Diversity of Structure and Function of DNA Polymerase (gp43) of T4-Related Bacteriophages
V. M. Petrov and J. D. Karam*
Tulane University Health Sciences Center, Department of Biochemistry SL43, 1430 Tulane Avenue, New Orleans, LA 70112-2699, USA; fax: 504-988-1611; E-mail: karamoff@tulane.edu* To whom correspondence should be addressed.
Received July 29, 2004
The replication DNA polymerase (gp43) of the bacteriophage T4 is a member of the pol B family of DNA polymerases, which are found in all divisions of life in the biosphere. The enzyme is a modularly organized protein that has several activities in one polypeptide chain (~900 amino acid residues). These include two catalytic functions, POL (polymerase) and EXO (3´-exonuclease), and specific binding activities to DNA, the mRNA for gp43, deoxyribonucleotides (dNTPs), and other T4 replication proteins. The gene for this multifunctional enzyme (gene 43) has been preserved in evolution of the diverse group of T4-like phages in nature, but has diverged in sequence, organization, and specificity of the binding functions of the gene product. We describe here examples of T4-like phages where DNA rearrangements have created split forms of gene 43 consisting of two cistrons instead of one. These gene 43 variants specify separate gp43A (N-terminal) and gp43B (C-terminal) subunits of a split form of gp43. Compared to the monocistronic form, the interruption in contiguity of the gene 43 reading frame maps in a highly diverged sequence separating the code for essential components of two major modules of this pol B enzyme, the FINGERS and PALM domains, which contain the dNTP binding pocket and POL catalytic residues of the enzyme. We discuss the biological implications of these gp43 splits and compare them to other types of pol B splits in nature. Our studies suggest that DNA mobile elements may allow genetic information for pol B modules to be exchanged between organisms.
KEY WORDS: DNA polymerase, pol B family, gp43, molecular evolution, DNA binding proteins, RNA binding proteins, DNA replication, multifunctional proteins, modular proteins, microbial diversity, bacteriophages
The DNA genome of bacteriophage T4 is about 170,000 base pairs (bp) long and encodes all of the proteins that are required for phage DNA replication in the T4-infected Escherichia coli host. Among these, the T4 encoded DNA polymerase (gp43, product of phage gene 43) plays two broadly essential roles in phage replication; (a) it determines the accuracy of replicative DNA synthesis (fidelity of replication) and (b) it participates in a number of interactions with nucleotides, nucleic acids, and other biological molecules that are needed for assembly and maintenance of the phage DNA replication complex in vivo [1]. The high fidelity of T4 gp43 resides in two catalytic activities of the 898 amino-acid protein: the polymerase (or POL) function, which determines the accuracy of nucleotide base selection for DNA synthesis, and the 3´-exonuclease (or EXO) function, which removes misincorporated nucleotides when the POL function fails to make the correct selection. The interplay between the POL and EXO functions of this enzyme limits replication errors in vivo to about 10-8 misincorporations per bp per genome duplication.
The ability of T4 gp43 to maintain high fidelity is dependent on the control of gene 43 expression and interactions of the protein with other replication proteins of the phage DNA replication complex. In recent years, we have used phylogenetic tools to identify the regulatory mechanisms for gp43 biosynthesis and the structural features of this protein that allow for its specific interactions with nucleic acids and other biological macromolecules. We summarize here some of our observations from examining the gene 43 regions of several phylogenetic relatives of T4 (T4-like phages) that grow in diverse bacterial species. The T4-like phages resemble T4 in their morphology and their dsDNA genomes contain many genes that are homologous to T4 genes; however, these genomes exhibit a variety of differences from the T4 genome in size, content of genes, and sequence divergence between homologous genes. More than 200 such phages have been isolated from different environmental sources over the last 50-60 years [2]. The coli-phage T4 and its “T-even” relatives (T2 and T6) are the best studied [3], but the advent of high-throughput genome sequencing is making it possible to rapidly learn about the high degree of biochemical diversity that is encoded by T4-like phages that grow in Aeromonas, Acinetobacter, Vibrio, or other bacterial species that do not share the same environmental habitat as the natural host for the T-even phages. The genome sequences of these T4 relatives provide a database (http://phage.bioc.tulane.edu), which allows us to examine the functional significance of sequence divergence between homologous proteins that evolved in separate bacterial domains. The DNA polymerase gp43 of these phages is a case in point that we will describe in this report.
We have observed that gene 43 has been preserved in evolution of the T4-like phages, although it has undergone considerable variation in sequence, regulation, and molecular form of the gene product, the phage DNA polymerase. We discuss here the biological implications of the divergence in this essential phage induced enzyme and draw parallels to the evolution of gp43-like enzymes in other organisms, particularly the archaea. Our studies suggest that gp43 and related DNA polymerases in nature are endowed with a structural plasticity that can accommodate large changes in their modular form without loss of the essential biological functions. These changes often lead to new specificities of the enzyme for nucleic acids and partner proteins in the phage DNA replication complex.
MATERIALS AND METHODS
Bacterial and phage strains. The T4-like coli-phages RB49 and RB69 were obtained from W. B. Wood (University of Colorado, Boulder). These and other “RB” series coli-phages are currently archived by B. Guttmann (Evergreen State College, Olympia, Washington, USA). The T4-like Aeromonas salmonicida phages 25, 31, 65, and 44RR and the T4-like Acinetobacter johnsonii phage 133 were obtained from H. Ackermann at the Felix D'Herelle Reference Center for Bacterial Viruses at Laval University (Quebec, Canada). Sequence information on these phages can be found at http://phage.bioc.tulane.edu and NCBI http://www.ncbi.nlm.nih.gov.
Cloning phage genes in E. coli plasmids and measuring plasmid-mediated expression. The methods for cloning phage genes of interest in bacterial plasmids and for expression under control of a plasmid-borne T7 promoter (Fig. 3) have been described previously [4, 5] and will be elaborated further elsewhere (Petrov et al., in preparation). The E. coli host used for all expression experiments was CAJ70, which has also been described previously [4].
Other methods. Methods of phage genome sequence determinations and annotation are outlined at http://phage.bioc.tulane.edu and will be described in detail elsewhere (Nolan et al., in preparation). Other experimental details are included in the figure legends.
RESULTS OF INVESTIGATION
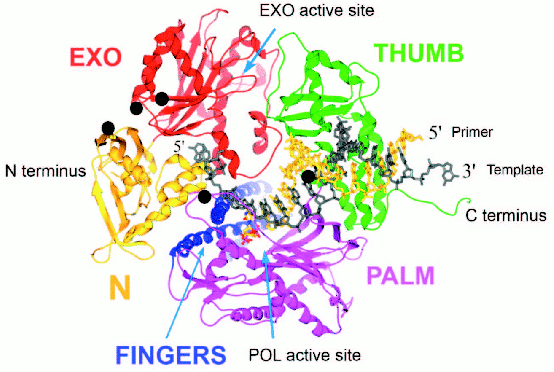
T4 gp43 is a pol B-like DNA polymerase. The amino acid sequence of T4 gp43 has been deduced from DNA and mRNA sequencing data and from analysis of proteolytic fragments of the purified protein [6]. The enzyme shares many similarities with E. coli DNA polymerase II (E. coli pol B) and other pol B-like (pol B family) DNA polymerases of the eukaryotes and archaea [7]. In studies where we compared T4 gp43 with the enzyme from its phylogenetic relative coli-phage RB69, we established that gp43 is modularly organized into domains of function that can be exchanged between the T4 and RB69 enzymes (domain swaps) without loss of biological activity and despite sequence variations between the homologous domains of the two gp43 variants [4]. The modular structure of this phage-induced enzyme was later confirmed directly through X-ray crystallographic studies that were conducted with RB69 gp43 [8]. This structure, which is represented in Fig. 1 (see color insert), was the first to be determined for a pol B family DNA polymerase and has been serving as a reference for structure-function relationships of this class of enzymes in general. Three other pol B DNA polymerases, all from the archaea, were subsequently crystallized and their structures found to be almost superimposable with that of RB69 gp43 [9-11]. The ribbon diagram in Fig. 1 shows RB69 gp43 in complex with primer-template DNA [12] and also highlights regions that have been implicated in mRNA binding [13]. We use this structure as a reference to discuss other molecular forms of gp43 that we have discovered among the T4-like phages.
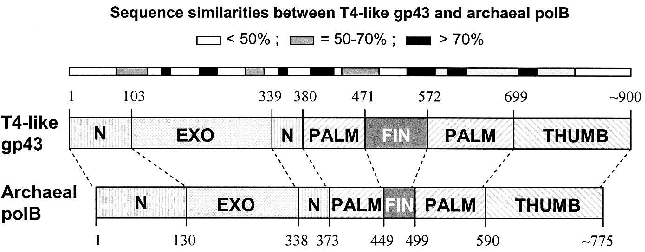
Similarities among pol B DNA polymerases. The typical pol B family DNA polymerase consists of five structural domains [9-11, 13, 14]: N, EXO, PALM, FINGERS, and THUMB (Fig. 1). The EXO domain interrupts sequence contiguity of the N domain and the FINGERS domain interrupts sequence contiguity of the PALM domain. In pair-wise sequence comparisons, we have observed that divergence between RB69 gp43 and T4 gp43 [4], or any other gp43 variant from the T4-like phages posted in http://phage.bioc.tulane.edu and GenBank, is clustered in segments of the protein that connect highly conserved sequences within the five domains or subdomains of the structure. As expected, we have observed a lower level of sequence divergence between gp43 variants than between the RB69 gp43 reference and the pol B enzymes of known structure. These differences are summarized in Fig. 2 and suggest that the diversity in primary structure between gp43 of the T4-like phages and pol B enzymes of other organisms reflects the different evolutionary paths taken by members of this enzyme family from common progenitors. We have examined the most highly diverged gp43 variants within the T4-like phage family for clues to the evolutionary events that have contributed to the divergence in structural features and functions of pol B enzymes in general.Fig. 1. A ribbon diagram of the structure of RB69 gp43 complexed with primer-template DNA. The diagram is modified from Franklin et al. [13] to highlight sites on the gp43 molecule (black dots) that have been implicated in binding the mRNA target for the protein [12]. The same structural framework has been observed for archaeal DNA polymerases belonging to the pol B family [9-11].
Diversity of the nucleic acid binding functions of gp43. In T4 and RB69, gp43 bears two nucleic acid binding functions, one to DNA and the other to the specific mRNA that encodes the protein [15]. The DNA binding function is essential for DNA replication and has been preserved as a function among gp43 variants, whereas the RNA binding function has diverged in specificity and may have either become extinct or evolved different biological roles in different T4-like phages. The DNA binding function of gp43 is not specific to the nucleotide sequence, which might be expected since this enzyme must be able to read all sequences of the phage genome during replication. However, like all other DNA-dependent DNA polymerases, gp43 binds to a specific type of DNA structure, the primer-template DNA complex, which consists of double-stranded (dsDNA) and single-stranded (ssDNA) portions (Fig. 1). In RB69 gp43, it has been determined that the dsDNA portion binds in a groove within the THUMB domain and the ssDNA (template) portion binds in a groove formed by the intersection between the N and EXO domains of the protein [13, 14]. Amino acid residues that determine primer-template binding are highly conserved among pol B enzymes, especially among the ~30 gp43 variants that we have examined from T4-like phages. Also conserved are the residues and protein structural features that determine binding of the dNTP substrate near the POL catalytic site of these enzymes. These observations underscore the segmental nature of amino acid sequence conservation among pol B enzymes across biological species and the modular organization of these enzymes (Fig. 2).Fig. 2. Diagrammatic comparison between the modular organizations of gp43 and pol B DNA polymerases. The bar at the top shows the clusters of divergence in amino acid sequence between the phage and archaeal enzymes. The highest degree of divergence (<30% similarity) occurs in the segment corresponding to the tip of the FINGERS domain (Figs. 1 and 4).
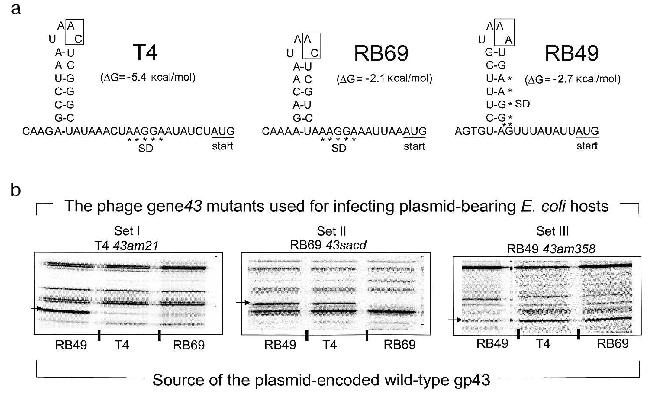
In contrast to the DNA binding function of gp43, the mRNA binding function appears to be highly susceptible to evolutionary divergence. In T4, RB69, and a very large number of other T4-like coli-phages isolated from the wild, the RNA target for gp43 includes a stem-and-loop (RNA hairpin) structure that is located in the translation initiation region of the mRNA for the protein [15]. In these phages, gp43 binds to the specific RNA site and down-regulates its own biosynthesis (translational autorepression) during assembly of the phage DNA replication complex in the phage-infected bacterial host. Among the T4-like phage genomes that we have examined so far, we have identified three distinguishable gp43-mRNA binding specificities, one represented by phage T4, one seen in phage RB69, and one in phage RB49. The RNA targets for the three gp43 variants from these phages are diagrammed in Fig. 3a. The T4-specific target appears to be the most common among natural isolates of T4-like phages, although all observed recurrences of this target are among phages that share the same hosts as T4. The RB69 target has so far been encountered only once and the RB49 target twice in our screening of T4-like genomes from natural sources.
In previous work, we observed that RB69 gp43 is able bind and repress the mRNA of T4 gp43, but that T4 gp43 is narrowly specific to its own mRNA [5]. As shown in Fig. 3b, RB49 gp43 is also narrowly specific to its own mRNA and is not repressed by either T4 gp43 or RB69 gp43. Based on such observations and other studies that examined interactions of T4 gp43 with mutant and in vitro-selected mRNA targets [16-19], we have surmised that three properties of the RNA target determine specificity of the gp43-mRNA interaction: (a) positioning of purine and pyrimidine residues in the RNA hairpin structure; (b) the sequence of two nucleotides in the unpaired segments of the RNA hairpin (boxed in Fig. 3); and (c) stabilizing nucleotide sequence-independent interactions of the protein with segments of the mRNA that are external to the RNA hairpin [16]. Together, these three criteria may contribute to the evolution of a wide spectrum of RNA targets for gp43 without necessarily requiring a major divergence in protein structure. In experiments that mapped sites on RB69 gp43 that interact with RNA, we concluded that multiple contacts in four of the five domains of the protein are required, with most contacts involving amino acid residues or sequence motifs that are highly conserved among gp43 variants [12]. Thus, the conserved structural features of this pol B DNA polymerase appear to have a plasticity to use alternative contacts with potential RNA targets that can accommodate evolutionary change in the target sequence without loss of the ability to maintain the protein-RNA interaction necessary for translational repression.Fig. 3. The divergent mRNA targets for translational repression by gp43. Panel (a) diagrams the RNA hairpin structures that serve as binding targets for the DNA polymerases (gp43s) of coli-phages T4, RB69, and RB49, respectively. The boxed nucleotides are essential for determination of the specificity of the RNA to its gp43 partner. Panel (b) shows results of an experiment in which the translational repressor (mRNA binding) specificities of plasmid-encoded T4 gp43, RB69 gp43, and RB49 gp43 could be distinguished from one another. In this experiment, phage mutants carrying chain-termination mutations in their respective gene 43 (T4 43am21, RB69 43sacd, or RB49 43am358) were used to infect E. coli CAJ70 hosts expressing a cloned wild-type gene 43 from T4, RB69, or RB49. To determine if the plasmid encoded gp43 could repress the synthesis of the phage-induced mutant gp43, the infected cells were labeled with 35S-labeled methionine at 10-15 min after infection and extracts were prepared and analyzed by SDS-PAGE and autoradiography, as described previously [5]. The horizontal arrows at the side of each set of autoradiograms point to the positions of the phage-induced mutant gp43 synthesized in each set of phage infections. The experiment demonstrated that plasmid-encoded T4 gp43 (set I) and RB49 gp43 (set III) are narrowly specific to mRNA from the T4 43am21 and RB49 43am358 infections, respectively. In contrast, plasmid-encoded RB69 gp43 can repress mRNA from both RB69 43sacd and T4 43am21 (set II). In some T4-like phages, gp43 does not repress its own translation.
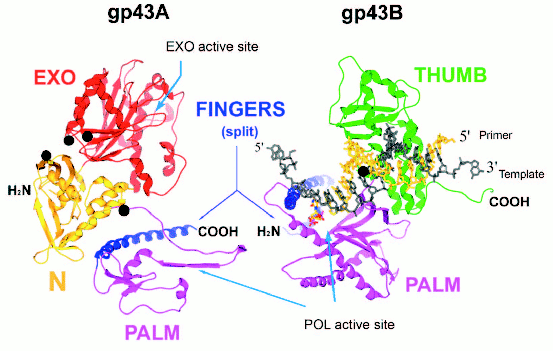
Split forms of gp43 in nature. Plasticity of the gp43 structure is also reflected in the diversity of molecular forms of this enzyme found in nature. We have discovered split (or two-cistron) variants of gene 43 in four different T4-like Aeromonas salmonicida phages and one T4-like Acinetobacter johnsonii phage (manuscript in preparation). In all of these cases, the splits are located in an ~170-bp sequence within gene 43 that is highly diverged among the T4-like phages and that encodes the tip of the FINGERS domain of gp43 (Fig. 4, see color insert). The two cistrons, 43A and 43B, that result from each split configuration are translated separately into N-terminal (gp43A) and C-terminal (gp43B) subunits, respectively, that together correspond to the full-length of the typical single-subunit gp43 encoded by monocistronic forms of the gene. Figure 4 diagrams the putative gp43A and gp43B subunits of these split enzymes, using the RB69 gp43 structure shown in Fig. 1 as a framework. We note that in such split-gp43 forms the amino acid determinants for the POL function are located in both subunits, which would have to physically interact in order to form an RB69 gp43-like structure. It is not yet known if gp43A-gp43B interactions are mediated by the same factors that assist the folding of the single-subunit gp43 of RB69, T4, or other T4-like phages. It is also important to point out that the split-gene 43 variants from phages 25, 31, 65, 44RR, and 133 lack the type of RNA hairpin that serves as a translational target for gp43 in the T4, RB69, and RB49 biological systems (Fig. 3). In the case of phage 44RR, we have determined that the split-gp43 does not control its own translation (manuscript in preparation). These observations raise the possibility that some gp43 variants either lack RNA binding activity altogether or that have selected for other RNA ligands in evolution that control the activity, but not the biosynthesis of the enzyme. In the diagram we present in Fig. 4, we highlight gp43 sites (based on the RB69 gp43 structure, Fig. 1) that have been implicated in RNA binding in our previous studies [12]. Conceivably, specific RNA ligands bind to these sites and play a role in bringing gp43A and gp43B subunits together during assembly of the dimeric enzyme.
Fig. 4. A representation of the two gp43 subunits, gp43A and gp43B, of a split gp43. The two subunits are configured to match the RB69 gp43 structure depicted in Fig. 1. Folding of the gp43A and gp43B subunits into each other would reconstitute the POL active site of the enzyme.
DISCUSSION
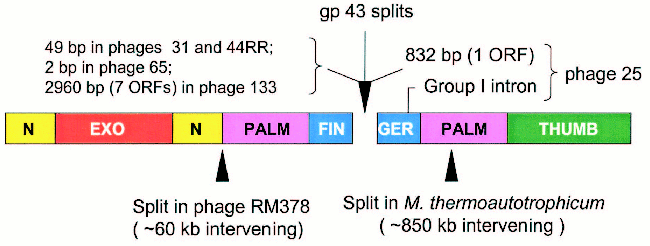
Figure 5 (see color insert) summarizes the characteristics of the regions that separate the 43A and 43B cistrons in the five cases encountered so far. As noted, the 43A-43B intercistronic sequences vary among these naturally occurring gene 43 configurations, probably reflecting their different evolutionary histories. In the configuration seen in Aeromonas phage 65, only two nucleotides separate the 43A and 43B reading frames from each other, suggesting that the split in phage 65 gene 43 originated through only a few base substitutions that created translation termination and re-initiation signals in the middle of a monocistronic progenitor. At the other extreme, the 43A and 43B cistrons of phage 133 are separated from each other by about 3000 bp of DNA that are predicted to encode as many as seven polypeptides of unknown function. Phages 44RR and 31 have identical splits, with separations of 50 bp between the two cistrons, and phage 25 contains a group 1 intron in the 43B cistron and the ORF for a homing endonuclease in the 43A-43B intercistronic region. That is, although the positions of the splits in gene 43 are closely similar among these phages, there is a high degree of diversity in intervening sequences between the separated components of the gene. These observations indicate that the modular organization of gp43 allows for the assembly of an active enzyme from independently synthesized structural domains.
Several examples of naturally occurring split DNA polymerases have been described in the literature [20-24], although the enzyme of Methanobacterium thermoautotrophicum deltaH is the only published pol B example outside the T4-like phages [22]. Another example, which is posted in the database at the National Center for Biotechnology Information (GenBank Ac. No. NC_004735), is the pol B enzyme of Rhodothermus marinus phage RM378. The positions of the methanobacterial and RM378 splits are entered in Fig. 5 to compare them to the positions of the gp43 splits in the five T4-like phages. The existence of split and fused variants in the same phylogenetic lineage for pol B enzymes probably attests to the adaptability of the modular structure of these DNA polymerases to the wide range of DNA rearrangements and other mutational events that can be imposed on protein structure by evolutionary processes. In the case of gene 43 of the T4-like phages, the highly divergent sequence that encodes the tip of the FINGERS domain (Fig. 1) seems to be an ideal target for illegitimate recombination, insertions, frameshift mutation, or other base-pair substitutions that might lead to translational termination to the upstream and new initiation signals to the downstream. Among the natural gene 43 splits, that we have observed (Fig. 5), the configuration seen in Aeromonas phage 65 involves closely linked translation termination-reinitiation signals that may have arisen from a small number of base-pair substitutions in the gp43 fingertip region of a monocistronic progenitor. The close genetic linkage between the 43A and 43B cistrons of the other three Aeromonas phages examined also suggests that these originated from monocistronic forms of gene 43. Sequence similarities among the split genes of these three phages and the presence of additional genetic elements in one of them (phage 25) are consistent with the notion that the fingertip-encoding segment of gene 43 is prone to invasion by mobile DNA.Fig. 5. Intervening sequences in split pol B genes. The diagram shows the positions of the gp43 splits that we have observed in five T4-like phages and compares these to the positions of other splits that have been observed in the pol B enzymes of R. marinus phage RM378 and M. thermoautotrophicum.
There are many examples of insertion elements, particularly mobile introns and inteins, in DNA polymerase genes in nature [25-31]. In the archaea, self-splicing inteins have been detected at three locations of pol B enzymes [25, 26, 32] that are within highly conserved sequence motifs of the PALM domain or PALM-FINGERS junction, where primer-template DNA is positioned for primer extension by the enzyme [8, 13]. Conceivably, such elements can mediate the translocation of DNA polymerase domains within a genome or between related genomes. The variable distances and sequences we observe between the 43A and 43B functional units of T4-related phages, also suggest that genetic information for gp43 modules can move around in a genome or between related genomes through standard biochemical mechanisms of DNA rearrangements. Such genetic rearrangements would create new environments for regulation of the genetic units that specify the structure of the multifunctional DNA polymerase. In the T4-like A. johnsonii phage 133, the R. marinus phage RM378, and the bacterium M. thermotrophicum, the pol B cistrons are separated from each other by several thousand base pairs of genomic DNA in each case (Fig. 5). So, genetic distance between complementing pol B cistrons is not a critical factor in determining the ability of the protein subunits to assemble into biologically active enzymes. However, it is not known if subunit-subunit interactions are aided by specific factors. In RB69 gp43, the determinants for specific mRNA binding map in segments of the protein that correspond to both the gp43A and gp43B components diagrammed in Fig. 4. Conceivably, the gp43A and gp43B subunits of some T4-like phages can also bind RNA, but that this ligand may serve as a facilitator of gp43A-gp43B interactions rather than as a regulator of protein synthesis. We are investigating this possibility.
This report is dedicated to the memory of Professor Boris F. Poglazov, D.Sc., whose leadership in the world of science inspired many of us. The work described here was supported by grants from the National Institutes of Health (GM54627) and National Science Foundation (MCB-138236) of the USA.
Addendum: We acknowledge the help received from Drs. Carine Desplat and Henry Krisch (CNRS, Toulouse, France) who shared with us sequence information from their studies of phage RB49 before publication (Desplats, C., et al., 2002, J. Bacteriol., 184, No. 10, 2789-2804) and an unpublished sequence from a 3´-terminal fragment of phage 44RR2.8t gene 43.
REFERENCES
1.Karam, J. D., and Konigsberg, W. H. (2000) in
Progress in Nucleic Acids Research and Molecular Biology
(Moldave, K., ed.) Vol. 64, Academic Press, San Diego, pp. 65-96.
2.Ackermann, H. W., and Krisch, H. M. (1997) Arch.
Virol., 142, 2329-2345.
3.Karam, J. D., et al. (eds.) (1994) Molecular
Biology of Bacteriophage T4, American Society for Microbiology,
Washington, DC.
4.Wang, C. C., Yeh, L. S., and Karam, J. D. (1995)
J. Biol. Chem., 270, 26558-26564.
5.Wang, C. C., Pavlov, A., and Karam, J. D. (1997)
J. Biol. Chem., 272, 17703-17710.
6.Spicer, E. K., Rush, J., Fung, C., Reha-Krantz, L.
J., Karam, J. D., and Konigsberg, W. H. (1988) J. Biol. Chem.,
263, 7478-7486.
7.Braithwaite, D. K., and Ito, J. (1993) Nucleic
Acids Res., 21, 787-802.
8.Wang, J., Sattar, A. K., Wang, C. C., Karam, J. D.,
Konigsberg, W. H., and Steitz, T. A. (1997) Cell, 89,
1087-1099.
9.Hopfner, K. P., Eichinger, A., Engh, R. A., Laue,
F., Ankenbauer, W., Huber, R., and Angerer, B. (1999) Proc. Natl.
Acad. Sci. USA, 96, 3600-3605.
10.Rodriguez, A. C., Park, H. W., Mao, C., and
Beese, L. S. (2000) J. Mol. Biol., 299, 447-462.
11.Zhao, Y., Jeruzalmi, D., Moarefi, I., Leighton,
L., Lasken, R., and Kuriyan, J. (1999) Structure Fold. Des.,
7, 1189-1199.
12.Petrov, V. M., Ng, S. S., and Karam, J. D. (2002)
J. Biol. Chem., 277, 33041-33048.
13.Franklin, M. C., Wang, J., and Steitz, T. A.
(2001) Cell, 105, 657-667.
14.Shamoo, Y., and Steitz, T. A. (1999) Cell,
99, 155-166.
15.Miller, E. S., Karam, J. D., and Spicer, E.
(1994) in Molecular Biology of Bacteriophage T4 (Karam, J. D.,
et al., eds.) American Society for Microbiolgy, Washington, DC, pp.
193-208.
16.Pavlov, A. R., and Karam, J. D. (2000) Nucleic
Acids Res., 28, 4657-4664.
17.Andrake, M. D., and Karam, J. D. (1991)
Genetics, 128, 203-213.
18.Tuerk, C., and Gold, L. (1990) Science,
249, 505-510.
19.Petrov, V. M., and Karam, J. D. (2002) Nucleic
Acids Res., 30, 3341-3348.
20.Gorbalenya, A. E. (1998) Nucleic Acids
Res., 26, 1741-1748.
21.Wu, H., Hu, Z., and Liu, X. Q. (1998) Proc.
Natl. Acad. Sci. USA, 95, 9226-9231.
22.Kelman, Z., Pietrokovski, S., and Hurwitz, J.
(1999) J. Biol. Chem., 274, 28751-28761.
23.Smith, D. R., Doucette-Stamm, L. A., Deloughery,
C., Lee, H., Dubois, J., Aldredge, T., Bashirzadeh, R., Blakely, D.,
Cook, R., Gilbert, K., et al. (1997) J. Bacteriol., 179,
7135-7155.
24.Huang, L., Ishii, K. K., Zuccola, H., Gehring, A.
M., Hwang, C. B., Hogle, J., and Coen, D. M. (1999) Proc. Natl.
Acad. Sci. USA, 96, 447-452.
25.Perler, F. B., Olsen, G. J., and Adam, E. (1997)
Nucleic Acids Res., 25, 1087-1093.
26.Perler, F. B. (2002) Nucleic Acids Res.,
30, 383-384.
27.Liu, X. Q. (2000) Annu. Rev. Genet.,
34, 61-76.
28.Goodrich-Blair, H., Scarlato, V., Gott, J. M.,
Xu, M. Q., and Shub, D. A. (1990) Cell, 63, 417-424.
29.Goodrich-Blair, H., and Shub, D. A. (1994)
Nucleic Acids Res., 22, 3715-3721.
30.Zhang, Y., Adams, B., Sun, L., Burbank, D. E.,
and van Etten, J. L. (2001) Virology, 285, 313-321.
31.Binas, M., and Johnson, A. M. (1998) Int. J.
Parasitol., 28, 1033-1040.
32.Cann, I. K., and Ishino, Y. (1999)
Genetics, 152, 1249-1267.