Subtilisin Protein Inhibitor from Potato Tubers
T. A. Revina, A. S. Speranskaya, G. V. Kladnitskaya, A. B. Shevelev, and T. A. Valueva*
Bach Institute of Biochemistry, Russian Academy of Sciences, Leninskii pr. 33, Moscow 119071, Russia; fax: (7-095) 954-2732; E-mail: valueva@inbi.ras.ru* To whom correspondence should be addressed.
Received May 18, 2004
A protein with molecular weight of 21 kD denoted as PKSI has been isolated from potato tubers (Solanum tuberosum L., cv. Istrinskii). The isolation procedure includes precipitation with (NH4)2SO4, gel chromatography on Sephadex G-75, and ion-exchange chromatography on CM-Sepharose CL-6B. The protein effectively inhibits the activity of subtilisin Carlsberg (Ki = 1.67 ± 0.2 nM) by stoichiometric complexing with the enzyme at the molar ratio of 1 : 1. The inhibitor has no effect on trypsin, chymotrypsin, and the cysteine proteinase papain. The N-terminal sequence of the protein consists of 19 amino acid residues and is highly homologous to sequences of the known inhibitors from group C of the subfamily of potato Kunitz-type proteinase inhibitors (PKPIs-C). By cloning PCR products from the genomic DNA of potato, a gene denoted as PKPI-C2 was isolated and sequenced. The N-terminal sequence (residues from 15 to 33) of the protein encoded by the PKPI-C2 gene is identical to the N-terminal sequence (residues from 1 to 19) of the isolated protein PKSI. Thus, the inhibitor PKSI is very likely encoded by this gene.
KEY WORDS: Kunitz-type proteinase inhibitors, subtilisin, potato tubers
Abbreviations: Z-AAL-pNa) N-carbobenzoxy-L-alanyl-L-alanyl-L-leucine-p-nitroanilide; Suc-F-pNa) N-succinyl-L-phenylalanine-p-nitroanilide; BAPA) Nalpha-benzoyl-L-arginine-p-nitroanilide; Pth) phenylthiohydantoin.
Protein inhibitors of proteolytic enzymes are found in plants from
various systematic groups. High contents of inhibitors of proteinases
are found in many plants of the Solanaceae family [1], in particular, these inhibitors are from 20 to 50%
of all water-soluble proteins in potato tubes [2].
These inhibitors include proteins with molecular weights from 20 to
24 kD, which interact with serine [3-7], aspartic [7-9], and cysteine proteinases [7,
10, 11]. Many of these
proteinases have been characterized in detail and, based on their
primary structure, assigned to the superfamily of Kunitz-type soybean
inhibitors of trypsin [3-11].
Proteinase inhibitors from potato tubers were proposed to form a
separate subfamily denoted as potato Kunitz-type proteinase inhibitors
(PKPIs) [12]. Furthermore, proteins of the PKPIs
subfamily were divided into three structural groups denoted as PKPIs-A,
PKPIs-B, and PKPIs-C. Proteins of each group are characterized by the
N-terminal conservative amino acid sequence consisting of the first ten
residues [12, 13]. PKPIs are
especially interesting mainly because they seem to be involved in plant
protection against unfavorable effects of the environment. Thus, these
inhibitors are accumulated in potato leaves and tubers in response to
mechanical wounding [14, 15],
UV-radiation [16], and lesion by insects [15] and phytopathogenic microorganisms [11, 15, 17]. Many phytopathogenic fungi are known to produce
extracellular proteinases [18, 19]. The data available now suggest that these
proteinases should play an active role in development of diseases [20-22]. Thus, the cell-free
preparation from suspension of spores and bursting cysts of
Phytophthora infestans injected into potato leaves caused
necrosis of the plant tissue, and the proteolytic activity of the
preparation correlated with its necrotic effect [21]. Nonpathogenic mutants of the fungus
Pyrenopeziza brassicae (a leaf pathogen affecting plants of the
Cruciferae family) are cannot produce extracellular cysteine
proteinase. The recovery of pathogenicity in such mutants was
accompanied by recovery of their ability for producing the enzyme [22].
In response to aggression by proteinases, protein inhibitors are synthesized in plants that can suppress the activities of enzymes of phytopathogenic microorganisms [23]. This phenomenon was first recorded in tomatoes infected with P. infestans [24]. And the increase in contents of trypsin and chymotrypsin inhibitors correlated with the plant resistance to the pathogen [24]. Later the accumulation of protein inhibitors of serine proteinases with molecular weights of 20-24 kD was shown in potato tubers in response to mechanical wounding and infection with P. infestans [11, 17].
We have shown [19] that the oomycete P. infestans (Mont.) de Bary grown on medium containing thermostable proteins from potato tubers produces a complex of subtilisin-specific serine exoproteinases. Therefore, the finding in potato tubers of PKPIs specific for subtilisin is especially interesting.
The purpose of the present work was to isolate, characterize, and determine the amino acid sequence of the subtilisin inhibitor found in cultivar Istrinskii potato tubers.
MATERIALS AND METHODS
Potato tubers (Solanum tuberosum L., cv. Istrinskii) were used. The tubers were stored in the dark at 4°C for 2-4 months. DNA was isolated as described in [25] from young sprouts developed during two weeks on growth of the tubers in a wet chamber under conditions of natural illumination and room temperature. For cloning, Escherichia coli strain BMH 71-19 (mutS) from Promega (USA) and the vector pGEM-T-Easy (+) from Promega were used. Chromogenic substrates were as follows: N-carbobenzoxy-L-alanyl-L-alanyl-L-leucine-p-nitroanilide (Z-AAL-pNa), N-succinyl-L-phenylalanine-p-nitroanilide (Suc-F-pNa), and Nalpha-benzoyl-L-arginine-p-nitroanilide (BAPA) from Calbiochem (USA); alpha-chymotrypsin (EC 3.4.21.1) from Serva (Germany); trypsin (EC 3.4.21.4) from Spofa (Czechia) additionally recrystallized from MgSO4; subtilisin Carlsberg (EC 3.4.21.14) from Sigma (USA); papain (EC 3.4.22.1) from Calbiochem. Low-molecular-weight marker proteins for SDS-PAGE and marker proteins for HPLC were from Pharmacia (Sweden).
The sap from potato tubers was acidified with 5 N HCl to pH 5.0-5.5, and starch was removed by centrifugation at 3000g on a K-26 D centrifuge (Germany). The supernatant fluid was supplemented with dry (NH4)2SO4 to 75% saturation and left overnight at 4°C to produce the precipitate. The precipitate was separated by centrifugation as described, dissolved in a minimal volume of 0.05 M NH4HCO3, and applied onto a column (2.5 × 90 cm) with Sephadex G-75. The gel was equilibrated and the proteins were eluted also with 0.05 M NH4HCO3. Fractions containing proteins with molecular weight of about 20 kD were combined and lyophilized.
The preparation (35 mg) of proteins with molecular weight of about 20 kD was applied onto a column (0.5 × 15 cm) with CM-Sepharose CL-6B equilibrated with 0.02 M acetate buffer (pH 4.4). The column was washed with two volumes of the starting buffer and then with 0.2 M acetate buffer (pH 4.4) until the complete removal of a substance with absorption at 280 nm. The protein bound to the ion-exchanger was eluted with a linear gradient of NaCl (0-0.5 M) in 0.2 M acetate buffer (pH 4.4). Fractions containing the activity of subtilisin inhibitor were combined, desalted on a column with Sephadex G-25, and lyophilized.
Electrophoresis in the presence of SDS and beta-mercaptoethanol was performed in 20% polyacrylamide gel as described in [26]. The gels were stained with 0.1% Coomassie R-250 in 25% ethanol containing 5% formaldehyde.
Molecular weights of the inhibitor and its complexes with subtilisin were determined by HPLC on a Bio Sep-Sec-S-200 column (Beckman Coulter, USA). The gel was equilibrated and the proteins were eluted with 0.02 M KH2PO4 buffer (pH 7.0) containing 0.1 M NaCl.
The protein concentration was determined by the method described in [27] with BSA as the standard.
The proteins were sequenced on a 470A gas sequencer (Applied Biosystems, USA) by the standard program 02 RPTH [28]. Phenylhydantoin (Pth) derivatives of amino acids were identified by reverse-phase HPLC on a column with C18 sorbent (Applied Biosystems) in 20% acetonitrile. Pth derivatives of amino acids were detected with a Cratos model 757 flow absorbance monitor (USA).
Activity of the inhibitor was assessed by its effect on activities of the corresponding enzymes: subtilisin, chymotrypsin, trypsin, and papain. The activities of the enzymes were assessed by the rate of hydrolysis of casein [29] and chromogenic substrates Z-AAL-pNa (for subtilisin), Suc-F-pNa (for chymotrypsin), and BAPA (for trypsin and papain) [30, 31]. The concentration of active subtilisin was determined by titration with the subtilisin and chymotrypsin inhibitor from maize seeds (its concentration was predetermined by titration with chymotrypsin with known active site concentration). Concentration of the isolated inhibitor and the equilibrium dissociation constant (Ki) of its complex with subtilisin were determined upon incubation of the enzyme with increasing concentrations of the inhibitor. The apparent Ki value was calculated using the Enzfitter computer program [32].
Molecular cloning, isolation of plasmid DNA, and restriction analysis were performed as described in [33]. The primers 5´(GGGAGCTCAATCCAATTGTCCT(C/G)CC(C/A)(A/T)CAACT)3´ and 5´(GGGTCGACCTACGCCTTGATGAACACAAATGGAA)3´ denoted as PKPI-C10 and PKPI-C13, respectively, were constructed.
The polymerase chain reaction (PCR) was performed on a Tertsik MS-16 amplifier, model 2 (DNA-technology, Russia) with the genomic DNA and the primers PKPI-C10 and PKPI-C13, using DNA polymerase Taq (Bionem, Russia). The concentration of DNA-template in the reaction mixture was 0.01 µg/µl. The PCR was performed under the following conditions: 94°C for 1 min; then five cycles: 94°C for 30 sec, 56°C for 10 sec, 72°C for 10 sec; and 25 cycles: 94°C for 5 sec, 60°C for 5 sec, 72°C for 30 sec. The amplified DNA fragments were separated by electrophoresis in 1% agarose gel and isolated using a DNA extraction kit (Fermentas, Lithuania).
The amplified DNA fragments flanked with the primers PKPI-C10 and PKPI-C13 were cloned according to the producer's instructions into a vector from a pGem-T-Easy kit for cloning of amplifiers. The cloned fragments were sequenced using standard primers pUC/M13 (Promega).
RESULTS AND DISCUSSION
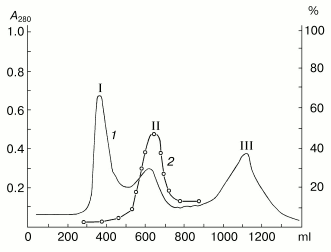
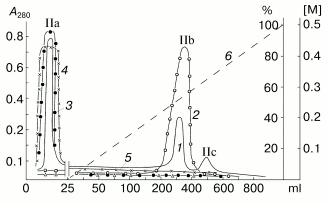
Results of separation on Sephadex G-75 of the proteins precipitated from the potato sap with (NH4)2SO4 at 75% saturation are shown in Fig. 1. These proteins are represented by at least three fractions with molecular weights of about 40, 20, and 5 kD, which is in a good correlation with our previous data [34]. The inhibitor activity to subtilisin was detected only in fraction II containing proteins with the molecular weight of about 20 kD. Then the proteins of fraction II were separated by ion-exchange chromatography on CM-Sepharose CL-6B at pH 4.4 (Fig. 2). Fraction II was separated into at least three protein components, and only the component IIb had the activity of the subtilisin inhibitor. Note, that the protein component IIb failed to suppress activities of trypsin, chymotrypsin, and papain.
Fig. 1. Gel chromatography of proteins from potato tubers on Sephadex G-75. I, II, III are protein components with molecular weights of about 40, 20, and 5 kD, respectively. 1) A280; 2) activity (%) of the subtilisin inhibitor.
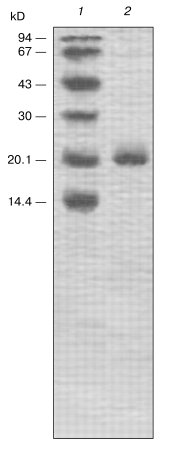
SDS-PAGE in the presence of a reductant (Fig. 3) showed that the chromatographic fraction IIb contained a 21-kD homogenous protein and was not contaminated with proteins with molecular weights of 20-24 kD. Thus, the molecule of the isolated protein denoted as the potato Kunitz-type subtilisin inhibitor (PKSI) was concluded to consist of a single polypeptide chain.Fig. 2. Ion-exchange chromatography on CM-Sepharose CL-6B of component II prepared by gel chromatography. IIa-IIc are protein components. 1) A280; 2-5) activities (%) of inhibitors of subtilisin, chymotrypsin, trypsin, and papain, respectively; 6) NaCl (M).
The homogeneity of the protein PKSI was confirmed by determination of its N-terminal amino acid sequence. The first 19 amino acid residues were identified by 19 stages of automated degradation according to Edman (Fig. 4). Figure 4 allows us to compare the N-terminal sequences of the protein PKSI and some proteins isolated earlier from potato tubers which were assigned to group C of the PKPI subfamily [3, 4, 10, 35, 36]. The identity of the first ten amino acid residues in the N-terminal sequences of these proteins is evident. These conservative residues are specific for members of the PKPI subfamily of structural group PKPI-C [12, 13]. Thus, it is reasonable to assign the protein PKSI to group C of this subfamily of inhibitors. Moreover, the N-terminal sequences of the protein PKSI and the inhibitor of cysteine proteinases (PCPI) [10] are quite identical but differ from the sequence of the subtilisin inhibitor (PKI-2) [3], trypsin inhibitor (PTI) [35], inhibitor of serine proteinases (PI-22) [4], and inhibitor of serine proteinases (p34021) [36] by substitutions in position 13: Asn -->_Lys, His, and Gln (PKI-2, PTI, and PI-22, respectively), and in position 17: Gly -->_Val (p34021).Fig. 3. SDS-PAGE of component IIb (2) prepared by ion-exchange chromatography. 1) Marker proteins (from top down): phosphorylase b, BSA, ovalbumin, carboanhydrase, the Kunitz-type soybean inhibitor, lactalbumin; molecular weights of marker proteins in kD are indicated on the left.
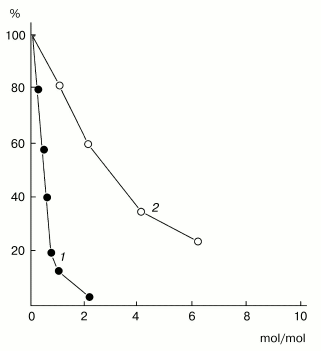
Figure 5 shows the effect of the protein PKSI on the activity of subtilisin Carlsberg. PKSI effectively inhibited the activity of the enzyme, and the inhibition degree depended on the substrate used for determination of the residual enzymatic activity. The activity of subtilisin was assessed by the rate of hydrolysis of a peptide substrate, Z-AAL-pNa, and the suppression of the enzymatic activity depended linearly on the amount of the inhibitor added, virtually up to 90% inhibition (Fig. 5, curve 1). By calculations, 1 mol of inhibitor reacted stoichiometrically with 1 mol of subtilisin. The equilibrium dissociation constant (Ki) was calculated from data on suppression of activity of the enzyme (14 nM) with varied amounts of the inhibitor during hydrolysis of 0.03 mM Z-AAL-pNa. The calculations gave Ki = 1.67 ± 0.2 nM.Fig. 4. Comparison of the N-terminal amino acid sequences of the protein PKSI and other proteins of the PKPI subfamily: the different residues are hatched. Inhibitors from potato tubers: 1) PKSI; 2) inhibitor of subtilisin, PKI-2 [3]; 3) inhibitor of trypsin, PTI [35]; inhibitor of serine proteinases, PI-22 [4]; 5) protein inhibitor of serine proteinases, p34021 [36]; 6) inhibitor of cysteine proteinases, PCPI-8.3 [10].
However, when the enzyme activity was assessed by the rate of hydrolysis of a protein substrate, casein, the inhibition depended non-stoichiometrically on the amount of the inhibitor, and more than a threefold excess of the inhibitor was required to provide 50% inhibition (Fig. 5, curve 2). The non-stoichiometric interaction in this case seemed to be a result of dissociation of the enzyme-inhibitor complex in the presence of the protein substrate [37]. A similar phenomenon was earlier recorded for the inhibitor of trypsin which was isolated by us from the same potato cultivar [34] and also for a number of inhibitors of serine proteinases from the superfamily of Kunitz-type soybean inhibitor of trypsin: inhibitors from Psophocarpus tetragonolobus L., Gleditchia triacanthos L., and Acacia elata L. [38-40].Fig. 5. Effect of the protein from potato tubers on the activity of subtilisin Carlsberg: 1, 2) enzyme activity was determined with Z-AAL-pNa and casein as substrates, respectively.
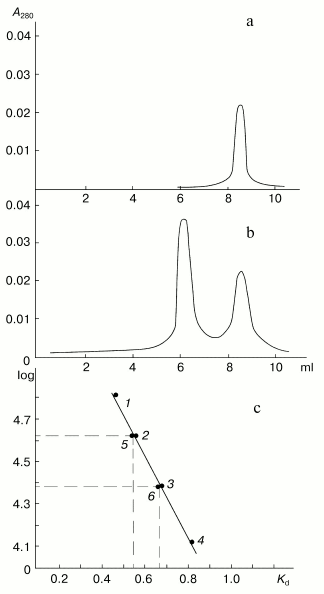
Figure 6 shows results of HPLC of the inhibitor and the mixture of subtilisin with excess inhibitor PKSI. The inhibitor was eluted as a single homogenous protein component (Fig. 6a). Its calculated molecular weight of 21 ± 1 kD corresponded to the value determined by SDS-PAGE. The coincidence of the molecular weight values obtained by two different approaches supports the earlier hypothesis that the protein molecule should consist of a single polypeptide chain. The mixture of the enzyme and PKSI also contained a component with higher molecular weight (Fig. 6b). The molecular weight of this component was 43 ± 1 kD, i.e., it was virtually equal to the sum of the molecular weights of PKSI and subtilisin (Fig. 6c). This result also confirms the above conclusion that the inhibitor complexes with subtilisin at the ratio of 1 mol per 1 mol.
Thus, isolated protein PKSI specifically inhibited subtilisin and formed with the enzyme a stoichiometric complex (1 mol/mol), which was stable under conditions of HPLC. However, this complex could dissociate in the presence of a protein substrate.Fig. 6. HPLC of the protein PKSI and its complex with subtilisin: a) the free protein (0.5 mg); b) mixture containing the protein (1.0 mg) and subtilisin (0.5 mg); c) dependence of log of molecular weight of proteins on their elution from the Bio Sep-Sec-S-200 column; 1-4) marker proteins: BSA, ovalbumin, Kunitz-type soybean inhibitor, RNase, respectively; 5) complex of protein PKSI with subtilisin; 6) protein PKSI.
As mentioned, protein PKSI was assigned to structural group PKPI-C. Gene sequences of this structural group of proteins present in the GenBank database (National Center of Biotechnological Information (NCBI), USA) were analyzed, and primers PKPI-C10 and PKPI-C13 were elaborated and constructed (see “Materials and Methods”). The PCR performed with these primers on the template of the genomic DNA isolated from potato sprouts resulted in a fragment of expected size containing 600 n.p. Cloning of this fragment resulted in a sufficient number of plasmid clones, most of which were sequenced. The recovered amino acid sequence of the protein encoded by the gene denoted as PKPI-C2 is shown in Fig. 7. The N-terminal sequence (residues from 15 to 33) of the protein encoded by the gene PKPI-C2 was identical to the N-terminal sequence (residues from 1 to 19) of protein PKSI isolated by us. This strongly suggests that the inhibitor under study is encoded by this gene and has the amino acid sequence presented in Fig. 7. To compare amino acid sequences of the protein encoded by PKPI-C2 and other known proteins from the subfamily PKPI-C, a search was performed in the GenBank database (NCBI). As a result, a great similarity was found between the sequence of the protein encoded by the gene PKPI-C2 and sequences of four known proteins from group PKPI-C. Note that amino acid sequences of proteins PCPI-8, PCPI-10, and p34021, as well as the amino acid sequence of the protein encoded by gene PKPI-C2 were recovered by nucleotide sequences of genes encoding these proteins [13, 36]. And the sequence of the protein PCPI-8.3 isolated from potato tubers was established by approaches of classical protein chemistry [10].
PKPIs are synthesized in the plant cell as precursors and then located in vacuoles [12, 13]. The N-terminal part of the precursor is supposed to be a transmembrane key segment, which includes the endoplasmic signal sequence containing the conservative motif Asn-Pro-Ile-Val-Leu-Pro encoding the vacuole-sorting signal [12, 41]. This signal peptide together with seven amino acid residues is detached posttranslationally at the site Asn14-Leu15, after the protein has been transferred into the vacuole [42]. Thus, the first 19 amino acid residues of the isolated protein PKSI (Fig. 4) were fully coincidental with the N-terminal sequence of the mature inhibitor encoded by the gene PKPI-C2.Fig. 7. Amino acid sequences of the protein encoded by PKPI-C2 (1) compared to those of other proteins of the PKPI group C: 2, 3) inhibitors of cysteine proteinases, PCPI-8 and PCPI-10, respectively [13]; 4) inhibitor of cysteine proteinases, PCPI-8.3 [10]; 5) protein inhibitor of serine proteinases, p34021 [36]. The distinguishable residues are hatched, the sequence of the signal peptide with the vacuolar-sorting information is delimited, and v shows the region of posttranslational modification of the precursor after the protein is transferred into the vacuole; * and ** indicate disulfide bonds.
Primary structures of the protein encoded by PKPI-C2 and of the protein PCPI-10 are different only by residues Ser7-->Thr7, and of the protein PCPI-8 by residues Ser7-->Thr7 and Asn169-->Thr169. However, the differences between amino acid sequences of the protein encoded by PKPI-C2 and the proteins PCPI-8.3 and p34021 are 10 and 20%, respectively (corresponding to 17 and 38 different residues, respectively) [10, 36]. This confirms the hypothesis that proteins of the C group are encoded by different allelic genes of the same locus. However, the protein PSPI isolated by us was different from these inhibitors in the specificity of its effect on enzymes. Thus, unlikely the protein p34021 acting a number of serine proteinases [36], protein PSPI specifically inhibited subtilisin. The lack of effect on cysteine proteinases also suggested the difference between PSPI and proteins PCPI-8, PCPI-10, and PCPI-8.3 interacting with these enzymes [10, 35]. Note, that proteins with molecular weight of about 20 kD failed to suppress the activity of papain (Fig. 2). This indicated that cv. Istrinskii potato tubers lacked cysteine proteinase inhibitors of the PKPI-type. Further studies on features of the protein produced by expression of the PKPI-C2 gene are expected to show the degree of identity of this protein with protein PSPI.
This work was supported by the Russian Foundation for Basic Research (project Nos. 04-04-48644 and 04-04-49694).
REFERENCES
1.Mosolov, V. V., and Valueva, T. A. (1993) Plant
Protein Inhibitors of Proteolytic Enzymes [in Russian], VINITI,
Moscow.
2.Pouvreau, L., Gruppen, H., Piersma, S. R., van den
Brock, L. A. M., van Koningsveld, G. M., and Voragen, A. G. J. (2001)
J. Agric. Food Chem., 49, 2864-2874.
3.Walsh, T. A., and Twitchell, W. P. (1991) Plant
Physiol., 97, 15-18.
4.Suh, S.-G., Stiekema, W. J., and Hannapel, D. J.
(1991) Planta, 184, 423-430.
5.Valueva, T. A., Revina, T. A., and Mosolov, V. V.
(1997) Biochemistry (Moscow), 62, 1367-1374.
6.Valueva, T. A., Revina, T. A., Mosolov, V. V., and
Mentele, R. (2000) Biol. Chem. Hoppe-Seyler, 381,
1215-1221.
7.Pouvreau, L., Gruppen, H., van Koningsveld, G. A.,
van den Brock, L. A. M., and Voragen, A. G. J. (2003) J. Agric. Food
Chem., 51, 5001-5005.
8.Mares, M., Meloun, B., Pavlik, M., Kostka, V., and
Baudys, M. (1989) FEBS Lett., 251, 94-98.
9.Srukelj, B., Pungercar, J., Mesko, P.,
Barlic-Magana, D., Gubensek, F., Kregar, J., and Turk, V. (1992)
Biol. Chem. Hoppe-Seyler, 373, 477-482.
10.10. Krizaj, M., Drobnic-Kosorok, J.,
Brzin, J., Jerata, R., and Turk, V. (1993) FEBS Lett.,
333, 15-20.
11.Valueva, T. A., Revina, T. A., Kladnitskaya, G.
V., and Mosolov, V. V. (1998) FEBS Lett., 426,
131-134.
12.Ishikawa, A., Ohta, S., Matsuoka, K., Hattori,
T., and Nakamura, K. (1994) Plant Cell Physiol., 35,
303-312.
13.Gruden, K., Strukelj, B., Ravnikar, M.,
Poljsak-Prijatelj, M., Mavric, I., Brzin, J., Pungercar, J., and
Kregar, I. (1997) Plant Mol. Biol., 34, 317-323.
14.Valueva, T. A., Revina, T. A., Gvozdeva, E. L.,
Gerasimova, N. G., Il'inskaya, L. I., and Ozeretskovskaya, O. L. (2001)
Prikl. Biokhim. Mikrobiol., 37, 601-606.
15.Bergey, D. R., Howe, G. A., and Ryan, C. A.
(1996) Proc. Natl. Acad. Sci. USA, 93, 12053-12058.
16.Conconi, A., Smerdson, M. J., Howe, G. A., and
Ryan, C. A. (1996) Nature, 383, 826-829.
17.Valueva, T. A., Revina, T. A., Gvozdeva, E. L.,
Gerasimova, N. G., and Ozeretskovskaya, O. L. (2003) Bioorg.
Khim., 29, 499-504.
18.Valueva, T. A., and Mosolov, V. V. (2002) Usp.
Biol. Khim., 42, 193-216.
19.Gvozdeva, E. L., Ievleva, E. V., Gerasimova, N.
G., Ozeretskovskaya, O. L., and Valueva, T. A. (2004) Prikl.
Biokhim. Mikrobiol., 40, 194-200.
20.Movahedy, S., and Heale, J. (1990) Physiol.
Mol. Plant Pathol., 37, 303-324.
21.Ball, A. M., Ashby, A. M., Daniels, M. J.,
Ingram, D. S., and Johnstone, K. (1991) Physiol. Mol. Plant
Pathol., 38, 147-161.
22.Paris, R., and Lamattina, L. (1999) Eur. J.
Plant Pathol., 105, 753-760.
23.Ryan, C. A. (1990) Annu. Rev.
Phytopathol., 28, 425-449.
24.Peng, J. H., and Black, L. L. (1976)
Phytopathol., 66, 958-963.
25.Murakami, K., Hattori, T., and Nakamura, K.
(1986) Plant Mol. Biol., 7, 343-355.
26.Laemmli, U. K. (1970) Nature, 227,
680-685.
27.Appenroth, K. J., and Angsten, H. (1987)
Biochem. Physiol. Pflanzen, 182, 85-89.
28.Levina, N. B., Slepak, V. Z., Kiseleva, O. G.,
Shemyakin, V. V., and Khokhlachev, A. A. (1989) Bioorg. Khim.,
15, 24-31.
29.Northrop, J. H., Kunitz, M., and Herriott, R.
(1950) in Crystalline Enzymes [Russian translation], IL Press,
Moscow.
30.Lyublinskaya, L. A., Yakusheva, L. D., and
Stepanov, V. M. (1977) Bioorg. Khim., 3, 273-279.
31.Kakade, M. L., Simons, N., and Liener, I. E.
(1969) Cereal Chem., 46, 518-526.
32.Knight, C. G. (1986) in Proteinase
Inhibitors (Barrett, A. J., and Salvesen, G. R., eds.) Elsevier,
Amsterdam, pp. 23-51.
33.Speranskaya, A. S., Dorokhin, A. V., Novikova, S.
I., Shevelev, A. B., and Valueva, T. A. (2003) Genetika,
39, 1490-1497.
34.Revina, T. A., Valueva, T. A., Ermolova, N. V.,
Kladnitskaya, G. V., and Mosolov, V. V. (1995) Biochemistry
(Moscow), 60, 1411-1416.
35.Yamagishi, K., Mitsumori, C., and Kikuta, Y.
(1991) Plant Mol. Biol., 17, 287-288.
36.Stiekema, W. J., Heidekamp, F., Dirkse, W. G.,
Bekum, J. V., Haan, P. D., Bosch, C. T., and Louserse, J. D. (1988)
Plant Mol. Biol., 11, 255-269.
37.Bieth, J. (1974) in Proteinase Inhibitors
(Fritz, H., Tschesche, H., Greene, L. J., and Truscheit, E., eds.)
Springer-Verlag, Berlin, pp. 463-469.
38.Kortt, A. A. (1980) Biochim. Biophys.
Acta, 624, 371-382.
39.Mosolov, V. V., Valueva, T. A., and Kolosova, G.
V. (1982) Biokhimiya, 47, 2015-2021.
40.Kortt, A. A., and Jermyn, M. A. (1981) Eur. J.
Biochem., 115, 551-557.
41.Matsouka, K., Matsumoto, S., Hattori, T.,
Machida, Y., and Nakamura, K. (1990) J. Biol. Chem., 265,
19750-19757.
42.Matsouka, K., and Nakamura, K. (1991) Proc.
Natl. Acad. Sci. USA, 88, 834-838.