviations: AD) Alzheimer's disease; APP) beta-amyloid precursor protein; EMBL) European Molecular Biology Laboratory; EST) expressed sequence tags; I-CliPs) intramembrane-cleaving proteases; ISREC) Swiss Institute for Experimental Cancer Research; KEGG) Kyoto Encyclopedia of Genes and Genomes; NCBI) National Center for Biotechnology Information; ORF) open reading frame; PA-domain) protease associated domain; PCR) polymerase chain reaction; PSEN1) presenilin 1; PSEN2) presenilin 2; RIP) regulated intramembrane proteolysis; TIGR) The Institute for Genomic Research; UPR) unfolded-protein response.
Endoproteolysis of proteins is a universal process involved in
regulation of cell activity, including cell death (necrosis and
apoptosis), regeneration processes, systemic and cell response to
infections, malignant transformation, and stress factors. The possible
existence of intramembrane proteolysis occurring inside plasma
membranes or intracellular membranes, such as Golgi apparatus and
endoplasmic reticulum, is one of the “hot spots” of current
research experimentation and discussion. Due to evident physicochemical
properties of bilayer lipid cellular membranes, endoproteolysis in the
highly hydrophobic intramembrane compartments seemed to be an unlikely
event. However, two previously unknown homologous genes, termed
presenilin 1 (PSEN1 /PS1) and presenilin 2 (PSEN2/PS2), encoding
proteins with multiple transmembrane domains, have been found and
characterized [1-3].
AD-associated mutations in these genes affect normal proteolysis of the
beta-amyloid precursor protein (APP), resulting in increased
endoproteolysis in the transmembrane domain of APP followed by
hyperproduction of insoluble amyloid Abeta42, the main component
of senile plaques in Alzheimer's disease [4-7]. Subsequent analysis revealed that complete loss of
function (knock-out) of the PSEN1 gene in mice, or homologous genes in
invertebrates, decreases or abolishes endoproteolysis of intramembrane
domains of APP and Notch1, the principal receptor of signal
transduction responsible for cell “fate” decisions in early
neurogenesis and hemopoiesis [8-13]. However, it has been recently reported that the
inactivation of PSEN1 and PSEN2 genes may not completely abolish
production of Abeta40,42 amyloid in mouse fibroblasts [14]. Mutations of evolutionary conservative aspartate
residues (Asp257 and Asp385), glycine (Gly384), and proline (Pro433)
residues cause a loss of proteolytic function of PSEN1 [15-18].
Lack of homology between PSEN1/PSEN2 and known eukaryotic proteases does not allow a conclusion to be drawn as to the precise function of presenilins: whether these proteins are gamma-secretases/proteases with intramembrane catalytic activity, or polytopic proteins as cofactors required for the endoproteolysis. Identification of other proteins with similar structure would contribute to the elucidation of molecular genetic mechanisms of intramembrane proteolysis in mammals and other eukaryotes and possibly, prokaryotes. The latter is important for the understanding of basic processes in the regulation of proteolysis in cellular systems, pathogenic mechanisms, and, ultimately, pharmacological intervention for treatment of certain “proteolysis”-related diseases. Abnormal proteolytic cleavage may result in: 1) accumulation of pathogenic insoluble proteins and their processed complexes, implied, e.g., in Alzheimer's disease and other dementias, Parkinson's disease, multiple sclerosis, ischemic heart disease, ischemic insult, etc.; 2) abnormal inter- and intracellular signal transduction, leading to impairments of the early development of the central nervous system, metabolic diseases, and, possibly, to cancer and psychiatric diseases.
Here, we report the discovery of new families of polytopic transmembrane proteins, characterized by important structural similarities with eukaryotic presenilins.
MATERIALS AND METHODS
A series of redundant oligonucleotide primers were used for searching presenilin homologs. Analyses of nucleotide and protein sequences for homology with presenilins were carried out on servers NCBI (USA), TIGR (USA), KEGG (Japan), and EMBL (Germany) using the BLAST database search programs. The hydrophobicity of proteins was analyzed by hydropathy plots (Weizmann Institute of Science, Israel) using the Kyte-Doolittle algorithm (x-1, window length 17). Transmembrane topology prediction was performed by PSORT II (Japan) and Tmpred (ISREC, Switzerland). ClustalW algorithm and the editor program GeneDoc were used for multiple alignment.
The expression of hIMP1 was analyzed by polymerase chain reaction (PCR) on human cDNA panels (Clontech, USA) using the following primers: 5´-TCCTGCACCTCAACAATGTC, 5´-AGGACAGGAAAACCGATGC. The size of the PCR product was about 400 bp. Primers for human glyceraldehyde 3-phosphate dehydrogenase gene (G3PDH, Clontech) were used to obtain control PCR products. The size of the control gene PCR product was about 1 kb.
hIMP1 cDNAs were cloned from leukocyte transcript templates into plasmid
vectors pcDNA3 and pcDNA4/myc-His B, pcDNA6/V5-HisA (Invitrogen, USA)
using the following pairs of primers:
5´-TTTTTGAATTCGAACCCTTCCTGTTGCCTTA,
5´-ACTTATTCATGAAGGGGCTGATG; 5´-ACACCATCAGCCCCTTCAT,
5´-AGGACAGGAAAACCGATGC; 5´-TTACCATCTTCATCATGCACATCT,
5´-TTTTTCTCGAGCCTGGCCATGGGATGAGT (pcDNA3),
5´-TTTTTCTCGAGCGTTTCTCTTTCTTCTCCAGC (pcDNA4/myc-His B),
5´-TTTTTCTCGAGTTTCTCTTTCTTCTCCAGC (pcDNA6/V5-HisA).
The full length copies of hIMP2,3,4,5 cDNAs were also cloned using the following PCR mixture : 2.5 mM MgCl2, 67 mM Tris-HCl, pH 8.4, 16.6 mM (NH4)2SO4, 200 µg/ml BSA, 200 µM dNTPs, 20 pM primers, 5 units of Pfu polymerase. Amplification was performed through 29-30 cycles of denaturation at 94°C for 40 sec, annealing at 57°C for 40 sec, and extension at 72°C for 50 sec. Nucleotide sequences cDNA transcripts of hIMP1 and other IMPAS members were determined by sequencing of the cloned PCR-products. For expression in mammalian cells, the full length hIMP1 cDNA was cloned into expression vectors within a reading frame with a myc-tag (pcDNA4myc-His B) and a V5-tag (pcDNA6/V5-His A).
Human embryonic kidney cells HEK293 were incubated at 37°C, 5% CO2 in DMEM medium (GibcoBRL, USA), containing 10% fetal bovine serum (FBS, GibcoBRL), 2 mM L-glutamine, and 1% penicillin/streptomycin. Transient transfection of HEK293 cells was performed using lipofectamine (LipofectAMINE PLUS Reagent, Gibco BRL). After 48 h, the cells were washed twice with cold phosphate buffered saline (PBS, ICN, USA). The washed cells were lysed at 4°C for 1 h in modified RIPA buffer containing 50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 tablet of inhibitor cocktail “Complete” (Mini, EDTA-free, Roche, France) per 10 ml of buffer. Cell lysates were collected and stored at -70°C.
Cell lysates were analyzed by electrophoresis in 12% polyacrylamide gel in the presence of sodium dodecyl sulfate (SDS-PAGE) according to Laemmli [19].
Protein transfer onto PVDF membrane (Amersham Pharmacia Biotech, England) was carried out in TTB-buffer (25 mM Tris, 192 mM glycine, and 15% methanol, pH 8.3) for 12 h at 30 V.
Western blot hybridization was carried out using primary monoclonal antibodies against myc- and V5-epitopes (Sigma, Invitrogen) and corresponding secondary antibodies. The hybridization signals were detected by ECL Western blotting detection reagents (Amersham Pharmacia Biotech). To identify endogenous hIMP1 protein, the 17-amino acid peptide corresponding to the N-terminal sequence of hIMP1 was synthesized. Polyclonal antibodies were obtained after six immunizations of rabbits for 3 months (Biosource International, USA). Antibodies were purified by affinity chromatography using the protocol of the supplier (Biosource International).
Stable transfected cell lines HEK293 and PC12 with hIMP1 cDNA constructs were obtained using transfection, followed by standard antibiotic selection procedure.
RESULTS
Discovery of new protein families sharing similarity with presenilins. We have used several approaches to search for proteins similar to presenilins.
1. In a preliminary study, PSEN1 cDNA probes were used for direct blotting hybridization with HindIII and EcoRI fragments of human genomic DNA, for Northern hybridization with total or poly-A RNA from human neocortex, and in situ hybridization with human chromosomes. In another series of experiments, the oligonucleotide primers for exons of PSEN1 and PSEN2 encoding the most conservative protein domains were used in RT-PCR analysis of cDNAs from various human tissues. These experiments revealed that the methods employed can detect only genomic fragments corresponding to PSEN1 and PSEN2 genes. Thus, it seems unlikely that any additional genes characterized by high DNA homology to presenilins exist in the human genome.
2. Hypothetical genes which might encode stretches of amino acid sequences homologous to presenilins were searched for using PCR with redundant primers. In this approach, we synthesized a series (more than 12) of redundant oligonucleotide primers (“universal primers”) to the most conservative amino acid sequences of the presenilin family and an evolutionary distant presenilin homolog, SPE4, found only in Caenorhabtidis elegans. Using combinations of these oligonucleotide primers and low temperature annealing conditions at 50-52°C, we were able to obtain only PCR-products corresponding to human PSEN1, PSEN2. Although presenilin orthologs from evolutionarily distant organisms (Drosophila melanogaster and Helix lucorum) were successfully isolated with this set of primers, no additional homologs were identified.
3. We compared amino acid sequences of various domains of human presenilins with all amino acid sequences available in EST and Genomic databases (see “Materials and Methods” section). This search revealed significant similarities only between sequences of orthologic or paralogic presenilin homologs. Such homologs were identified in all main eukaryotic taxons, excluding yeast, where we failed to detect presenilin sequences.
Comparison of protein domains of presenilins revealed that the hydrophilic loop between conservative transmembrane domains six and seven is the most divergent part of the protein. It is characterized by various lengths in presenilin orthologs and paralogs, with the longest loop (>200 residues) in presenilin isolated from the snail Helix lucorum (Riazanskya and Rogaev, unpublished). The existence of such variability in the loop complicates the alignment of amino acid sequences and search for distant presenilin homologs. Identification of the most ancient and/or primitive orthologs could facilitate the search for phylogenically distant protein families sharing with presenilins similar amino acid sequences, provided that such homologous sequences actually exist. The search for putative presenilin homologs in the Protista EST database (tblastn) found EST C84790 of Dictyostelium discoideum. The homology with two human PSEN1 regions was 41/87 (47%), 45/92 (48%) Sum p(2) = 3.9 e-40. EST C84790 contained an incomplete 5´-end of the open reading frame (ORF). The predicted partial amino acid sequence is characterized by 1) presence of domains with conservative aspartate residues; 2) a hydrophilic loop between these domains that is significantly shorter than in all other described presenilins; and 3) terminal PAL-motif that is invariant in all presenilins. Apparently, this sequence is an ortholog and, potentially, the most ancient archetype of presenilins. This sequence was used for the subsequent search of homologs in the EST Dictyostelium discoideum database (TIGR; tblastn). It was found that PSEN Dictyostelium discoideum shares 28% (44/155) homology with the translated sequence EST C89899 (p = 0.011). The sequence homology included regions of conservative aspartate residues, short loop, and terminal PAL-motif. Subsequent analysis revealed that this EST is a member of a family of genes encoding a new superfamily of proteins which has not been previously described in the literature. Polytopic transmembrane proteins of unknown function(s) are the predicted products of these genes. Homologous genes have been found in many organisms including yeast, plants, invertebrates, and vertebrates. Because these proteins share significant similarity with presenilins in domains implicated in intramembrane protease associated activity, we termed these proteins as IMPAS (IMP). Interestingly, members of the IMPAS gene family have been found in yeast, which have no presenilins (Saccharomyces cerevisiae, Schizosaccharomyces pombe). At least two homologous genes were found in Caenorhabditis elegans and Drosophila melanogaster. Thus rapid elucidation of effects of inactivation of IMPASes using these invertebrate models can be anticipated.
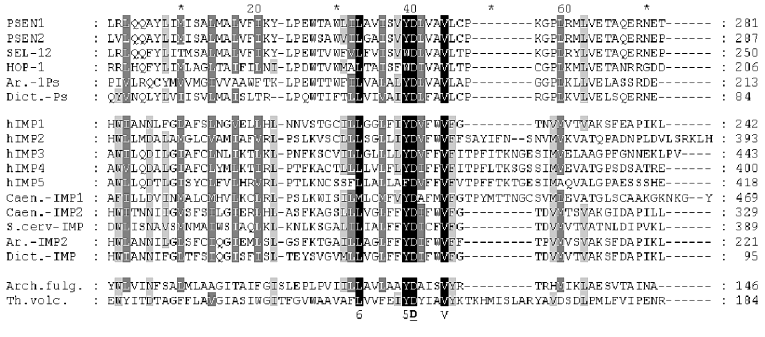
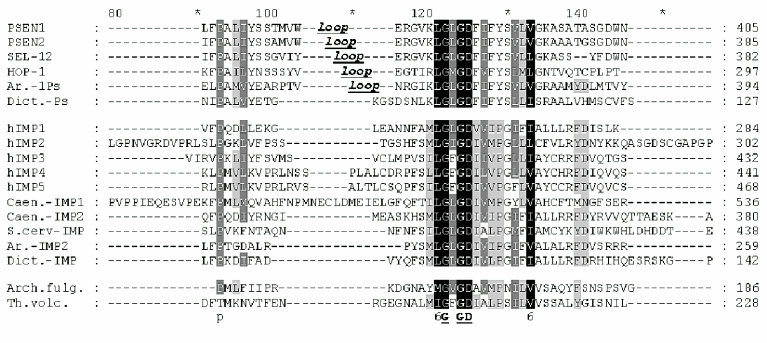
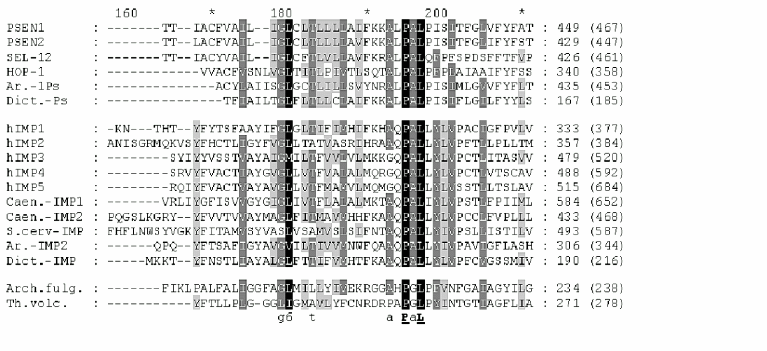
All IMPAS proteins have conservative sequences around the first ((Y/F)D(V/I/A)(F/Y/C)(F/W/M)VF) and the second (6(L/I)G(L/I/F)GD(I/V)(6/A)6PG) aspartate residues and also QPAL(L/I)Y(L/I)(V/S)(P/S) motif in the C-terminal part ( similar amino acids 1-DN, 2-EQ, 3-ST, 4-KR, 5-FYW, 6-LIVM; Blosum62). The distance between these two conservative aspartates in paralogs and orthologs of IMPASes from different species varied from 40 to 80 residues.
The search (tblastn, TIGR) of Homo sapiens EST sequences sharing homology with Dictyostelium discoideum IMPAS identified EST for four different genes: hIMP1 (THC503065), 50% (102/204), p = 4.7 e-49; hIMP2 (THC496042), 38% (51/134), p = 2.1 e-15; hIMP3 (THC483349), 33% (63/186), p = 7.4 e-16; hIMP4 (THC544530), 31% (62/194), p = 1.3 e-14. Using hIMP3,4 we found partial EST (THC488437) for hIMP5. Subsequent analysis of EST sequences by means of the search program Human Genome Blast revealed that genes hIMP1-5 are located on chromosomes 20, 12, 15, 19, and 17, respectively. We cloned and sequenced several IMPAS cDNAs from human blood lymphocytes and brain cells (prepared for publication).
The search (tblastn, NCBI) of sequences homologous to Dictyostelium discoideum IMPAS (Dict.-IMP) revealed genomic sequences of various Archaea encoding similar polytopic transmembrane proteins. The Arhaea's proteins shared the following homology with Dict.-IMP: 26% (45/169), p = 2 e-04, Archaeoglobus fulgidus; 27% (33/122), p = 0.004, Thermoplasma acidophilum; 26% (34/129), p = 0.059, Thermoplasma volcanium. The protein sequences corresponded to genes with unknown functions: AF1952, Ta0062, and TVN0018. The regions of protein homology with presenilins and IMPASes of eukaryotes included two various aspartate residues and a PxL-motif in the C-terminal region. We termed these genes and their protein products as “membrases” by their probable localization in inner bacterial membranes.
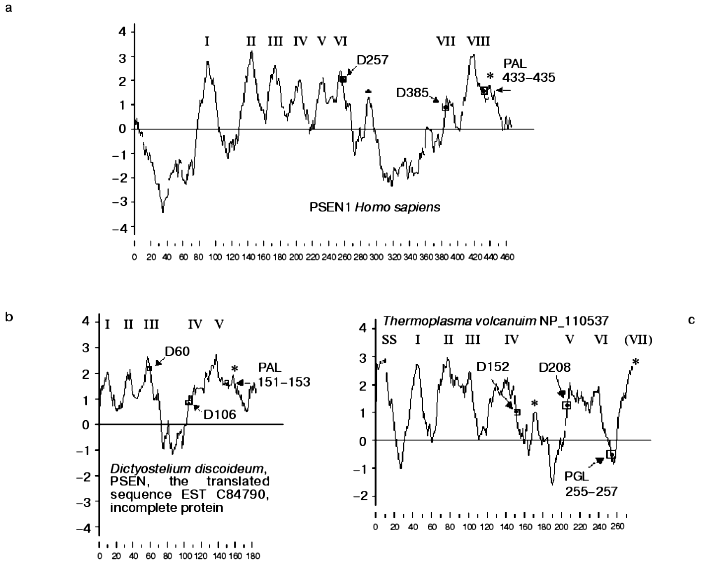
Similarity of presenilins, IMPASes, and membrases. Multiple alignments of amino acid sequences of members of presenilin, IMPAS, and membrase families (Fig. 1) revealed the following consensus sequences for the most conservative regions: 1) 6xxxxx5DxxxV (the region of the first aspartate); 2) 6GxGDxxxxxxx6 (the region of the second aspartate) and 3) PxL (the terminal motif, where x is any amino acid). Eukaryotic IMPASes and archaebacterial membrases are proteins with multiple (from 8-9 up to 10-11) hydrophobic domains (Fig. 2, c and d). Many IMPASes have N-terminal signal sequences. The comparison of hydrophobicity plots of presenilins, membrases, and IMPASes (Fig. 2) also revealed similarities in general structure and location of conservative amino acid residues. They may be summarized in a scheme of possible protein topology which was elaborated on the basis of the previously developed presenilin model [20] in the following order: 1) cytoplasmic N-terminal hydrophilic end, including N-terminal signal sequences in some IMPASes and membrases; 2) several transmembrane domains (n 1-3) for Arhaea's membrases and yeast IMPASes, (n 1-5) for IMPASes of other eukaryotes and presenilins; 3) transmembrane domain (n 4 or n 6), carrying the first conservative aspartate; 4) hydrophobic (para-) membrane domain, which is potentially associated with membranes and that is typical for presenilins and, possibly, for membrases; a hydrophilic cytoplasmic loop found in most presenilins; 5) transmembrane domain (n 5 or n 7), carrying the conservative motif of the second aspartate; 6) transmembrane domain (n 6 or n 8); 7) the conservative PxL motif, hydrophobic para-membrane or transmembrane C-terminal domain. Thus, members of these gene/protein families (presenilins, IMPASes, and membrases) are characterized by: 1) the presence of multiple hydrophobic/transmembrane domains suggesting its multi-pass membrane localization; 2) the presence of consensus sequences around conservative aspartates and PxL-terminal motif; 3) the location of conservative aspartate residues in adjacent transmembrane domains. Whether presenilins and IMPAses are homologous i.e., derived from a single ancestral precursor or analogous proteins, which share similarities only in some domains due to convergent evolution, requires more detailed evolutionary analysis.
Fig. 1. Multiple alignment (ClustalW) demonstrating similarity of conservative aspartate and PxL domains in protein families of eu- and prokaryotes. Eukaryote presenilins are represented by PSEN1,2 in Homo sapiens; SEL-12, HOP-1 in Caenorhabditis elegans; Ar.-1Ps in Arabidopsis thaliana AAF99776; Dict.-Ps in Dictyostelium discoideum EST C84790. Eukaryote IMPASes are represented by hIMP1-5 in Homo sapiens; Caen.-IMP1, Caen.-IMP2 in Caenorhabditis elegans CAB02277, CAA92975; S.cerv-IMP in Saccharomyces cerevisiae CAA81940; Ar.-IMP2 in Arabidopsis thaliana AAC34490; Dict.-IMP in Dictyostelium discoideum EST C89899; Archaea membrases include Arch. fulg. in Archaeoglobus fulgidus AAB89302; Th.volc. in Thermoplasma volcanium NP 110537; Th.ac. in Thermoplasma acidophilum CAC11210. Groups of similar amino acid are: 1-DN, 2-EQ, 3-ST, 4-KR, 5-FYW, 6-LIVM (Blosum62). Amino acids of 100, 80, and 60% homology are marked by black , dark gray, light gray and black letters, respectively. The total length of the protein is indicated in brackets. The region of the hydrophilic loop in presenilins is underlined.
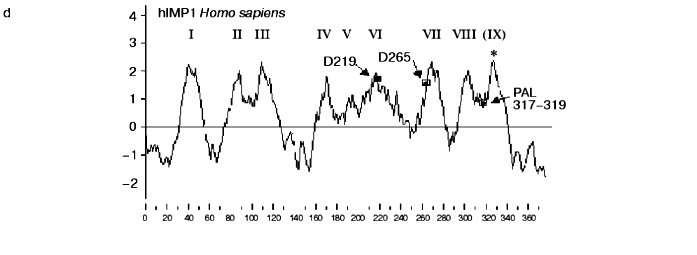
PA-domain. The analysis of protein sequences of IMPASes by the RPS-Blast search program using the NCBI database for conservative protein domains revealed that some members of this family (including hIMP3,4,5) contain a PA-domain ( protease-associated domain) in the N-terminal part. This domain was described as a typical motif for protease families [21]. The PA-domain was previously described in plant alveolar receptor BP-80 as the sequence which shares certain homology with the domain of mammalian transferrin receptor [21]. This sequence was also found in proteases of various families, including bacterial and plant subtilases; and Zn-dependent metalloproteases of M20, M28, and M33 families. The location of the PA domain varies and may be outside of the catalytic domain [21].Fig. 2. Protein hydrophobicity profiles(determined using Kyte-Doolittle method, x-1, 17): a) PSEN1, Homo sapiens; b) PSEN, Dictyostelium discoideum, the translated sequence EST C84790, incomplete protein; c) membrase, NP 11537, Thermoplasma volcanium;d) hIMP1, Homo sapiens. Abscissa indicates amino acid number, ordinate is hydrophobicity index. Transmembrane domains are indicated by Roman numerals. Arrows show the positions of conservative aspartates and PxL motif. Asterisks indicate additional hydrophobic domains. SS is signaling sequence.
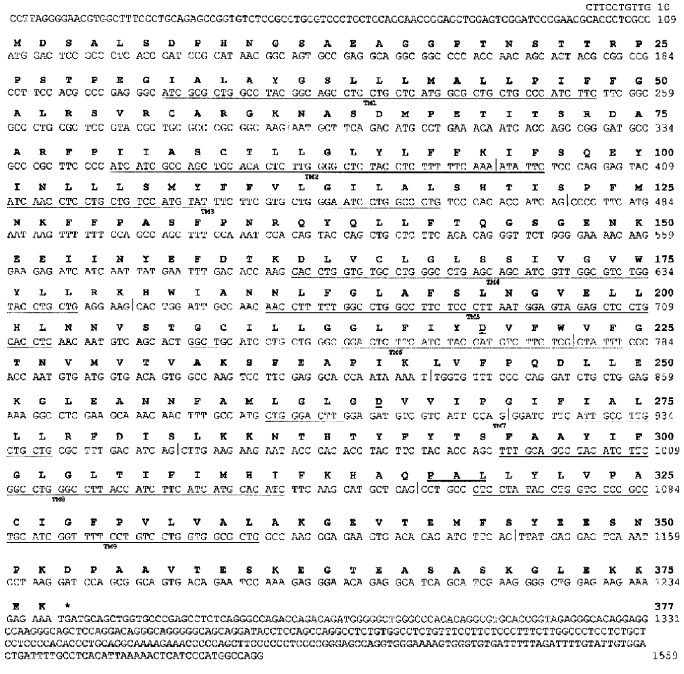
Cloning of hIMP1 gene and study of its expression. The hIMP1 transcripts were isolated from human blood cells and hippocampus , and corresponding cDNAs were cloned into pcDNA3 vector, followed by subsequent sequencing (Fig. 3). The hIMP1 ORF contains 1134 bp and consists of sequences of 11 exons encoding the protein of 377 residues. The predicted polytopic transmembrane protein has nine hydrophobic domains (Figs. 2d and 3).
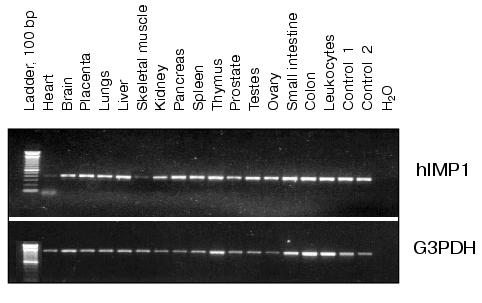
The analysis of hIMP1 expression by RT-PCR using human cDNAs as templates (Clontech) revealed abundant hIMP1 expression in many human organs and tissues and reduced expression in heart and skeletal muscles (Fig. 4).Fig. 3. Nucleotide and deduced amino acid sequences of the hIMP1 gene transcript isolated from blood and human brain cells. Vertical bar indicates exon border. Asterisk shows stop codon. Conservative aspartate residues, PAL, and putative transmembrane domains (TM1-9) are underlined.
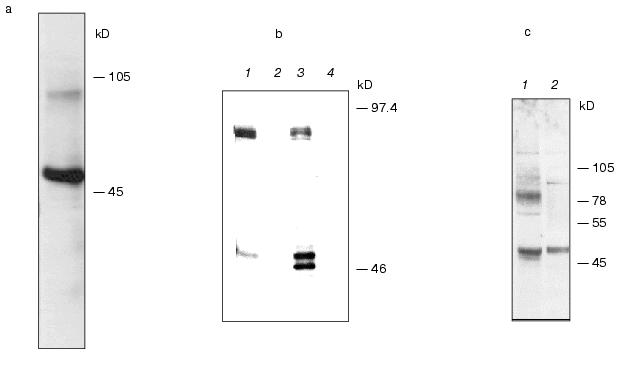
To express hIMP1 protein in mammalian cells, HEK293 cells were transfected with plasmid containing hIMP1 cDNA in vectors pcDNA4/myc-HisB, pcDNA6/V5-HisA. The protein product of 45 kD and a high molecular weight product (about 100 kD) was detected by immunoblots with antibodies against myc- and V5-epitopes (Fig. 5a). Similar results were obtained using stably transfected human (HEK293) and rat (PC12) cells (Fig. 5b). The use of specific antibodies generated to the N-terminal part of hIMP1 revealed an expected endogenous product in cellular cultures and human brain cells (Fig. 5c).Fig. 4. Study of hIMP1 gene expression in various human organs. The sizes of RT-PCR products are: ~400 bp (hIMP1), ~1000 bp (human G3PDH). Positive controls 1 and 2 are mixtures of cDNA from various tissues.
Fig. 5. Western blot hybridization of lysate proteins isolated from cells transfected with hIMP1 cDNA in pcDNA6/V5-HisA vector. The proteins were detected with antibodies against V5-epitope: a) transient transfection of HEK293 cells; b) stable transfected HEK293 and PC12 cells (HEK293 hIMP1-pcDNA6/V5-HisA (1); HEK293 (2); PC12 hIMP1-pcDNA6/V5-HisA (3); PC12 (4)). Endogenous expression of IMPASes was detected in human brain and HEK293 cells using antibodies against the N-terminal part of hIMP1: c) HEK293 (1); human brain (2).
DISCUSSION
We describe here novel conservative families of proteins characterized by unique structural similarity to presenilins. Presenilins are critical molecules for intramembranous cleavage of Notch1 and APP and other Type I proteins with single transmembrane domains. It is conceivable that IMPAS and membrase proteins represent previously unknown aspartate proteases or proteolytic cofactors which have physiological activities similar to presenilins or interacting with a presenilin-related pathway. In this respect, it may be noticed that the critical aspartate residues invariant in all members of presenilin family and type 4 prepilin peptidases (polytopic bacterial aspartate proteases [17, 22]) are also typical for IMPASes and membrases. Moreover, besides conservative aspartates, the prepilin peptidases also contain a conservative PxL motif in the C-terminal region. The existence of an extended hydrophilic loop between these aspartates in presenilins, and lack of this loop in IMPASes might underline structural differences between these families of proteins. However, we found that the most ancient presenilin identified in Dictyostelium discoideum also has no or a very short hydrophilic loop in the corresponding region. The full-length endogenous and, in part, transfected presenilin 1 and 2 proteins are rapidly processed in cells via endoproteolytic cleavage at the hydrophilic loop. The endoproteolysis of IMPASes between transmembrane domains 6 and 7 which might result in N- and C-terminal fragments was not detectable in our experiments with transfected hIMP1 constructs in mammalian cells.
It will be of interest to define further what specific mechanisms determine or, alternatively, abolish this endoproteolytic processing in presenilin and IMPAS proteins. The PA-domain is suggested to be required for binding with specific substrates in cytoplasm or intermembrane compartments. Identification of a PA-domain in some members of the IMPAS protein family indicates its possible relation to proteases. Presenilins were the first identified multi-pass proteins with putative intramembrane protease activity. Although the precise mechanisms of proteolytic cleavage inside the lipid bilayer remains elusive, there is increasing experimental evidence for the existence of a large class of intramembrane cleaving proteases (I-CliPs) which play an important role in certain cellular processes [23, 24]. To date several examples of regulated intramembrane proteolysis (RIP) have been described: 1) proteolysis of the type I transmembrane proteins with lumen C-terminus (APP, Notch, Ire) cleaved by aspartate proteases, such as presenilins; 2) proteolysis of the type II transmembrane proteins with cytoplasmic N-terminus. These include the transcription factor regulating cholesterol biosynthesis (SREBP) and ATF6 (transcription factor UPR). The proteolysis of these substrates is regulated by polytopic Zn-dependent metalloproteinase S2P (see for review [24]). Recently, a unique serine protease Rhomboid-1 has also been found. This enzyme is involved in proteolytic cleavage of intramembrane domain Spitz, a ligand for Drosophila epidermal growth factor [25]. Since presenilins have been considered as the only “possible” aspartate proteases with multiple transmembrane structure, we suggest that IMPAS and membrase families represent a novel class of proteins regulating intra-(or para-)membrane proteolysis of yet unknown cellular proteins. The alternative hypothesis is that IMPASes regulate transportation of putative proteases and/or its substrates to corresponding membrane or submembrane compartments where endoprotolytic cleavage occurs. Since IMPASes and presenilins resemble the structure of ion channel proteins, it is also possible that these proteins regulate membrane ion channels and, via this regulation, membrane associated protease activity.
Identification of proteolytic substrates for IMPASes and the comparative study of the proteolytic processes regulated by IMPASes and presenilins is an important direction for future study. Certain data, e.g., detection of products of gamma-secretase cleavage of APP in PSEN1,2 in double knock-out PSEN1 and 2 mouse fibroblasts [14], and the presence of pharmacologically distinct gamma-secretase activities with respect to APP substrate [26]) raise the possibility that, in addition to presenilins, other intramembrane proteases interacting with presenilin activity may exist in mammalian cells. The results presented here and in our previous preliminary report [27] provide evidence for the existence of a large class of transmembrane polytopic eu- and prokaryotic proteins with similar structures that include the presenilin family. Whether IMPASes activity can interact with the processing of APP and Notch, and whether the IMPAS proteins are involved in regulation of normal embryogenesis or development of Alzheimer's disease, as described for presenilins, is a subject for further investigations.
This work was supported by grants from INTAS (YSF-00-4208), INTAS-RFBR (95-087), Howard Hughes Medical Institute, European Commission Inco-Copernicus, NIH FIRCA, Russian Foundation for Basic Research, Russian Research Program “Human Genome”, and the RF President grant.
REFERENCES
1.Sherrington, R., Rogaev, E. I., Liang, Y., Rogaeva,
E. A., Levesque, G., Ikeda, M., Chi, H., Lin, C., Li, G., and Holman,
K. (1995) Nature, 375, 754-760.
2.Rogaev, E. I., Sherrington, R., Rogaeva, E. A.,
Levesque, G., Ikeda, M., Liang, Y., Chi, H., Lin, C., Holman, K., and
Tsuda, T. (1995) Nature, 376, 775-778.
3.Rogaev, E. I., Sherrington, R., Wu, C., Levesque,
G., Liang, Y., Rogaeva, E. A., Ikeda, M., Holman, K., Lin, C., Lukiw,
W. J., de Jong, P. J., Fraser, P. E., Rommens, J. M., and
George-Hyslop, P. (1997) Genomics, 40, 415-424.
4.Borchelt, D. R., Thinakaran, G., Eckman, C. B.,
Lee, M. K., Davenport, F., Ratovitsky, T., Prada, C. M., Kim, G.,
Seekins, S., Yager, D., Slunt, H. H., Wang, R., Seeger, M., Levey, A.
I., Gandy, S. E., Copeland, N. G., Jenkins, N. A., Price, D. L.,
Younkin, S. G., and Sisodia, S. S. (1996) Neuron,17,
1005-1013.
5.Duff, K., Eckman, C., Zehr, C., Yu, X., Prada, C.
M., Perez-tur, J., Hutton, M., Buee, L., Harigaya, Y., Yager, D.,
Morgan, D., Gordon, M. N., Holcomb, L., Refolo, L., Zenk, B., Hardy,
J., and Younkin, S. (1996) Nature, 383, 710-713.
6.Lemere, C. A., Lopera, F., Kosik, K. S., Lendon, C.
L., Ossa, J., Saido, T. C., Yamaguchi, H., Ruiz, A., Martinez, A.,
Madrigal, L., Hincapie, L., Arango, J. C., Anthony, D. C., Koo, E. H.,
Goate, A. M., Selkoe, D. J., and Arango, J. C. (1996) Nat. Med.,
2, 1146-1150.
7.Citron, M., Westaway, D., Xia, W., Carlson, G.,
Diehl, T., Levesque, G., Johnson-Wood, K., Lee, M., Seubert, P., Davis,
A., Kholodenko, D., Motter, R., Sherrington, R., Perry, B., Yao, H.,
Strome, R., Lieberburg, I., Rommens, J., Kim, S., Schenk, D., Fraser,
P., St. George, H. P., and Selkoe, D. J. (1997) Nat. Med.,
3, 67-72.
8.Shen, J., Bronson, R. T., Chen, D. F., Xia, W.,
Selkoe, D. J., and Tonegawa, S. (1997) Cell,89,
629-639.
9.Wong, P. C., Zheng, H., Chen, H., Becher, M. W.,
Sirinathsinghji, D. J., Trumbauer, M. E., Chen, H. Y., Price, D. L.,
van der Ploeg, L. H., and Sisodia, S. S. (1997)
Nature,387, 288-292.
10.De Strooper, B., Saftig, P., Craessaerts, K.,
Vanderstichele, H., Guhde, G., Annaert, W., von Figura, K., and van
Leuven, F. (1998) Nature, 391, 387-390.
11.De Strooper, B., Annaert, W., Cupers, P., Saftig,
P., Craessaerts, K., Mumm, J. S., Schroeter, E. H., Schrijvers, V.,
Wolfe, M. S., Ray, W. J., Goate, A., and Kopan, R. (1999)
Nature, 398, 518-522.
12.Struhl, G., and Greenwald, I. (1999)
Nature, 398, 522-525.
13.Ye, Y., Lukinova, N., and Fortini, M. E. (1999)
Nature, 398, 525-529.
14.Armogida, M., Petit, A., Vincent, B., Scarzello,
S., da Costa, C. A., and Checler, F. (2001) Nat. Cell Biol.,
3, 1030-1033.
15.Wolfe, M. S., Xia, W., Ostaszewski, B. L., Diehl,
T. S., Kimberly, W. T., and Selkoe, D. J. (1999) Nature,
398, 513-517.
16.Kimberly, W. T., Xia, W., Rahmati, T., Wolfe, M.
S., and Selkoe, D. J. (2000) J. Biol. Chem., 275,
3173-3178.
17.Steiner, H., Kostka, M., Romig, H., Basset, G.,
Pesold, B., Hardy, J., Capell, A., Meyn, L., Grim, M. L., Baumeister,
R., Fechteler, K., and Haass, C. (2000) Nat. Cell Biol.,
2, 848-851.
18.Tomita, T., Watabiki, T., Takikawa, R.,
Morohashi, Y., Takasugi, N., Kopan, R., de Strooper, B., and Iwatsubo,
T. (2001) J. Biol. Chem., 276, 33273-33281.
19.Laemmli, U. K. (1970) Nature,227,
680-685.
20.Li, X., and Greenwald, I. (1998) Proc. Natl.
Acad. Sci. USA, 95, 7109-7114.
21.Mahon, P., and Bateman, A. (2000) Protein
Sci., 9, 1930-1934.
22.LaPointe, C. F., and Taylor, R. K. (2000) J.
Biol. Chem., 275, 1502-1510.
23.Wolfe, M. S., de los Angeles, J., Miller, D. D.,
Xia, W., and Selkoe, D. J. (1999) Biochemistry, 38,
11223-11230.
24.Brown, M. S., Ye, J., Rawson, R. B., and
Goldstein, J. L. (2000) Cell, 100, 391-398.
25.Urban, S., Lee, J. R., and Freeman, M. (2001)
Cell, 107, 173-182.
26.McLendon, C., Xin, T., Ziani-Cherif, C., Murphy,
M. P., Findlay, K. A., Lewis, P. A., Pinnix, I., Sambamurti, K., Wang,
R., Fauq, A., and Golde, T. E. (2000) FASEB J., 14,
2383-2386.
27.Rogaev, E. I., Grigorenko, A. P., Ryazanskaya,
N., Sherbatich, T., Molyaka, Y., Korovaitseva, G. I., and
Dvoryanchikov, G. (2001) Human Genome Meeting. Program and Abstract
Book, Edinburgh, Scotland, April 19-22, ER No. 101.