REVIEW: Bacterial and Archaeal Flagella as Prokaryotic Motility Organelles
A. L. Metlina
Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow 119992, Russia; fax: (7-095) 939-3181; E-mail: metlina@belozersky.msu.ru
Received July 6, 2004
The properties and molecular organization of flagella--the bacterial and archaeal motility organelles--are reviewed. The organization of these functional motility elements of prokaryotic organisms belonging to different kingdoms is compared. A mechanism for both in vivo and in vitro assembly of bacterial flagellum filaments (BFFs) is discussed, and similarity is supposed between flagellin and actin with regard to their polymeric forms (BFF and F-actin). Our own data on intracellular fixation of the Halobacterium salinarium flagellum are presented. Comparative characteristics of intracellular fixation of bacterial and archaeal flagella are also described.
KEY WORDS: bacterial flagellum, archaeal flagellum, flagellin, biological motility
Abbreviations: BFF) bacterial flagellum filament; AFF) archaeal flagellum filament; TNM-flagellin) tetranitromethane-modified flagellin; S1) myosin subfragment 1; PEG) polyethylene glycol.
The locomotion of procaryotic cells, i.e., eubacteria and archaea, is
provided by a special molecular mechanism distinct from that known in
all eucaryotic cells; the latter is based on the interaction of at
least two proteins and on the energy of nucleoside triphosphate
hydrolysis. The ability of bacterial and archaeal cells for rapid and
directed movement in a medium depends on the presence of the flagellum,
the special motility organelle, whose structural and functional
proteins are encoded by ~50 genes in bacteria [1,
2]. In archaea, only the genes encoding flagellins
are known [3, 4]. Studies of
the molecular organization and properties of the bacterial flagellum as
a motility organelle began long before the studies on the
archaebacterial motility system. The bacterial flagellum proved to be a
discrete system composed of three main components: basal body
localized within the cell wall, prolonged external spiral
protein filament composed of flagellin subunits, and flexible
joint, so-called “hook”, connecting these two parts
[5, 6]. Transmembrane
electrochemical proton gradient was found in 1970s as the energy source
driving the bacterial flagellum [7-10]. In some marine bacteria, sodium ions fulfill
this role of protons [11-14].
The structure embedded in the bacterial cell wall and known as a proton
driven machine powers external semi-rigid helical flagellin filament
and generates the hydrodynamic force that directionally pushes the cell
in the medium.
Once this fundamental information had been obtained, the fact becomes obvious that bacterial locomotion is fundamentally different from other biological motility systems; however, the structure, protein composition, and functioning of this extraordinary propulsion aggregate remained unknown for a long time.
Boris Fedorovich Poglazov, to whose commemoration this issue of Biochemistry (Moscow) is devoted, was the first Russian scientist interested in the structure and properties of motility organelles in bacteria. Poglazov's and my studies (1969-1974) were the pioneer investigations in this field. First, they were devoted to the properties of flagellin, the protein of the external helical filaments of the bacterial flagellum (BFF), including its self-association. A series of B. F. Poglazov's studies was devoted to the problem of self-association in biological systems [15-18], wherein he proposed a conception of spontaneous and ordered molecular aggregation resulting in formation of biological structures. In modern science, the problem of biopolymer (including bacterial flagellum) formation has been elevated to a higher level after the revealing of molecular chaperones, special proteins facilitating assembly of protein-containing structures. It is this problem that is highlighted in one of last publications of B. F. Poglazov, who gave an explanatory view of building of large supramolecular systems involving chaperons [19]. Our results on BFF assembly are to be considered in frames of earlier works on self-association of protein structures [20, 21], in which the assembly of supramolecular protein complexes is regarded as self-association, a spontaneous process without participation of any other macromolecules or exogenous energy sources. Recently, the role of chaperons in assembly of the bacterial flagellum has come under scrutiny since its distinct intracellular protein components, such as FliJ or FliI, were found to be possible chaperons participating both in export and assembly of structural components of the bacterial flagellum [22-24]. Possible involvement of chaperons in the archaeal flagellum assembly was hypothesized in the work of some Russian investigators [25].
In the initial steps of our work in the 1960s, the phenomenon of “crowding” was not taken into account in the studies on BFF self-association in vitro [26, 27]. In the living cell, various macromolecules can form zones of high local concentration, and this part of cell volume may prove to be inaccessible for other molecules, thus implying serious energetic consequences for assembly of other biopolymers. Well in advance of active discussion of biological macromolecule functioning under crowding, we developed an experimental model using polyethylene glycol (PEG) for the in vitro study of BFF self-association [28-31]. This study is to be considered as one of the first attempts to investigate the process of supramolecular structure assembly under crowding conditions imitating those observed in the living cell.
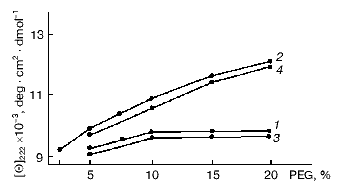
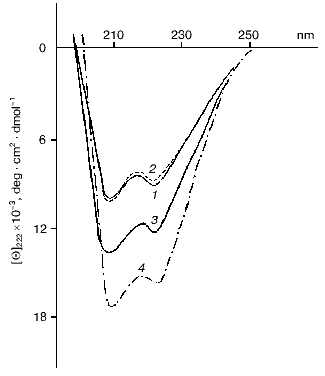
The fraction of alpha-helices in the flagellin molecule increases from ~30% (monomer) to ~50% (polymer) in the course of BFF self-assembly [28, 32-35]. Japanese authors proposed a two-step mechanism of BFF association: in the first stage partial alteration of flagellin conformation occurs allowing incorporation of flagellin molecules into the polymer at the following stage [36, 37]. Since no direct experimental confirmation of the proposed scheme was provided, we attempted to develop a model of flagellin polymerization to demonstrate real conformational change in flagellin subunit preceding the formation of its polymeric structure. With this aim, we combined inhibition of the polymerization process with conditions shifting equilibrium of the flagellin-BFF system to polymer (BFF) formation [29-31]. To inhibit flagellin polymerization we used the protein at concentration substantially lower than the threshold (10-20 µg/ml) [35] and flagellin whose tyrosine residues were modified by tetranitromethane (TNM-flagellin) [31, 38]. The flagellin modified in this way retains properties of the intact protein but completely loses its polymerizing ability. At low concentrations of intact flagellin polymerization is not completely disabled, whereas the use of TNM-flagellin excludes the possibility of conformational fixation of the protein in its polymeric form. The agents used for shifting the system equilibrium to polymer formation were PEG, which was first used for BFF and ensures virtually instant polymerization of flagellin [29], and ammonium sulfate, which accelerates BFF assembly but with weaker effect [39]. Under ordinary conditions, it takes about 18-20 h for flagellin polymerization over a wide pH range (3.5-9.5). The conformation of Bacillus brevis var. G.-B.p+ flagellum flagellin was estimated from CD spectra in the range from 260 to 190 nm, as well as from change in molar ellipticity [Theta]222 [40]. Figure 1 demonstrates the ability of BFF monomers to change their conformation before incorporation into the polymer. A similar pattern was discovered when flagellin was polymerized in the presence of ammonium sulfate. Note that salting out substances, which shift the equilibrium to polymer formation, do not elevate the [Theta]222 value to the values corresponding to flagellin incorporated into the polymer (both intact and reconstituted BFF) (Fig. 2). So, only partial elevation of the alpha-helix fraction is detected, but the fraction as alpha-helices in the flagellin incorporated into flagella is not achieved. Gradual elevation in either PEG or ammonium sulfate concentration in the pH range of 4.0-10.0 results in the same gradual elevation of [Theta]222 (Fig. 1). Knowing the length of BFF, one can determine the velocity of their growth in the presence of PEG. Simple calculations give the BFF elongation rate of 600 ± 40 nm/min (these data are for Bacillus subtilis 168 BFF) in the presence of 10% PEG2000 at the protein concentration of 2.5 mg/ml. According to optical diffraction data the flagellum 10 µm in length is composed of 22,000 flagellin molecules, i.e., 22 flagellin molecules per second are incorporated into B. subtilis 168 flagellum in 10% PEG solution. This elongation rate is eightfold higher than that in a thin layer of solution, which was determined by dark-field microscopy [41]. At so high growth rate, the flagellum 10 µm in length can be reconstituted in 15 min, which is in agreement with the flagellar growth rate on the B. subtilis 168 cells [42]. It follows from the above that PEG allows flagellin polymerization rate in vitro comparable to that of BFF on the surface of a living cell.
Fig. 1. Effect of PEG on molar ellipticity [Theta]222 of flagellin at pH 3.0 (1) and 6.0 (2) and TNM-flagellin at pH 3.0 (3) and 6.0 (4).
A possible cause of changes in flagellin conformation in the presence of agents accelerating polymerization is a change in the hydration shell of the protein. As a polymer, PEG possesses, on one hand, exclusive properties providing microscopic aggregation of proteins in solution [43] and, on the other hand, the ability to cause intramolecular fluctuations resulting in water structure disturbances which must prominently destabilize the protein hydration shell. Since the effect of one protein on the conformation of another protein is realized through changes in water structure, PEG may be regarded as an agent influencing the intracellular environment of flagellin molecules.Fig. 2. CD spectra of flagellin (1) and TNM-flagellin (2) of Bacillus brevis var. G.-B.p+ flagellum fibrils (pH 6.0) in the absence (1, 2) and presence of either 20% PEG or 0.8 M ammonium sulfate (3); 4) short fragments of intact flagellum fibrils.
The collision of flagellin with growing flagellum tips has been previously shown to result in disturbance of its hydration shell and consequent conformational change [44]. Our results have confirmed this fact by demonstrating the possibility of conformational change in flagellin subunits in the presence of salting out agent before their incorporation into the growing flagellum tips. Since flagellin polymerizes in a remarkable manner at the distant tip of the bacterial flagellum [45, 46], the high growth rate of these flagella in vivo is apparently determined by appropriate conformation, which flagellin acquires on its way along the inner channel in tight surrounding of protein molecules inside the BFF. PEG provides similar conditions for the bacterial flagellum flagellin in vitro.
The results of our studies in the 1970s and considerations put forward about possible mechanism of flagellin polymerization [31, 38] are in good agreement with recent data on BFF association in vivo. The BFF elongation is known to be unusual since flagellin subunits are attached to the growing flagellum not at its end proximal to the cell surface, but at the distal end fixed by the capping element HAP2 (hook-associated protein 2) or “cap” (FliD) [47, 48]. It turns out that the “cap” not only prevents the “leakage” of flagellin molecules from the distal end of BFF as considered earlier [48], but also induces necessary conformational changes in flagellin subunits on their polymerization. In particular, the 3-D reconstitution based on electron cryomicroscopy imaging has demonstrated that the “cap” localized at the distal tip of growing flagellin filament builds-in the flagellin molecules into growing filament and fits them to the required conformation [49]. The “cap” is a pentameric flat discoid structure, which comes into contact with flagellin monomers arriving through the inner channel and facilitates their conformational change that is necessary for their incorporation into the polymer. In our model system, PEG apparently facilitates the transition of flagellin monomers to the conformation resembling that of flagellin molecules passing the inner channel of the flagellum to interact with “cap” at its distal tip. The subsequent events going on in the living cell cannot be modeled in vitro. That is, the multistage process of filament assembly from the flagellin molecules existing in some intermediate conformation is facilitated by the “cap” [49]. The incorporation of these conformationally adapted flagellin molecules into the polymer is likely the second polymerization step, which was proposed earlier [36, 37].
Our model for bacterial flagellin polymerization has been indirectly supported in a recent study in which X-ray diffraction on a crystal of virtually pure flagellin (such crystals were prepared for the first time) confirmed and elaborated many earlier results concerning structural transitions in BFF [50]. Among other results it was shown that tyrosine stabilizes the folding of three flagellin domains in the R-type BFF conformation because the molecular volume of tyrosine is considerably greater than that of aspartate, which constitutes the major portion of the flagellin molecule. We have modified tyrosine residues of flagellin and used the modified flagellin in studies. It is possible that our modification of tyrosine residues of flagellin subunits in flagellum filaments of B. brevis var. G.-B.p+ affects the site stabilizing the polymer.
In 1971 so-called “intact bacterial flagella” consisting of BFF, the external part of the flagellum, and its intracellular part (the basal body), joined by the flexible pin-joint (“hook”) was first isolated from Escherichia coli and B. subtilis cells [5]. This bacterial structure survives conditions and biochemical treatment during its isolation from the cell, but it is not the entire functional organelle providing bacterial locomotion. The second important benchmark in the study of bacterial motility was achieved in the mid 1970s, when a series of unsuccessful attempts to find ATPase in bacterial flagella led to the discovery of the true energy source for the bacterial flagellum rotation, namely the proton (or sodium ion) transmembrane gradient [7-10]. Thus, bacterial flagella were quasi outstanding from the common view on the structure and energy supply of biological motility systems. However, close inspection of various motility systems reveals some similarity. They all contain helical biopolymers (F-actin, microtubules, or BFF) composed of large numbers of equivalent monomers. The literature shows the possibility of interaction between motility proteins. In particular, an interaction between myosin and tubulin was discovered [51]. BFF vaguely resembles F-actin, which is present in apparently in all cell types. We have raised the issue of the place of bacterial flagella among other motility systems and obtained interesting results.
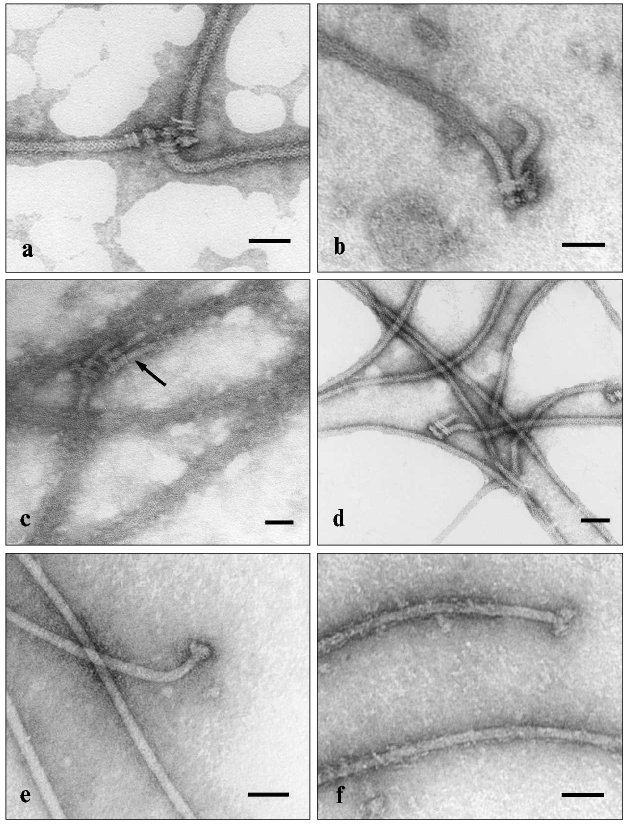
One of techniques for identification of actin is the decoration of F-actin by water-soluble myosin fragments, such as heavy meromyosin and myosin subfragment 1 (S1), followed by electron microscopy of the structures formed. The method is based on the specific interaction between actin and myosin. The subfragment S1 represents isolated myosin heads with preserved active sites but lacks the myosin rods. Using S1, we have revealed the ability of BFF to bind S1 [52-54] (Fig. 3). The electron microscopic images clearly show that flagellin filaments of E. coli and B. brevis flagella bind S1, which becomes evenly distributed along the flagellum filament, thus enlarging its diameter. This is an intrinsic property of BFF flagellin, since neither the hook nor basal body becomes decorated. The specificity of the found effect is supported by rapid dissociation of the complex when 1 mM ATP is added (in the presence of 5 mM MgCl2) (Fig. 3d). The effect is characteristic for the interaction of actin with S1; therefore, the influence of ATP on the BFF-S1 complex virtually excludes the possibility of nonspecific adhesion of S1 on BFF.
The ability to bind S1 appears to be a common property of BFF in both gram-positive and gram-negative bacteria, since BFF of E. coli, B. brevis, and B. subtilis were subjected to decoration, although to different extents. S1 decorates not only BFF as a component of “intact flagellum”, but also mechanically detached BFF from the cell and BFF attached to the whole bacterial cell. Moreover, both decorated and non-decorated BFFs, as well as decorated and non-decorated sites on the same BFF can be found in the same sample. This can result from impaired structure of BFF due to some impacts (physical and chemical factors, proteolysis, etc.). However, the most attractive supposition is that the difference in decoration of flagella with S1 reflects a series of polymorphic states of BFF and corresponding changes in flagellin conformation, whose appearance controls the bacterial cell reorientation in the surrounding medium. A detailed description of our experimental data on ability of bacterial flagella to specifically bind the S1 protein of skeletal muscle myosin lies outside this short review and can be found elsewhere [52-54]. I shall only enumerate the data obtained. We have found that BFF co-precipitates with myosin, inhibits both its Ca2+- and K+-EDTA-ATPase activities, and competes with F-actin for binding sites of myosin. It is notable that flagellin only expresses its ability to interact with myosin when associated in the polymeric structure in the composition of supramolecular polymeric BFF, whereas monomeric flagellin does not express this ability. A similar feature is characteristic of actin [55, 56]. Apparently, only the polymeric form of actin (F-actin) can specifically interact with myosin. Moreover, the myosin-binding site of actin is formed by two adjacent actin monomers [55]. The same model can be proposed for the interaction of BFF flagellin with myosin. It is worth noting that myosin can only interact with BFF, but not with the hook, another protein structure of the bacterial flagellum. So, the nature of the protein rather than its polymeric state is essential in this interaction.Fig. 3. Electron microscopic images of intact flagella of E. coli MS 1350 (a-d) and B. brevis var. G.-B.p+ (e, f) in the absence (a, e) or presence of myosin subfragment 1 (b, c, f); d) flagella in the presence of S1 after addition of 1 mM Mg2+-ATP. The scale length is 50 nm. Arrows indicate the “hooks” which are not decorated by S1.
Unlike F-actin, BFF binds myosin slowly. Moreover, F-actin, when bound to myosin, completely deprives BFF of this possibility. These data provide evidence for low affinity of BFF compared with F-actin to myosin. On the other hand, the data on competition of BFF and F-actin for binding sites of myosin are suggestive of adjacent, if not identical, BFF-binding and actin-binding sites localized in myosin heads. The N-termini of B. subtilis 168 flagellin [57] and rabbit skeletal muscle actin [58] have similar sequences of 18 amino acid residues [53]. These sites of homology are in positions 11 through 28 of actin and 28 through 45 of flagellin. They are enriched with amino acids destabilizing alpha-helix and possibly are stretched fragments of primary sequences of these proteins. Naturally, certain resemblance of these protein fragments is not sufficient for far-reaching conclusions, but it allows some explanation for the ability of BFF to interact with myosin. Unfortunately, these preliminary data, which were obtained more than two decades ago, remain without a progress on the basis of modern approaches; nevertheless, they deserve attention and discussion and require further research.
Myosin is localized inside the cell, whereas BFF is outside it, so it seems unlikely that the interaction between myosin and BFF has any physiological implication. It is probably a result of some structural resemblance between actin and flagellin or common characteristics of their polymeric forms. What is the reason for the similarity between actin and flagellin, such evolutionarily diverse proteins? Until quite recently it was thought that cytoskeletons formed by actin and tubulin and characteristic of eukaryotic cells are absent in bacteria. Yet, proteins homologous to actin (MreB and ParM [59-62]) and tubulin (FtsZ protein [63-65]) were recently found in bacteria. The novel data on bacterial cytoskeleton proteins are encouraging for further studies on bacterial motility mechanisms involving these intracellular proteins. These data on existence of actin- and tubulin-like proteins in bacterial cells have arisen from sophisticated methodology and are essential for understanding of our own results. Previous reports on bacterial actin-like protein, which can bind to muscle myosin [66, 67] and interact with bacterial myosin-like protein [68], has had no further progress. Back in 1982, we guessed that the possible cause of resemblance between flagellin and skeletal muscle actin might be their common genesis. Actin, which is present in all cell types including (as it becomes apparent) bacterial cells, is a conservative polyfunctional protein. One can suppose that some evolutionary ancestor, which was able to form well-ordered structure, could well give birth not only to actin itself, but also to tubulin and flagellin. During evolution, these proteins have maintained their ability to interact with myosin, an ability that was previously reported for tubulin [51]. In the course of evolution, this actin predecessor was involved in actin-myosin-dependent motility systems of eukaryotic cells and later changed insignificantly because it never got outside the cell. Alternatively, flagellin “trespassed” beyond the cell and took part in the formation of the bacterial motility apparatus. During evolution, bacteria had to adapt to various environmental conditions, which cannot but have influenced flagellin of BFF localized outside the cell. As a result, flagellin underwent significant changes during evolution, so that it's molecular mass as well as antigen specificity differs in different bacteria.
The following example is also illustrative of the resemblance of proteins that can be found in different biological systems. Recently, a certain homology was discovered between FliI protein of the basal body of the bacterial flagellum and the beta-subunit of bacterial FoF1 ATPase, as well as between vacuolar and archaebacterial ATPases [69]. It has been hypothesized that these ATPases diverged from their common ancestor at the same time. Moreover, when homology between MreB and ParM on one hand and eukaryotic actin on the other hand is discussed in the literature, it is hypothesized that pro- and eukaryotes had a common ancestor possessing a protein whose characteristics were common with both eukaryotic actin and bacterial actin-like proteins [70].
Naturally, the above conclusions upon the ability of BFF to bind S1 are not conclusive. Other considerations can also be suggested to explain this phenomenon, though our interpretation seems to be the most attractive.
For a long time of studies of the structure and properties of bacterial flagellum as motility organelle have been applied to the archaebacterial flagellum as well. Once the existence of three evolution lines had been proposed for the origin of living organisms--eucaryotes, eubacteria, and archaebacteria [71, 72], interest has appeared in the archaeal motility organs that are apparently like bacterial flagella. In the 1980s, a significant difference was demonstrated between the properties of bacterial and archaeal flagellum filaments [73-78]. The thickness of the archaeal flagellum filament (AFF) is half that of the BFF [79], and, unlike the BFF, it consists of several glycosylated flagellins [80-82], which are found in AFF in different quantities [83, 84]. The main difference between archaeal and bacterial flagella is obvious from the analysis of the full genome structures of many archaeal flagellates [85]. Genes encoding bacterial proteins or involved in regulation of the bacterial flagellum assembly are not found in the archaeal genome. The intracellular component of the archaeal flagellum contains no structure that can be regarded as similar to the basal body of the bacterial flagellum.
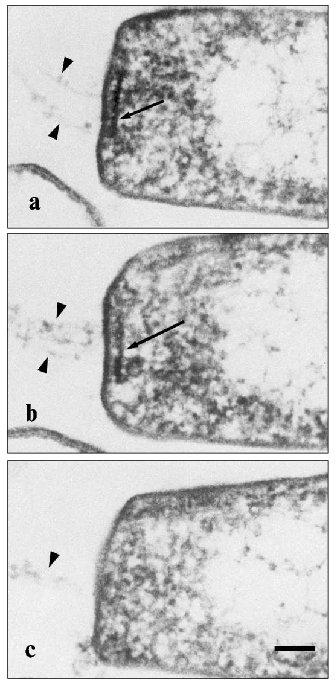
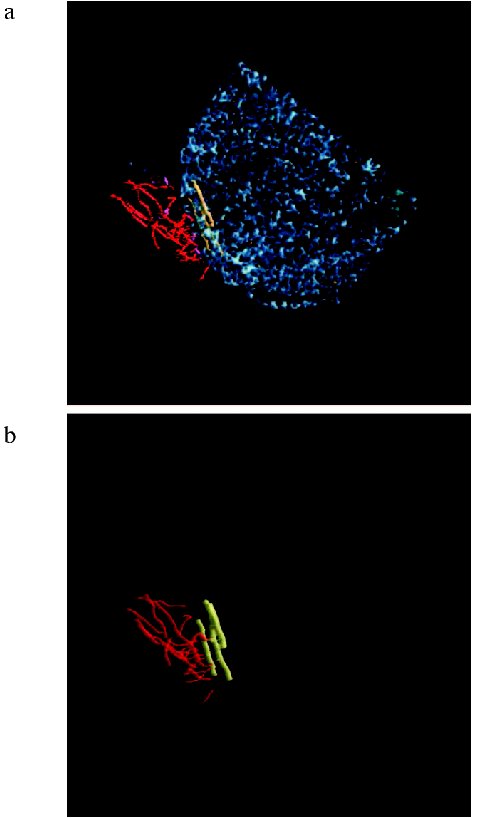
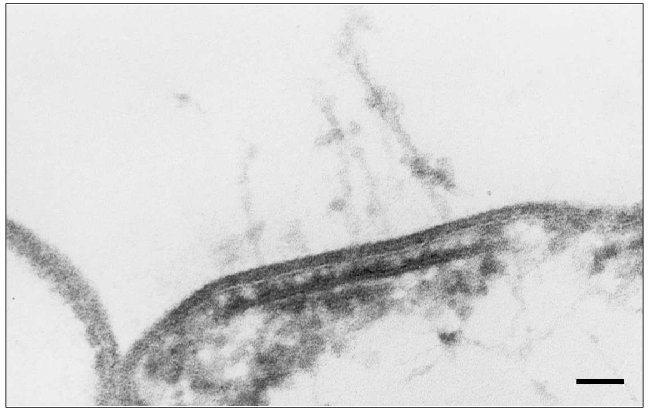
We pioneered in the description of a plateau-like intracellular structure below the cytoplasmic membrane on which the Halobacterium salinarium flagellum is based [86]. The existence of a structure like this was suggested by Alam and Oesterhelt in 1984 [73]. On longitudinal shearing of the cell, this structure is seen as an electron dense line 250-300 nm in length and 20-25 nm thick which underlies the flagellum bundle and is located parallel with the plane of the plasma membrane. Analysis of serial sections (Fig. 4) showed that these lines represent cross-sections of a discoid plate, and this plate is always located at the cell pole exactly in the site of the flagellum bundle entry at the distance of ~20 nm from plasma membrane. On average, this electron dense line can be observed in five to six serial sections of 60-70 nm thick, and its maximum extent is detected in the median sections. Bipolar cells with two flagellum bundles have two corresponding electron-dense lines. We termed this structure the “discoid lamellar structure” (DLS) [86]. The pole of the H. salinarium cell was reconstituted on the basis of ultra-thin serial sections (Fig. 5, see color insert) (the experiments were performed jointly with I. I. Kireev, T. M. Novikova, and E. V. Sheval; Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University).
Fig. 4. Ultra-thin serial sections of the Halobacterium salinarium VKMB-1231 cell pole. Cells were fixed with glutaraldehyde/osmium tetroxide fixative and sliced in ~60-80-nm-thick sections. Arrows indicate the discoid lamellar structures in the roots of flagellum bundles; spikes indicate individual flagellum fibrils. The scale length is 100 nm.
When cut directly across DLS, the sections viewed at high magnification show that this plate is a complex structure composed of several layers differing in affinity to osmium tetroxide [86] (Fig. 6). Both morphology and ordering of these layers are consistent with current view of membranes stained with osmium tetroxide and examined under the electron microscope [87]. Nonetheless, these data alone do not allow categorical assertion that the DLS is membrane-like, because a series of questions arises in this case.Fig. 5. a) Reconstruction of the Halobacterium salinarium cell pole from ultra-thin serial sections. The cell is drawn in blue, flagella in red, and discoid lamellar structure (DLS) in yellow; b) blue color is omitted for better resolution of DLS.
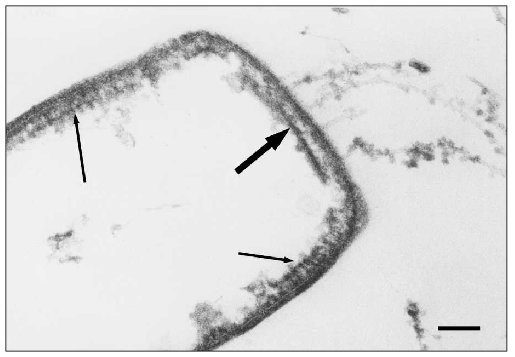
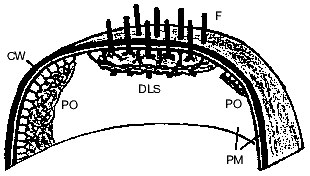
Along with the DLS, another well-ordered structure is found near the poles of halobacterial cells just underneath the plasma membrane [88]; this structure differs significantly from the DLS and resembles the “polar organelle” of eubacteria [89] (Fig. 7). Like the “polar organelle” [90, 91], this structure is positively stained in cytochemical determination of ATPase activity [88]. We found that DLS does not express ATPase activity. Thus, we have discovered two previously unknown structures in the H. salinarium cells, one of which (the DLS) in all likelihood is associated with the motility apparatus of this organism [86, 88, 92]. On the basis of these data we have offered a scheme for the H. salinarium cell pole (Fig. 8).Fig. 6. High-resolution image of the discoid lamellar structure on the Halobacterium salinarium VKMB-1231 cell pole. The scale length is 50 nm.
Fig. 7. Electron microscopic image of ultra-thin section of Halobacterium salinarium VKMB-1231 cell “shadow”. Bold arrow indicates DLS, and arrows indicate polar organelles. The scale length is 100 nm.
It is known that the archaeal and eubacterial cell walls are quite different, and the archaea live in extreme environments, so one can hardly expect that structures of their flagella are identical to the bacterial flagellum structures. Data indicating diverse (as compared to eubacteria) flagellum assembly mechanism in archaea also deserve considerable attention. Jarrell and coworkers were the first to direct attention to the homology between N-terminal hydrophobic region of archaeal flagellins and type IV pilins [93]. Type IV pilins are associated with proteins forming filamentous structures composed of spiral protein subunits and are responsible for various processes such as adhesion, conjugation, and twitching motility (a particular case of translocation) [94, 95]. This research group further proposed a model for archaeal flagellum assembly--from the proximal end with leader peptides of archaeal proflagellins [84]. Alternatively, in bacterial flagella the assembly of flagellin filaments starts from the distal end of the flagellum [45, 46, 96] via a type III protein secretion system [97, 98]. The proteins secreted by this system, which are found in some bacteria pathogenic for plants or animals, are characterized by lack of signaling sequences being cleft when penetrating the host cell.Fig. 8. A scheme for the Halobacterium salinarium cell pole. F, flagellum; CW, cell wall; PM, plasma membrane; DLS, discoid lamellar structure; PO, polar organelle. The depiction is not to scale.
The data described above suggest differences between bacteria and archaea in both structure and assembly mechanisms of their motility organelles. However, when studying the fundamentals of our wonderful world, we often face enigmatic phenomena. In the case of bacteria and archaea, they possess virtually identical sensorial systems of chemotaxis, despite distinct difference in their flagellum-based locomotion machinery [99]. Moreover, horizontal gene transfer may have occurred in evolution of bacterial and archaeal flagellin genes [100]. So, the bacterial and archaeal locomotion systems appear to be convenient models for investigations into evolutionary pathways of biological species.
It should be emphasized in the conclusion of this not so exhaustive review of molecular structure and properties of flagella that archaeal flagella are far less understood than bacterial flagella. This is true in regard to both external flagellin filaments and internal components of archaeal flagella. Whereas the BFF assembly may be considered as a classical example of a mechanism controlled by conformational adaptation between the assembled proteins with dynamic conformational changes of subunits and partial folding, we cannot yet say the same about the assembly of AFF. The latter is a not well-understood multiflagellin system that is formed in a different way than the BFF.
The present data on two types of locomotion organelles in prokaryotes allow the following conclusions: bacterial flagella are composed preferably of a single flagellin type whereas the archaeal flagella consist of several distinct flagellin types; unlike bacterial flagellins, archaeal flagellins are glycosylated; the diameter of archaeal flagella is about twofold less than bacterial flagella; bacterial flagellins have conservative N- and C-termini whereas only the N-termini of archaeal flagellins are thought to be conservative; despite similar amino acid composition no homology was found between bacterial and archaeal flagellins, however, archaeal flagellins are homologous to bacterial pilins; the mechanisms of bacterial and archaeal flagellum assembly seem to be different.
Several reviews [84, 95, 101] are recommended to reader especially interested in structure and properties of bacterial and archaeal motility organelles.
In conclusion, I would like to emphasize again that studies on bacterial and archaeal locomotion mechanisms could scarcely have developed in Russia without the contributions of Boris Fedorovich Poglazov, a man of multifarious vision and infinite scientific curiosity.
REFERENCES
1.Jones, C. J., and Aizawa, S. (1991) Adv. Microb.
Physiol., 32, 109-172.
2.Macnab, R. M. (1992) Annu. Rev. Genet.,
26, 131-158.
3.Jarrell, K. F., Bayley, D. P., and Faguy, D. M.
(1993) Curr. Top. Mol. Genet. (Life Sci. Adv.), 1,
15-31.
4.Jarrell, K. F., Bayley, D. P., Correia, J. D., and
Thomas, N. A. (1999) BioScience, 49, 279-281.
5.De Pamphilis, M. L., and Adler, J. (1971) J.
Bacteriol., 105, 376-407.
6.Iino, T. (1977) Annu. Rev. Genet.,
11, 161-182.
7.Larsen, S., Reader, R., Kort, E., Tso, W.-W., and
Adler, J. (1974) Nature, 249, 74-75.
8.Skulachev, V. P. (1975) Proc. 10th FEBS
Meet., pp. 225-238.
9.Bekyakova, T. N., Glagolev, A. N., and Skulachev,
V. P. (1976) Biokhimiya, 41, 1478-1483.
10.Skulachev, V. P. (1977) FEBS Lett.,
74, 1-9.
11.Hirota, N., Katada, M., and Imae, Y. (1981)
FEBS Lett., 132, 278-280.
12.Hirota, N., and Imae, Y. (1983) J. Biol.
Chem., 258, 10577-10581.
13.Chernyak, B. V., Dibrov, P. A., Glagolev, A. N.,
Sherman, M. Yu., and Skulachev, V. P. (1983) FEBS Lett.,
164, 38-42.
14.Bakeeva, L. E., Chumakov, K. M., Drachev, L. A.,
Metlina, A. L., and Skulachev, V. P. (1986) Biochim. Biophys.
Acta, 850, 466-472.
15.Poglazov, B. F. (1967) Uspekhi Sovr.
Biol., 63, 342.
16.Poglazov, B. F. (1970) Assembly of Biological
Structures [in Russian], Nauka, Moscow.
17.Poglazov, B. F. (1975) in Nucleic Acids and
Virion Proteins (Gendon, Yu. Z., Poglazov, B. F., and Tikhonenko,
T. I., eds.) [in Russian], Nauka, Moscow.
18.Poglazov, B. F. (1977) The Rules for Assembly
of Elementary Biological Structures, in 32nd Bach Reading
[in Russian], Nauka, Moscow.
19.Poglazov, B. F. (1996) Biochemistry
(Moscow), 61, 1377-1382.
20.Caspar, D. L. D., and Klug, A. (1962) Cold
Spring Harb. Sym., 27, 1-24.
21.Anfinsen, C. B. (1973) Science,
181, 223-230.
22.Minamino, T., and Macnab, R. (1999) J.
Bacteriol., 181, 1388-1394.
23.Minamino, T., Chu, R., Yamaguchi, S., and Macnab,
R. (2000) J. Bacteriol., 182, 4207-4215.
24.Zhu, K., Gonzalez-Pedrajo, B., and Macnab, R. M.
(2002) Biochemistry, 41, 9516-9524.
25.Kostyukova, A. S., Polosina, Ya. Yu.,
Pyatibratov, M. G., Tiktopulo, E. I., and Fedorov, O. V. (1994)
Dokl. RAN, 339, 544-546.
26.Ellis, R. J. (2001) Trends Biochem. Sci.,
26, 597-604.
27.Minton, A. P. (2001) J. Biol. Chem.,
276, 10577-10580.
28.Metlina, A. L., and Poglazov, B. F. (1970)
Biokhimiya, 35, 994-1001.
29.Novikov, V. V., Metlina, A. L., and Poglazov, B.
F. (1980) Patent of USSR No. 3008188/13 from October 17, 1980.
30.Novikov, V. V., Metlina, A. L., Mitina, N. A.,
Topchieva, I. N., and Poglazov, B. F. (1981) Biokhimiya,
46, 1333-1336.
31.Novikov, V. V., Metlina, A. L., and Poglazov, B.
F. (1994) Biochem. Mol. Biol. Int., 33, 723-728.
32.Metlina, A. L., and Poglazov, B. F. (1969)
Dokl. Akad. Nauk SSSR, 187, 459-462.
33.Klein, D., Foster, J., and Koffler, H. (1969)
Biochem. Biophys. Res. Commun., 36, 844-850.
34.Metlina, A. L. (1970) Study of Physical and
Chemical Properties of Bacillus brevis Flagellin: Candidate's
dissertation (in Biology), Lomonosov Moscow State University,
Moscow.
35.Uratani, Y., Asakura, S., and Imahori, K. (1972)
J. Mol. Biol., 67, 85-98.
36.Asakura, S. (1970) Adv. Biophys.,
1, 99-155.
37.Gerber, B. R., Asakura, S., and Oosawa, F. (1973)
J. Mol. Biol., 74, 467-487.
38.Novikov, V. V. (1982) Study of the Mechanism
of Bacillus brevis flagellum assembly: Candidate's dissertation (in
Biology), Bach Institute of Biochemistry, Moscow.
39.Wakabayashi, K., Hotani, H., and Asakura, S.
(1969) Biochim. Biophys. Acta, 175, 195-203.
40.Chen, W. H., Yang, G. T., and Martinez, H. M.
(1972) Biochemistry, 11, 4120-4131.
41.Ishikihara, A., and Hotani, H. (1980) J. Mol.
Biol., 139, 265-276.
42.Grant, G. F., and Simon, M. (1968) J.
Bacteriol., 95, 81-86.
43.Topchieva, I. N. (1980) Usp. Khim.,
49, 494-517.
44.Bode, W., Hinz, W., Jaenicke, R., and Blume, A.
(1974) Biophys. Struct. Mech., 1, 55-64.
45.Kerridge, D. (1959) Biochim. Biophys.
Acta, 31, 579-581.
46.Emerson, S. U., Tokyuasu, K., and Simon, M. I.
(1970) Science, 169, 190-192.
47.Homma, M., Iino, T., Kutsukake, K., and
Yamaguchi, S. (1986) Proc. Natl. Acad. Sci. USA, 83,
6169-6173.
48.Ikeda, T., Homma, M., Iino, T., Asakura, S., and
Kamiya, R. (1987) J. Bacteriol., 169, 1168-1173.
49.Yonekura, K., Maki, S., Morgan, D. G., DeRosier,
D. J., Vonderviszt, F., Imada, K., and Namba, K. (2000) Science,
290, 2148-2152.
50.Samatey, F. A., Imada, K., Nagashima, S.,
Vonderviszt, F., Kumasaka, T., Yamamoto, M., and Namba, K. (2001)
Nature, 410, 331-337.
51.Shimo-Oka, T., Hayashi, M., and Watanabe, J.
(1980) Biochemistry, 19, 4921-4926.
52.Novikova, N. A., Metlina, A. L., Levitskii, D.
I., and Poglazov, B. F. (1983) Dokl. Akad. Nauk SSSR,
269, 1252-1255.
53.Novikova, N. A. (1983) Interaction of
Bacterial Flagella with the Skeletal Muscle Myosin: Candidate's
dissertation (in Biology), Bach Institute of Biochemistry, Moscow.
54.Novikova, N. A., Levitsky, D. I., Metlina, A. L.,
and Poglazov, B. F. (2000) IUBMB Life, 50, 385-390.
55.Estes, J. E., and Gershman, L. C. (1978)
Biochemistry, 17, 2495-2499.
56.Lheureux, K., and Chaussepied, P. (1995)
Biochemistry, 34, 11435-11444.
57.De Lange, R. J., Chang, J. Y., Shaper, J. H., and
Glazer, A. N. (1976) J. Biol. Chem., 251, 705-711.
58.Elzinga, M., Collins, J. H., Kuchaell, V. M., and
Adelstein, R. S. (1973) Proc. Natl. Acad. Sci. USA, 70,
2687-2691.
59.Van den Ent, F., Amos, L. A., and Lowe, J. (2001)
Nature, 413, 39-44.
60.Van den Ent, F., Moller-Jensen, J., Amos, L. A.,
Gerdes, K., and Lowe, J. (2002) EMBO J., 21,
6935-6943.
61.Jones, L. J., Carballido-Lopez, R., and
Errington, J. (2001) Cell, 104, 913-922.
62.Egelman, E. H. (2003) Curr. Opin. Struct.
Biol., 13, 244-248.
63.Nogales, E., Downing, K. H., Amos, L. A., and
Lowe, J. (1998) Nat. Struct. Biol., 5, 451-458.
64.Erickson, H. P. (1995) Cell, 80,
367-370.
65.De Pereda, J. M., Leynadier, D., Evangelio, J.
A., Chacon, P., and Andreu, J. M. (1996) Biochemistry,
35, 14203-14215.
66.Nakamura, K., and Watanabe, S. (1978) J.
Biochem., 83, 1459-1470.
67.Somaya, A., and Tanaka, N. (1979) FEBS
Lett., 101, 166-168.
68.Nakamura, K., Takahashi, K., and Watanabe, S.
(1978) J. Biochem., 84, 1453-1458.
69.Volger, A. P., Homma, M., Irikura, V. M., and
Macnab, R. M. (1991) J. Bacteriol., 173, 3564-3572.
70.Doolittle, R. F., and York, A. L. (2002)
BioEssays, 24, 293-296.
71.Woese, C. R., and Fox, G. E. (1977) Proc.
Natl. Acad. Sci. USA, 74, 5088-5090.
72.Woese, C. R., Kandler, O., and Wheelis, M. L.
(1990) Proc. Natl. Acad. Sci. USA, 87, 4576-4579.
73.Alam, M., and Oesterhelt, D. (1984) J. Mol.
Biol., 176, 459-475.
74.Alam, M., and Oesterhelt, D. (1987) J. Mol.
Biol., 194, 495-499.
75.Sumper, M. (1987) Biochim. Biophys. Acta,
906, 69-79.
76.Gerl, L., and Sumper, M. (1988) J. Biol.
Chem., 263, 13246-13251.
77.Gerl, L., Deutzmann, R., and Sumper, M. (1989)
FEBS Lett., 244, 137-140.
78.Jarrell, K. F., Bayley, D. P., and Faguy, D. M.
(1993) Curr. Top. Mol. Genet. (Life Sci. Adv.), 1,
15-31.
79.Jarrell, K. F., and Koval, S. F. (1989) Crit.
Rev. Microbiol., 17, 53-87.
80.Wieland, F., Paul, G., and Sumper, M. (1985)
J. Biol. Chem., 260, 15180-15185.
81.Pyatibratov, M. G., Kostyukova, A. S., Tarasov,
V. Yu., and Fedorov, O. V. (1993) Dokl. RAN, 330,
116-119.
82.Serganova, I. S., Polosina, Ya. Yu., Kostyukova,
A. S., Metlina, A. L., Pyatibratov, M. G., and Fedorov, O. V. (1995)
Biochemistry (Moscow), 60, 953-958.
83.Kalmokoff, M. L., Koval, S. F., and Jarrell, K.
F. (1992) Arch. Microbiol., 157, 481-487.
84.Jarrell, K. F., Bayley, D. P., and Kostyukova, A.
S. (1996) J. Bacteriol., 179, 5057-5064.
85.Jarrell, K. F., Bayley, D. P., Correia, J. D.,
and Thomas, N. A. (1999) BioScience, 49, 530-541.
86.Bakeeva, L. E., Metlina, A. L., Novikova, T. M.,
and Speranskii, V. V. (1992) Dokl. RAN, 326, 914-915.
87.Sjostrand, F. (1960) Radiat. Res., Suppl.
2, 349-362.
88.Speranskii, V. V. (1996) Ultrastructure of
Flagellar Apparatus of Halophilic Archaea: Candidate's dissertation
(in Biology), Bach Institute of Biochemistry, Moscow.
89.Murray, R. G. E., and Birch-Andersen, A. (1963)
Can. J. Microbiol., 9, 393-401.
90.Wachstein, M., and Meisel, E. (1957) Amer. J.
Clin. Pathol., 27, 13-23.
91.Tauschel, H.-D. (1988) Meth. Microbiol.,
20, 237-259.
92.Speranskii, V. V., Metlina, A. L., Novikova, T.
M., and Bakeeva, L. E. (1996) Biofizika, 41, 159-163.
93.Faguy, D. M., Jarrell, K. F., Kuzio, J., and
Kalmokoff, M. L. (1994) Can. J. Microbiol., 40,
67-71.
94.Whitchurch, C. B., Hoobs, M., Livingston, S. P.,
Krishnapillai, V., and Mattick, J. S. (1991) Gene, 101,
33-44.
95.Bardy, S. L., Ng, Y. M., and Jarrell, K. F.
(2003) Microbiology, 149, 295-304.
96.Iino, T., Suzuki, H., and Yamaguchi, S. (1972)
Nat. New Biol., 237, 238-240.
97.Fan, F., Ohnishi, K., Francis, N. R., and Macnab,
R. M. (1997) Mol. Microbiol., 26, 1035-1046.
98.Hueck, C. J. (1998) Microbiol. Mol. Biol.
Rev., 62, 379-433.
99.Rudolph, J., and Oesterhelt, D. (1996) J. Mol.
Biol., 258, 548-554.
100.Serganova, I., Ksenzenko, V., Serganov, A.,
Meshcheryakova, I., Pyatibratov, M., Vakhrusheva, O., Metlina, A., and
Fedorov, O. (2002) J. Bacteriol., 184, 318-322.
101.Macnab, R. M. (1999) J. Bacteriol.,
181, 7149-7153.