REVIEW: Cell-Cell Interactions in Bacterial Populations
S. A. Voloshin and A. S. Kaprelyants*
Bach Institute of Biochemistry, Russian Academy of Sciences, Leninsky pr. 33, Moscow 119071, Russia; fax: (7-095) 954-2732; E-mail: arseny@inbi.ras.ru* To whom correspondence should be addressed.
Received July 9, 2004
In developing bacterial populations many essential processes, such as division, genetic transformation, sporulation, and synthesis of antibiotics and secondary metabolites, are regulated by intercellular communication mediated by secretion of signaling molecules, such as homoserine lactones and peptides. Another intercellular communication type, namely a physical contact between cells (cell aggregation), plays a key role in formation of biofilms or cellular consortia and in cell proliferation under unfavorable conditions. The mechanisms involved in these two types of bacterial communication are discussed in this review.
KEY WORDS: intercellular interaction, bacteria, pheromone, aggregation
Abbreviations: HL) homoserine lactone; AHL) N-acyl homoserine lactone.
For a long time the ability of individual bacterial cells for autonomous
proliferation in culture hide the relevance of intercellular
communications for many life processes in bacterial populations. Now
the novel idea that a bacterial population is a community of
interacting cells with possible differentiation in this supracellular
“organism” succeeds the old view on bacterial culture as a
homogeneous soup in which individual cells live and proliferate
independently of each other [1]. So, bacterial
populations can be brought into comparison with tissues of higher
organisms by their organization. It is well known that in tissues cells
interact both physically to form tissue structure and via signaling
molecules, such as the well-known communication molecules (cytokines)
diffusing in the intercellular space. Now a sufficient body of
experimental data suggests that both mechanisms are active in
developing bacterial populations as well. The goal of our review is to
define circumstances and processes in which intercellular interactions
occur and to summarize the data on mechanisms of these interactions.
INTERCELLULAR INTERACTIONS MEDIATED BY CHEMICAL FACTORS
In the 1970s, A. S. Khokhlov discovered a small molecule, so-called A-factor, which is secreted by a streptomycete and, when it accumulates in the medium, induces both sporulation and antibiotic production by its own cells. This finding that previously seemed to be a curious but limited phenomenon (see below about the A-factor), but in last decade a growing number of studies have revealed a number of communicatory molecules secreted into the medium with biological effect similar to that of A-factor. A variety of bacterial living processes appeared to depend on those molecules. All these substances demonstrated similar mode of action: they cause specific changes in the bacterial metabolism when they reach a critical concentration in the medium (that is, at a definite density of producing cells). This principle called “quorum sensing” involves the action of various chemical substances including low molecular weight secondary metabolites, peptides, lipids, and secreted proteins.
To emphasize the similarity of these signaling molecules in prokaryotes and eukaryotes, the term bacterial pheromones is often used. Their difference from other metabolites is that they: 1) are produced by the organism itself; 2) have very low effective concentration; 3) do not require cleavage for expression of their activity, although conversion of propheromone to pheromone and inactivation via partial degradation are possible [2].
Bacterial pheromones exert control over many essential processes in prokaryotes including sporulation, conjugation, bioluminescence, virulence, cell division, etc. [2, 3]. Bacteria derive definite benefits from this intracellular regulation allowing them to survive by adaptive plasticity, differentiation (if necessary) into distinct cell types, or gene exchange depending on total density of the producing population. The latter, in turn, depends on how fully the ecological niche is supplying the given microorganism with substrates and other growth factors.
Despite significant diversity of all known bacterial pheromones comprising the “language” of bacteria, we can specify some general features. First, they regard the “languages” by which gram-positive and gram-negative bacteria communicate each other. Gram-negative cells use low molecular weight substances, in particular N-acyl homoserine lactones (AHLs) (table) as pheromones [3]. AHLs regulate a variety of processes, such as beta-lactam antibiotic production by Erwinia carotovora, intercellular DNA transfer, cell aggregation, capsule formation, protein secretion, biofilm formation, nodule formation by nitrogen-fixing bacteria, and so on [4]. A classical example of such processes is bioluminescence, which is widespread in nature, particularly in marine bacteria such as Vibrio fischeri. This process is under the control of an operon preferably encoding enzymes. The key enzyme is LuxI responsible for the synthesis of specific molecular inducer N-3-oxohexanoyl homoserine lactone [5]. At early stages of population growth the autoinducer production is scarce until a threshold is achieved, which is required for activation of the gene luxR initiating the entire operon for the synthesis of luciferase and other proteins providing bioluminescence. The gene luxI is also activated, so the inducer concentration dramatically increases, and the synthesis accelerates greatly as a positive feedback loop is established [4, 5]. Significantly, modules of luxI-luxR homologs and closely related chemical inducers operate in different control circuits governing various processes, such as synthesis of antibiotics by E. carotovora and Streptomyces virginiae, conjugation with plasmid transfer in Agrobacterium tumifaciens, and virulence development in Pseudomonas aeruginosa [6, 7].
Unlike gram-negatives, gram-positive bacteria “talk” with each other via secretion of either small (sometimes modified) oligopeptides or proteins (see table). The processes controlled by these pheromones include genetic competence, conjugation, sporulation, virulence, etc. Unlike AHLs, which freely penetrate both the outer and plasmatic membranes and reach their intracellular targets (LuxR homologs), oligopeptides do not transfer across membranes, and their receptors are localized on the outer surface of the plasma membrane. The receptors then transmit the signal inside the cell via a two-component system composed of membrane-associated protein kinase and regulatory protein (see below for details). Another regulatory pathway provides oligopeptide transport inside the cell, where the pheromone expresses its activity.
Signaling molecules (pheromones) of bacteria
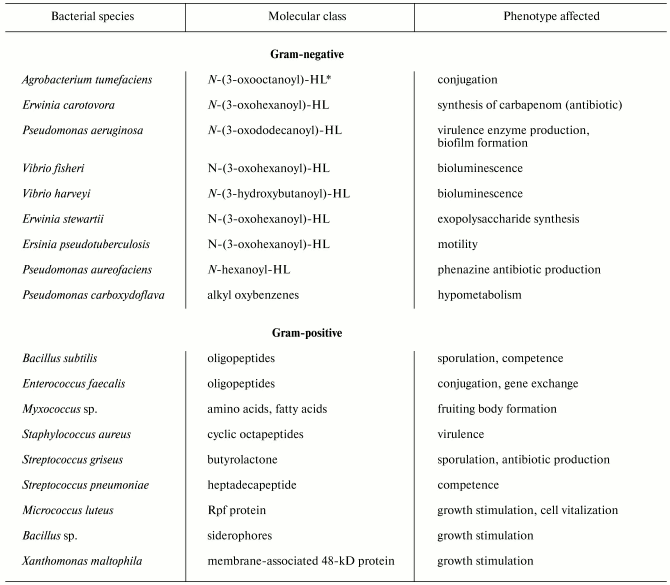
*HL, homoseryl lactone.
The difference between communication languages of gram-positives and gram-negatives is not always so great. Some bacteria use both languages. In particular, Vibrio harveyi has one AHL-based signaling system and another system using another effector, furanon, which is also secreted into the medium. Both systems are quorum-sensing and control bacterial bioluminescence [8]. In attempts to answer why bacteria have two systems, it was found that synthesis and secretion of furanons are much more widespread among bacteria than synthesis and secretion of AHLs. So, it has been hypothesized that furanons serve as a language for interspecific communication, whereas AHLs are rather intraspecific. Indeed, both gram-negative and gram-positive bacteria can synthesize furanons. This non-specific language “Furan” is supposedly used in bacterial communities, such as biofilms and consortia (in which V. harveyi often dwells). More specific AHL-based language is supposedly used in uniform bacterial populations [8].
Likewise, in cooperated formation of biofilms by P. aeruginosa and Bacillus cepatia the production of one autoinducer interferes with the production of another autoinducer, so that successful colonization of host organs is achieved in fibrosis [9]. Other cases are also reported, when a microorganism represses its neighbors belonging to other species due to secretion of enzymes inactivating their communicatory molecules [10].
We give below some well-understood examples of processes controlled by intercellular interactions via chemical signals.
Sporulation and competence in Bacillus subtilis. It is now well proven that secreted pheromones exert a quorum-sensing control over two very important processes in B. subtilis, namely competence, which is necessary for exogenous DNA uptake (transformation), and sporulation. In this case, peptides play the role of pheromones. In particular, the ComX pheromone responsible for competence is a decapeptide. The secreted pheromone binds to membrane-associated histidine protein kinase ComP, which is a ComX receptor. In turn, activated ComP undergoes autophosphorylation and transmits phosphate to the transcription factor ComA. Phosphorylated ComA activates a series of genes including the regulators comS, comK, and some other genes, which induce the genes encoding proteins responsible for transformation [11].
However, another backup mechanism was found in B. subtilis that also regulates competence and uses peptide pheromone. In this case, the bacteria produce and secrete the pentapeptide CSF (competence and sporulation factor). In spite of its small size, this peptide is multifunctional and fulfils at least three distinct functions. At low to medium concentration CSF is transported into the cytoplasm by the oligopeptide carrier Opp (oligopeptide permease), which is also known as ATP-binding cassette (ABC) transporter. In the cytoplasm, CSF induces the same genes participating in the first circuit of competence development (in particular, comS). The quorum-sensing mechanism is supposed to facilitate intraspecific gene exchange in these cases because the signal (pheromone) is species-specific, and DNA transfer increases with increase in culture density. At high to medium concentration, CSF both inhibits the expression of comS and induces sporulation. The latter process is associated with activation of the plasmatic phosphatase RapB. This protein dephosphorylates the phosphorylated factor SpoF involved in the regulatory cascade of sporulation. The factor SpoF transmits phosphate to the transcription factor SpoA, initiating sporulation processes. So, the same effector (pheromone) regulates two processes depending on its concentration in the medium. Although the role of signals for sporulation is not well understood, sporulation is supposed to be an advantageous strategy for survival at high cell density and nutritional deficiency [11].
The receptor-associated signaling of peptide type pheromones is rather universal among bacterial species and occurs at gene level via functioning of so-called peptide/kinase cassette. The above-mentioned B. subtilis competence regulation by the pheromone ComX involves expression of four tandemly ordered genes. The first gene encodes the protein ComQ involved in processing of ComX precursor (encoded by the second gene of the cassette), the third gene encodes the kinase/receptor ComP, and, lastly, the fourth gene encodes the transcription regulator ComA. The same kind of “signaling cassette” was found in Staphylococcus aureus, Streptococcus pneumoniae, and several other bacteria, in which it regulates not only competence, but some other processes [12, 13]. The second mechanism of signal transmission (transport of signaling peptide inside the cell) is also typical of gram-negative bacteria. For instance, plasmid transfer between the Enterococcus faecalis involves the intracellular transport of the peptide pheromone cCF10 secreted by recipient cells. This peptide penetrates the donor cell and finally stimulates both the conjugation protein synthesis and plasmid replication. Once the plasmid is transmitted from the donor to recipient cell, cCF10 blocks cell sensitivity to itself. So, the entire system including the extracellular pheromone provides the donor cell with ability to both detect the recipient cell and conjugate to it [14].
Regulatory role of A-factor in streptomycetes. Factor A is one of the butyryl lactones and the first autoregulator found in bacteria. It was discovered by Khokhlov and described as a secreted low molecular weight substance accumulating in cultures of streptomycin producer Streptomyces griseus. In a mutant S. griseus strain selected in Khokhlov's laboratory, a very small amount of A-factor (less than several nanomoles per liter) completely restored both streptomycin production and sporulation [15]. Since this pioneering discovery, similar autoregulators were also found in other streptomycetes producing antibiotics, such as virginiamycin and anthracyclins [16]. Like A-factor, these autoinducers are butyryl lactones. Their biosynthetic pathways in cells of streptomycetes are now well studied [16]. Very important progress in studies on signaling mechanism of these substances was achieved with discovery of their cellular receptor, whose existence was earlier postulated, because of very low actual concentration of the effector. The receptor of A-factor was found many years after the discovery of A-factor itself [17]. The receptor is a cytoplasmic homodimeric protein composed of two identical monomers, 276 amino acid residues each, encoded by the arpA gene. The spatial structure of this protein represents a pattern of alpha-helices interspersing non-ordered peptide regions, which is characteristic of DNA-binding proteins. The receptor binds to the adp (A-factor-dependent protein) gene promoter to repress transcription of this gene. Interaction of A-factor with its receptor results in release of the latter from the promoter site, thus initiating expression of adp, which in turn activates the genes strR and aphD controlling both streptomycin synthesis and resistance of the producer to this antibiotic. Unfortunately, our understanding of mechanisms providing A-factor-induced sporulation is still poor.
Cell division. A common notion is that convenient medium with corresponding set of nutrients, vitamins, and microelements and optimal physical conditions are sufficient for bacterial growth. Some microbiological methods (growth on solid media, dilutions to extinction, etc.) are based on the postulate that any viable cell can generate a population (either colony or liquid culture) of daughter cells in convenient medium with no requirement for specific growth factors [18]. However, there are experimental data indicating that bacterial growth is really under the control of autocrine growth factors secreted into the medium. Among these data, some peculiarities in behavior of bacteria in the growth lag-phase should be particularly mentioned. The number of viable cells in the inoculum is well known to influence the true lag-phase duration, thus indicating the presence of some inducers secreted by viable cells during lag-phase and accumulating in the medium [19]. However, the effect of inoculum amount on lag-phase duration can only be observed under certain circumstances. In particular, for Achromobacter delmarvae and Bacillus sp. [20] this effect was only observed in cultures growing in poor media. Interestingly, the effect of culture medium on the inoculum-dependent lag-phase was particularly expressed at minimum inoculum.
Many years of investigations into inoculum-dependent lag-phase have given little information about the chemical composition of secreted substances determining this phenomenon. Among few examples of substances whose structures are already determined, there are siderophores (schizokines), the iron carriers influencing inoculum-dependent lag-phase in Bacillus cultures [20], and N-(3-oxyhexanoyl)-homoseryl lactone, which addition results in significant shortening of lag-phase in Nitrosomonas europea cultures starved for six weeks before inoculation into fresh medium [21].
Bacterial sensitivity to some mammalian growth hormones may also provide indirect evidence for involvement of signaling molecules in cell division mechanisms of bacteria [22].
Rpf family proteins, the bacterial growth stimulators. It was found that growing Micrococcus luteus cells secrete a factor stimulating restoration of dormant micrococcal cells--Rpf (resuscitation promoting factor). The Rpf factor was purified to homogeneity and proved to be a protein with molecular weight of about 19 kD. At picomolar concentration Rpf caused several-order increase in number of cultured cells “vitalized” from the dormancy. Some time later, besides its resuscitating activity, the Rpf protein was found to play a more general role as a factor stimulating bacterial proliferation and so can be regarded as a “bacterial cytokine” [23]. In particular, Rpf stimulated proliferation of M. luteus cells either subjected to exhaustive washing on growing on very poor media. The gene encoding Rpf has been found and completely sequenced. This gene encodes a protein composed of 220 amino acid residues and carrying a leader N-terminal peptide (38 amino acid residues long) characteristic of gram-positive bacteria. The predicted mass of this protein is 19,148 daltons. Genes homologous to Rpf have been found in many microorganisms comprising a group of GC-rich bacteria, such as Mycobacterium tuberculosis (five genes), Mycobacterium leprae (two genes), Mycobacterium smegmatis (four genes), Mycobacterium bovis (BCG) (five genes), Corynobacterium glutamicum, and several species of the genus Streptomyces [23].
Like M. luteus Rpf, the Rpf family proteins express growth-stimulating activity towards their own producers, particularly when this growth was affected by some factors, such as in the case of continuously starved M. bovis cells [24]. Because of their very low actual concentration, the role of Rpf proteins in microbial growth is difficult to study; nevertheless, M. luteus null-mutants in the Rpf gene were selected, demonstrating the necessity in Rpf protein for the growth of micrococci [24]. The same results were found in experiments demonstrating the inhibitory effect of antibodies specific to the Rpf protein, which were raised against recombinant Rpf, on the growth of both M. luteus and M. tuberculosis [24]. Different Rpf forms obviously play transposable roles in the same microorganism, because unlike M. luteus possessing a single Rpf form, separate knockout of any Rpf form of M. tuberculosis has no effect on growth [25].
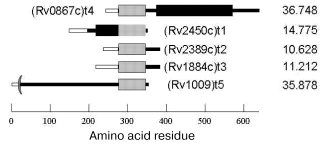
The spatial structure of Rpf family proteins predicted from their amino acid sequences is common for all members of this family. It contains three domains: a signal sequence responsible for both the protein transport across membrane and secretion and being cleaved in mature protein; a conservative domain, whose sequence is homologous through all the family; and a variable domain specific for each member (Fig. 1). The molecular weights of Rpf proteins vary from 10 to 30 kD.
The mechanism of the bacterial growth-stimulating effect of Rpf is still unclear. No functional similarity was revealed from direct comparison of Rpf primary structures with that of known proteins. However, recent reports suggest some kind of homology between the conservative domains of Rpf proteins and so-called lysozyme fold characteristic of several lytic enzymes involved in bacterial cell wall metabolism [26]. So, Rpf might to be an enzyme, whose actual mechanism is someway connected with enzymatic modification of the cell wall. Interestingly, the early spore-activating processes are associated with induction of lytic trans-glycosylases, the enzymes involved in cell wall remodeling and, like Rpf, structurally related to lysozyme [27].Fig. 1. Rpf family proteins of Mycobacterium tuberculosis: signal sequence (light rectangles), conservative Rpf domain (gray rectangles), proline-rich segment (black rectangles). On the right, molecular mass in daltons.
CELL-CELL INTERACTIONS MEDIATED BY PHYSICAL FACTORS
(INTERCELLULAR CONTACTS)
Bacterial cells growing in liquid media undergo regular physical interactions with each other, and when grow on solid media are in tight contact. In some species, such as myxobacteria, cell aggregation is essential for morphogenesis and differentiation. In particular, Myxococcus xanthus under improper conditions forms a multicellular fruiting body containing spores [28]. This differentiation is controlled by chemical signals, whose secretion is stage-specific. Among these signaling substances, the amino acid “cocktail” (A-factor), a hydrophobic protein with molecular weight of 17 kD (C-factor), and still unidentified E-factor were found. The A-factor is synthesized at high culture density in response to starvation, and both C- and E-factors are synthesized at later stages of fruiting body formation and sporulation. The C-factor is a membrane-associated protein providing the low-distance interaction of the cells comprising the formed fruiting body via binding with a specific surface receptor. At present, the genes involved in this complex process, as well as distinct sequences of regulatory events resulting in cell differentiation, are well understood [29].
More complex bacterial cell aggregates composed of several species, such as consortia of methanogenous and methylotrophic bacteria [30, 31] are also found in natural habitats. This kind of cooperation is beneficial for bacteria because the products secreted by one species can serve as substrates for another species. Sometimes such consortia form complex structures in which tens of microbial species are ordering in a multilayer pattern. These so-called bacterial mats can often contain algal and cyanobacterial cells producing organic substances. Some observations are indicative of bacterial cell aggregation induced and stimulated by certain environmental changes in active mud [32].
Biofilms. Biofilms are cell aggregates attached to various biotic and abiotic surfaces and represent one of the best-studied forms of bacterial associations [33].
Biofilms are formed as either mono- or multispecific biocenoses, in which the cells interact with each other to adapt themselves to varying environmental conditions. The role of biofilm formation is of particular importance for medical microbiology, because the cells comprising biofilms compared to free-living ones can hundredfold increase their resistance to antibiotics [34]. Multispecific biofilms dominate in many natural habitats, whereas monospecific biofilms are preferably found in organs of higher animals subjected to infection; for instance, Pseudomonas aeruginosa forms biofilms in the lung of infected humans. Along with P. aeruginosa, Vibrio cholerae is best studied among gram-negatives in regards to biofilm formation in infection [33]. Among gram-positives, these are Staphylococcus epidermidis, Staphylococcus aureus, and some enterococci [33]. In bacteria, biofilm formation and progression seem to be initiated by changes in environment, such as variations in substrate availability [33]. In particular, E. coli cells only form biofilm while the medium level of substrate is sufficient and become free-living when it drops. Thus, substrate availability can switch the developmental cycle in bacterial population between biofilm and free-living form [33]. The biofilms formed by P. aeruginosa are the best studied because of their particular importance for medicine. They are 3-D-structured and composed of microcolonies, in which some portion of cells may undergo lysis, which in turn is necessary for successful development of the biofilm [35]. Moreover, the quorum-sensing mechanism was found to govern biofilm development in this bacterium [34]. The quorum-sensing gene-deficient mutants could only form unusual mushroom-like bodies rather than normal biofilms. The progress achieved in our understanding of biofilms suggests that the normal process of biofilm formation requires bacterial adhesion to surface, movement along it, and ability to form 3-D structure [33]. Nonetheless, current studies are focused on medical and biotechnological problems, and the role of biofilms and other cell associations for bacteria themselves is not so well understood.
Bacterial colonies. The bacterial colony in which the cells are tightly associated is the most common example of bacterial aggregates a microbiologist deals with. The colony is a complex structure composed of nonuniform cells [1]. In E. coli colonies, a vertical stratification is well developed with distinct cell layers differently susceptible to vital staining [36, 37].
Many researchers have observed a system of air-containing cavities intercepted with cellular bulks inside the colonies [38]. This complex system eventually turns a colony into aggregate of partially separated nodes of condensation (microcolonies). The microcolonies consist of mucous matrix and are separated by open (often containing water) channels. It is a primitive analog of the circulatory system, which serves for supply with nutrients and removal of metabolites [38]. In colonies formed by the Alcaligenes sp. strain d2, pores and channels were found, as well as more specialized structures surrounded by singular “membrane” and containing extracellular heme proteins. This kind of structures seem to facilitate the transport of oxygen to the cells comprising colonies (aggregates), thus playing the role of a respiratory system [39, 40].
Both gram-positive and gram-negative bacteria form specific films of rather complex structure on the surface of their colonies [41]. In colonies of gram-negatives this film is a thin membrane, whereas the “skin” coating colonies of gram-positive bacteria displays three-layer structure with an intramural membrane and two adjoining amorphous layers inward and outward from it [41]. In colonies formed by Neisseria gonorrhoea the cells are connected by pili to form 3-D structure [42]. In some genera, such as Escherichia, Shigella, and Salmonella, the cells develop two types of joints [43]. The first is characterized by very tight adhesion with each cell interacting with several others to form 3-D structure; the second is an intercellular bridge between outer membranes of neighboring cells to form channels [43]. In colonies of gram-positives, such as Staphylococcus and Brevibacterium, the intercellular joints were also found, but they are structurally different from both types described above: they are formed due to integration of mureinic layers of two adjacent cells [43].
Bacterial aggregation in liquid media. Some of scientists suggest that many bacterial species maintain their “colonial” type of organization in liquid media as well [1]. A growing bulk of evidence indicates that bacteria need cell-cell communication, which is achieved via direct contact of neighboring cells, for their growth in liquid media. In some bacteria cell, aggregation is governed by specific signaling molecules secreted into the medium. In particular, the above-described processes associated with gene exchange (and correspondingly with cell aggregation) are controlled by quorum-sensing-type secretion of factors. In Lactobacillus acidophilus a group of proteins was studied, that are expressed on the surface of the cell wall and are involved in cell aggregation [44].
The extracellular matrix composed predominantly of polysaccharides also plays an important role in formation of cell aggregates. Cells secrete an exopolysaccharide-protein complex to encase themselves in a capsule of complex exopolymer structure and to form intercellular matrix [45]. Sophisticated capsules composed of several exopolysaccharides were found in streptococci and pneumococci [46]. Butterfield has isolated a bacterium Zoogloea ramigera from the active mud, which secretes gelatinous matrix coating the cells and forming flocculi [47]. It was also reported that poly-beta-oxobutyrate accumulation in Zoogloea ramigera cells correlates with cell aggregation in culture [48, 49], but it remains unclear whether these two processes are directly associated. Cell aggregation is not always associated with extracellular matrix formation. Many bacteria, such as Flavobacterium, can aggregate without formation of matrix or capsule [50]. Some data are indicative of positive effect of enhanced capsule growth on cell aggregation [51]. However, aggregation of some gram-negative cocci could only begin when the cells were artificially released from their capsules [52]. Some bacteria can produce exopolymers without visible capsule formation [45]. These capsule-lacking bacteria can also aggregate, possibly due to formation of cell-cell bridges consisting of some kind of polymer(s). A series of species belonging to the genera Nocardia, Proactinomyces, and Corynebacterium was found to develop structures resembling plasmodesma (structures characteristic of plant cells), which serve for connection of cells in aggregates [53]. Stanley and Rose have studied a series of Corynebacterium xerosis strains and concluded that aggregation is provided by surface proteins, whereas disaggregation is caused by proteolytic enzymes (resembling papain, which completely disintegrates aggregates) [54]. The same was observed in preparations of isolated cell walls. In Micrococcus halodenitrificans and Vibrio costicolus growing in nutritionally deficient media, DNA plays a role of connector: it is secreted into the medium to maintain cell aggregates [55]. A similar phenomenon was described in Pasteurella (Yersinia) pestis [56]. The cells of Staphylococcus aureus secrete excessive amounts of RNA and proteins participating in cell aggregation [57].
Some members of a group of GC-rich bacteria, such as nocardii, streptomycetes, and mycobacteria, are characterized by continuous formation of cell associates varying in size and differentiation, which occurs even in nutritionally optimized cultures. The cell aggregates formed by Mycobacterium bovis are encased in a specific envelope consisting of proteins, carbohydrates, and acidic residues, but not lipids, and are possibly associated with virulence of pathogenic strains [58].
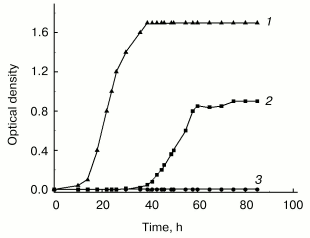
Although in this group of bacteria as well as in many other species cell aggregation to associates is well established, its biological role still remains unclear. Our own studies of GC-rich Rhodococcus rhodochrous were undertaken to elucidate this problem. These studies revealed the necessity of cell-cell contacts for normal growth under stress conditions. In poor medium the growth of R. rhodochrous depends on agitation--the cells were unable to grow in flasks subjected to vigorous agitation, whereas moderate agitation resulted in expressed growth of the culture (Fig. 2). This phenomenon of growth arrest is reversible, independent of aeration, and only confined to lag-phase (becomes invisible in the exponential phase) and to nutritionally deficient media. Moderately agitated cells form associates in lag-phase followed by transition to individual cells in the exponential phase. The inhibitory effect of vigorous agitation on the cell growth can be eliminated by addition of logarithmic phase supernatant of the same culture grown at moderate agitation. We hypothesize that cell-cell contacts are necessary for R. rhodochrous cell growth in poor medium, and vigorous agitation breaks them. Our data draw a parallel between bacterial growth under given conditions and micro(cryptic) growth within cell associate [59]. These peculiarities in R. rhodochrous growth are possibly indicative of a specific strategy which bacteria use for their growth and survival under conditions providing no chance for their autonomous growth, but allowing the growth of cells demonstrating “social behavior” (that is, joining into associates).
We are thankful to the Program “Molecular and Cell Biology” of the Russian Academy of Sciences, the Russian Foundation for Basic Research (project No. 03-04-49044), and the International Scientific and Technical Center (project No. 2201) for financial support.Fig. 2. Effect of agitation on the growth of Rhodococcus rhodochrous cells in poor (synthetic) and rich media: 1) vigorous agitation (250 rpm), rich medium (Broth E); 2) moderate agitation (100 rpm), synthetic medium (Saton's medium); 3) vigorous agitation (250 rpm), synthetic medium (Saton's medium). Cells were grown in Erlenmeyer flasks at 37°C.
REFERENCES
1.Oleskin, A. B., Botvinko, I. V., and Tsavkelova, E.
A. (2000) Microbiology (Moscow), 69, 249-265.
2.Kell, D. B., Kaprelyants, A. S., and Grafen, A.
(1995) Trends Ecol. Evol., 10, 126-129.
3.Williams, P., Baldwin, T. J., and Downie, J. A.
(1999) in Microbial Signalling and Communication (England, R.,
Hobbs, G., Bainton, N., and Roberts, D., eds.) Cambridge University
Press, Cambridge, pp. 1-32.
4.Greenberg, E. (1999) in Microbial Signalling and
Communication (England, R., Hobbs, G., Bainton, N., and Roberts,
D., eds.) Cambridge University Press, Cambridge, pp. 70-84.
5.Fuqua, C., Winans, S., and Greenberg, E. P. (1996)
Ann. Rev. Microbiol., 50, 727-751.
6.Salmond, G. P. C., Bycroft, B. W., Stewart, G., and
Williams, P. (1995) Mol. Microbiol., 16, 615-624.
7.Swift, S., Throup, J. P., Williams, P., Salmond, G.
P. C., and Stewart, G. S. A. B. (1996) Trends Biochem. Sci.,
21, 214-219.
8.Shauder, S., and Bassler, B. (2001) Genes
Dev., 15, 1468-1480.
9.Lewenza, S., Conway, B., Greenberg, E., and Sokol,
P. (1999) J. Bacteriol., 181, 748-756.
10.Dong, Y., Xu, J., Li, X., and Zhang, L. (2000)
Proc. Natl. Acad. Sci. USA, 97, 3526-3531.
11.Lazazzera, B., and Grossman, A. (1998) Trends
Microbiol., 6, 288-294.
12.Ji, G. Y., Beavis, R. C., and Novick, R. P.
(1995) Proc. Natl. Acad. Sci. USA, 92, 12055-12059.
13.Pestova, E. V., Havarstein, L. S., and Morrison,
D. (1996) Mol. Microbiol., 21, 853-862.
14.Dunny, G. M., Leonard, B. A. B., and Hedberg, P.
J. (1995) J. Bacteriol., 177, 871-876.
15.Khokhlov, A. S. (1988) Low-Molecular-Weight
Microbial Autoregulators [in Russian], Nauka, Moscow.
16.Yamada, Y. (1999) in Microbial Signalling and
Communication (England, R., Hobbs, G., Bainton, N., and Roberts,
D., eds.) Cambridge University Press, Cambridge, pp. 177-196.
17.Miyake, K., Kuzuyama, T., Horinouchi, S., and
Beppu, T. (1990) J. Bacteriol., 172, 3003-3008.
18.Postgate, J. (1967) in Advances in Microbial
Physiology (Rose, A. H., and Wilkinson, J., eds.) Vol. 1, Academic
Press, London, pp. 1-21.
19.Hinshelwood, C. N. (1946) The Chemical
Kinetics of the Bacterial Cell, The Clarendon Press, Oxford, pp.
49-53.
20.Solomon, J., Magnuson, R., Srivastava, A., and
Grossman, A. (1995) Genes Dev., 9, 547-558.
21.Batchelor, S. E., Cooper, M., Chhabra, S. R.,
Glover, L. A., Stewart, G. S. A. B., Williams, P., and Prosser, J. I.
(1997) Appl. Environ. Microbiol., 63, 2281-2286.
22.Kaprelyants, A. S., and Kell, D. B. (1996)
Trends Microbiol., 4, 237-242.
23.Mukamolova, G. V., Kaprelyants, A. S., Young, D.
I., Young, M., and Kell, D. B. (1998) Proc. Natl. Acad. Sci.
USA, 95, 8916-8921.
24.Mukamolova, G. V., Turapov, O. A., Young, D.
I., Kaprelyants, A. S., Kell, D. B., and Young, M. (2002) Mol.
Microbiol., 46, 623-635.
25.Tufariello, J., Jacobs, W., and Chan, J. (2004)
Infect. Immun., 72, 515-526.
26.Cohen-Gonsaud, M., Keep, N., Davies, A., Ward,
J., Henderson, B., and Labesse, G. (2004) Trends Biochem. Sci.,
29, 7-10.
27.Atrith, A., and Foster, S. (1999) Antonie Van
Leeuwenhoek Int. J., 75, 299-307.
28.Kaiser, D., and Losick, R. (1993) Cell,
73, 873-885.
29.Kaiser, D. (1999) in Microbial Signalling and
Communication (England, R., Hobs, G., Bainton, N., and Mcl.
Roberts, D., eds.) Cambridge University Press, Cambridge, pp.
138-160.
30.Oleskin, A. V., and Samuilov, V. D. (1994)
Technological Bioenergetics [in Russian], Moscow State
University, Moscow, p. 136.
31.Boetius, A., Ravenschlag, K., Schubert, C. J.,
Rickert, D., Widdel, F., Gieseke, A., Amann, R., Jorgensen, B. B.,
Witte, U., and Pfannkuche, O. (2000) Nature, 407,
623-626.
32.Bossier, P., and Verstraete, W. (1996) Appl.
Microbiol. Biotechnol., 45, 1-6.
33.O'Toole, G., Kaplan, H. B., and Kolter, R. (2000)
Ann. Rev. Microbiol., 54, 49-79.
34.Greenberg, E. P. (2003) Nature,
424, 134.
35.Webb, S. J., Thompson, S. J., Charlton, T.,
Tolker-Neilsen, T., Koch, B., Givskov, M., and Kjelleberg, S. (2003)
J. Bacteriol., 185, 4585-4592.
36.Kuznetsov, O. Yu. (1988) in Electron
Microscopy for the Studies on Induced Functional Changes in Cell
Structure [in Russian], Moscow State University Press, Moscow, pp.
89-92.
37.Shapiro, J. A. (1994) Sci. Progr.,
76, 399-424.
38.Costerton, J. W., Lewandowski, Z., Caldwell, D.
E., Korber, D. R., and Lappin-Scott, H. M. (1995) Ann. Rev.
Microbiol., 49, 711-745.
39.Duda, V. I., Vypov, M. G., Sorokin, V. V.,
Mityushina, L. L., and Lebedinskii, A. V. (1995) Mikrobiologiya,
64, 69-73.
40.Duda, V. I., Il'chenko, A. P., Dmitriev, V. V.,
Shorokhova, A. P., and Suzina, N. E. (1998) Microbiology
(Moscow), 67, 7-12.
41.Tetz, V. V., Rybalchenko, O. V., and Savkova, G.
A. (1993) J. Gen. Microbiol., 139, 855-858.
42.Todd, W. J., Wray, G. P., and Hitchcock, P. J.
(1984) J. Bacteriol., 159, 312-320.
43.Tetz, V. V., Rybalchenko, O. V., and Savkova, G.
A. (1990) J. Basic Microbiol., 30, 597-607.
44.Kos, B., Suskovic, J., Vukovic, S., Simpraga, M.,
Frece, J., and Matosic, S. (2003) J. Appl. Microbiol.,
94, 981-987.
45.Harris, R. H., and Mitchell, R. (1973) Ann.
Rev. Microbiol., 27, 27-50.
46.Scott, J. E., Thomlinson, A. M., and Prehm, P.
(2003) Exp. Cell Res., 285, 1-8.
47.Butterfield, C. T. (1935) US Pub. Health
Rep., 50, 671.
48.Crabtree, K., McCoy, E., Boyle, W. C., and
Rohlich, G. A. (1965) Appl. Microbiol., 13, 218-226.
49.Crabtree, K., Boyle, W. C., McCoy, E., and
Rohlich, G. A. (1966) J. Water Pollut. Contr. Fed., 38,
1968-1980.
50.Tezuka, Y. (1969) Appl. Microbiol.,
17, 222-226.
51.McKinney, R. E. (1952) Sew. Ind. Wastes,
24, 280-287.
52.Juni, E., and Heym, G. A. (1964) J.
Bacteriol., 87, 461-467.
53.Clark, J. B. (1958) J. Bacteriol.,
75, 400-402.
54.Stanley, S. O., and Rose, A. H. (1967) J. Gen.
Microbiol., 48, 9-23.
55.Smithies, W. R., and Gibbons, N. E. (1955)
Can. J. Microbiol., 1, 614-621.
56.Wessman, G. E., and Miller, D. J. (1966) Appl.
Microbiol., 14, 636-642.
57.Cripps, R. E., and Work, E. (1967) J. Gen.
Microbiol., 49, 127-137.
58.Devadoss, P., Klegerman, M. E., and Groves, M. J.
(1991) Microbios, 65, 111-125.
59.Voloshin, S. A., Shleeva, M. O., and
Kaprel'yants, A. S. (2005) Microbiology (Moscow), in press.