Can a Biologist Fix a Radio? -- or, What I Learned while Studying Apoptosis
Y. Lazebnik
Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724, USA; E-mail: lazebnik@cshl.org
This article by Yu. Lazebnik, "Can a Biologist Fix a Radio? -- or, What I Learned while Studying Apoptosis" has already been published in English (Cancer Cell, 2002, 2, 179-182) and in Russian (Uspekhi Gerontologii, 2003, No. 12, 166-171). Nevertheless, we have undertaken its secondary publication in our journal for two reasons: first, our journal has different readers, and, second, the great significance of this manifest of Yuri Lazebnik. The author in bright and clever form shows the emerging necessity to create formalized language designed to describe complicated systems of regulation of biochemical processes in living cells. The article is published with permission of Cancer Cell and Uspekhi Gerontologii.Editor in Chief of Biokhimiya/Biochemistry (Moscow) V. P. Skulachev
As a freshly minted Assistant Professor, I feared that everything in my field would be discovered before I even had a chance to set up my laboratory. Indeed, the field of apoptosis, which I had recently joined, was developing at a mind-boggling speed. Components of the previously mysterious process were being discovered almost weekly, frequent scientific meetings had little overlap in their contents, and it seemed that every issue of Cell, Nature, or Science had to have at least one paper on apoptosis. My fear led me to seek advice from David Papermaster (currently at the University of Connecticut), who I knew to be a person with pronounced common sense and extensive experience. David listened to my outpouring of primal fear and explained why I should not worry.
David said that every field he witnessed during his decades in biological research developed quite similarly. At the first stage, a small number of scientists would somewhat leisurely discuss a problem that would appear esoteric to others, such as whether cell cycle is controlled by an oscillator or whether cells can commit suicide. At this stage, the understanding of the problem increases slowly, and scientists are generally nice to each other, a few personal antipathies notwithstanding. Then, an unexpected observation, such as the discovery of cyclins or the finding that apoptosis failure can contribute to cancer, makes many realize that the previously mysterious process can be dissected with available tools and, importantly, that this effort may result in a miracle drug. At once, the field is converted into a Klondike gold rush with all the characteristic dynamics, mentality, and morals. A major driving force becomes the desire to find the nugget that will secure a place in textbooks, guarantee an unrelenting envy of peers, and, at last, solve all financial problems. The assumed proximity of this imaginary nugget easily attracts both financial and human resources, which results in a rapid expansion of the field. The understanding of the biological process increases accordingly and results in crystal clear models that often explain everything and point at targets for future miracle drugs. People at this stage are not necessarily nice, though, as anyone who has read about a gold rush can expect. This description fit the then current state of the apoptosis field rather well, which made me wonder why David was smiling so reassuringly. He took his time to explain.
At some point, David said, the field reaches a stage at which models, that seemed so complete, fall apart, predictions that were considered so obvious are found to be wrong, and attempts to develop wonder drugs largely fail. This stage is characterized by a sense of frustration at the complexity of the process, and by a sinking feeling that despite all that intense digging the promised cure-all may not materialize. In other words, the field hits the wall, even though the intensity of research remains unabated for a while, resulting in thousands of publications, many of which are contradictory or largely descriptive. The flood of publications is explained, in part, by the sheer amount of accumulated information (about 10,000 papers on apoptosis were published yearly over the last few years), which makes reviewers of the manuscripts as confused and overwhelmed as their authors. This stage can be summarized by the paradox that the more facts we learn the less we understand the process we study.
It becomes slowly apparent that even if the anticipated gold deposits exist, finding them is not guaranteed. At this stage, the Chinese saying that it is difficult to find a black cat in a dark room, especially if there is no cat, comes to mind too often. If you want to continue meaningful research at this time of widespread desperation, David said, learn how to make good tools and how to keep your mind clear under adverse circumstances. I am grateful to David for his advice, which gave me hope and, eventually, helped me to enjoy my research even after my field did reach the state he predicted.
At some point, I began to realize that David's paradox has a meaning that is deeper than a survival advice. Indeed, it was puzzling to me why this paradox manifested itself not only in studies of fundamental processes, such as apoptosis or cell cycle, but even in studies of individual proteins. For example, the mystery of what the tumor suppressor p53 actually does seems only to deepen as the number of publications about this protein rises above 23,000.
The notion that your work will create more confusion is not particularly stimulating, which made me look for guidance again. Joe Gall at the Carnegie Institution, who started to publish before I was born, and is an author of an excellent series of essays on history of biology [1], relieved my mental suffering by pointing out that a period of stagnation is eventually interrupted by a new development. As an example, he referred to the studies of cell death that took place in the 19th century, faded into oblivion, and re-emerged a century later with about 60,000 studies on the subject published during a single decade. Even though a prospect of a possible surge in activity in my field was relieving, I started to wonder whether anything could be done to expedite this event, which brought me to think about the nature of David's paradox. The generality of the paradox suggested some common fundamental flaw of how biologists approach problems.
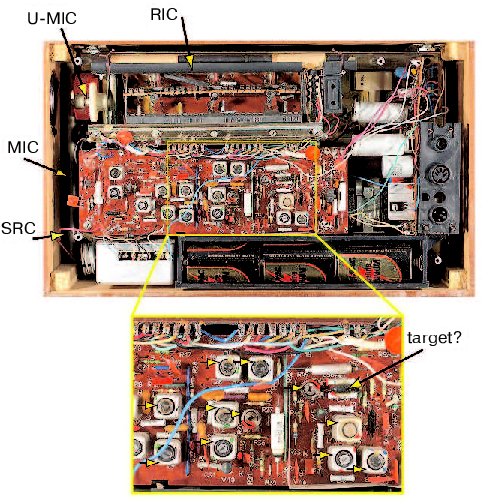
To understand what this flaw is, I decided to follow the advice of my high school mathematics teacher, who recommended testing an approach by applying it to a problem that has a known solution. To abstract from peculiarities of biological experimental systems, I looked for a problem that would involve a reasonably complex but well understood system. Eventually, I thought of the old broken transistor radio that my wife brought from Russia (Fig. 1, see color insert). Conceptually, a radio functions similarly to a signal transduction pathway in that both convert a signal from one form into another (a radio converts electromagnetic waves into sound waves). My radio has about a hundred various components, such as resistors, capacitors, and transistors, which is comparable to the number of molecules in a reasonably complex signal transduction pathway. I started to contemplate how biologists would determine why my radio does not work and how they would attempt to repair it. Because a majority of biologists pay little attention to physics, I had to assume that all we would know about the radio is that it is a box that is supposed to play music.
How would we begin? First, we would secure funds to obtain a large supply of identical functioning radios in order to dissect and compare them to the one that is broken. We would eventually find how to open the radios and will find objects of various shape, color, and size (Fig. 2, see color insert). We would describe and classify them into families according to their appearance. We would describe a family of square metal objects, a family of round brightly colored objects with two legs, round-shaped objects with three legs and so on. Because the objects would vary in color, we will investigate whether changing the colors affects the radio's performance. Although changing the colors would have only attenuating effects (the music is still playing but a trained ear of some people can discern some distortion), this approach will produce many publications and result in a lively debate.Fig. 1. The radio that has been used in this study.
A more successful approach will be to remove components one at a time or to use a variation of the method, in which a radio is shot at a close range with metal particles. In the latter case, radios that malfunction (have a “phenotype”) are selected to identify the component whose damage causes the phenotype. Although removing some components will have only an attenuating effect, a lucky postdoc will accidentally find a wire whose deficiency will stop the music completely. The jubilant fellow will name the wire Serendipitously Recovered Component (SRC) and then find that SRC is required because it is the only link between a long extendable object and the rest of the radio. The object will be appropriately named the Most Important Component (MIC) of the radio. A series of studies will definitively establish that MIC should be made of metal and the longer the object is the better, which would provide an evolutionary explanation for the finding that the object is extendable.Fig. 2. The insides of the radio. See text for description of the indicated components. The inset is an enlarged portion of the radio. The horizontal arrows indicate tunable components.
However, a persistent graduate student from another laboratory will discover another object that is required for the radio to work. To the delight of the discoverer, and the incredulity of the flourishing MIC field, the object will be made of graphite and changing its length will not affect the quality of the sound significantly. Moreover, the graduate student would convincingly demonstrate that MIC is not required for the radio to work, and will suitably name his object the Really Important Component (RIC). The heated controversy, as to whether MIC or RIC is more important, will be fueled by the accumulating evidence that some radios require MIC while other, apparently identical ones, need RIC. The fight will continue until a smart postdoctoral fellow will discover a switch, whose state determines whether MIC or RIC is required for playing music. Naturally, the switch will become the Undoubtedly Most Important Component (U-MIC). Inspired by these findings, an army of biologists will apply the pull-it-out approach to investigate the role of each and every component. Another army will crush the radios into small pieces to identify components that are on each of the pieces, thus providing evidence for interaction between these components. The idea that one can investigate a component by cutting its connections to other components one at a time or in a combination (“alanine scan mutagenesis”) will produce a wealth of information on the role of the connections.
Eventually, all components will be catalogued, connections between them will be described, and the consequences of removing each component or their combinations will be documented. This will be the time when the question, previously obscured by the excitement of productive research, would have to be asked: Can the information that we accumulated help us to repair the radio? It will turn out that sometimes it can, such as if a cylindrical object that is red in a working radio is black and smells like burnt paint in the broken radio (Fig. 2, inset, a component indicated as a target). Replacing the burned object with a red object will likely repair the radio.
The success of this approach explains the pharmaceutical industry's mantra: “Give me a target!”. This mantra reflects the belief in a miracle drug and assumes that there is a miracle target whose malfunction is solely responsible for the disease that needs to be cured.
However, if the radio has tunable components, such as those found in my old radio (indicated by yellow arrows in Fig. 2, inset) and in all live cells and organisms, the outcome will not be so promising. Indeed, the radio may not work because several components are not tuned properly, which is not reflected in their appearance or their connections. What is the probability that this radio will be fixed by our biologists? I might be overly pessimistic, but a textbook example of the monkey that can, in principle, type a Burns poem comes to mind. In other words, the radio will not play music unless that lucky chance meets a prepared mind.
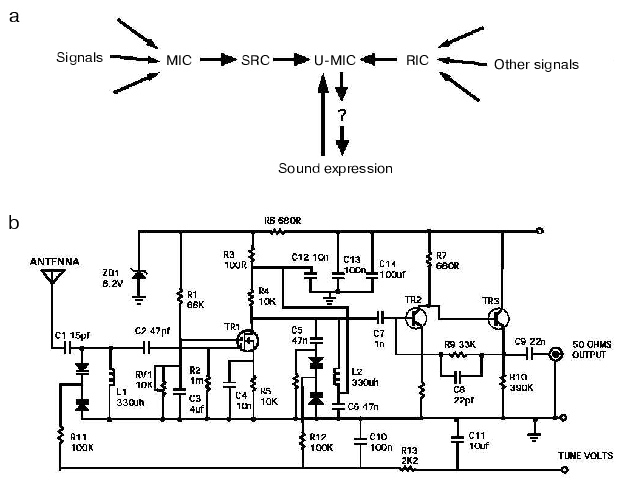
Yet, we know with near certainty that an engineer, or even a trained repairman could fix the radio. What makes the difference? I think the languages that these two groups use (Fig. 3, see color insert). Biologists summarize their results with the help of all-too-well recognizable diagrams, in which a favorite protein is placed in the middle and connected to everything else with two-way arrows. Even if a diagram makes overall sense (Fig. 3a), it is usually useless for a quantitative analysis, which limits its predictive or investigative value to a very narrow range. The language used by biologists for verbal communications is not better and is not unlike that used by stock market analysts. Both are vague (e.g., “a balance between pro- and anti-apoptotic bcl-2 proteins appears to control the cell viability, and seems to correlate in long-term with the ability to form tumors”) and avoid clear predictions.
These description and communication tools are in a glaring contrast with the language that has been used by engineers (compare Figs. 3a and 3b). Because the language (Fig. 3b) is standard (the elements and their connections are described according to invariable rules), any engineer trained in electronics would unambiguously understand a diagram describing the radio or any other electronic device. As a consequence, engineers can discuss the radio using terms that are understood unambiguously by the parties involved. Moreover, the commonality of the language allows engineers to identify familiar patterns or modules (a trigger, an amplifier, etc.) in a diagram of an unfamiliar device. Because the language is quantitative (a description of the radio includes the key parameters of each component, such as the capacity of a capacitor, and not necessarily its color, shape or size) it is suitable for a quantitative analysis, including modeling.Fig. 3. The tools used by biologists and engineers to describe processes of interest: a) the biologist view of a radio. See Fig. 2 and text for description of the indicated components; b) the engineer view of a radio (please note that the circuit diagram presented is not that of the radio used in the study; the diagram of the radio was lost, which, in part, explains why the radio remains broken).
I would like to argue that the absence of such language is the flaw of biological research that causes David's paradox. Indeed, even though the impotence of purely experimental approaches might be a bit exaggerated in my radio metaphor, it is common knowledge that the human brain can keep track of only so many variables. It is also common experience that once the number of components in a system reaches a certain threshold, understanding the system without formal analytical tools requires geniuses, who are so rare even outside biology. In engineering, the scarcity of geniuses is compensated, at least in part, by a formal language that successfully unites the efforts of many individuals, thus achieving a desired effect, be that design of a new aircraft or of a computer program. In biology, we use several arguments to convince ourselves that problems that require calculus can be solved with arithmetic if one tries hard enough and does another series of experiments.
One of these arguments postulates that the cell is too complex to use engineering approaches. I disagree with this argument for two reasons. First, the radio analogy suggests that an approach that is inefficient in analyzing a simple system is unlikely to be more useful if the system is more complex. Second, the complexity is a term that is inversely related to the degree of understanding. Indeed, the insides of even my simple radio would overwhelm an average biologist (this notion has been proven experimentally), but would be an open book to an engineer. The engineers seem to be undeterred by the complexity of the problems they face and solve them by systematically applying formal approaches that take advantage of the ever-expanding computer power. As a result, such complex systems as an aircraft can be designed and tested completely in silico, and computer-simulated characters in movies and video games can be made so eerily life-like. Perhaps, if the effort spent on formalizing description of biological processes would be close to that spent on designing video games, the cells would appear less complex and more accessible to therapeutic intervention.
A related argument is that engineering approaches are not applicable to cells because these little wonders are fundamentally different from objects studied by engineers. What is so special about cells is not usually specified, but it is implied that real biologists feel the difference. I consider this argument as a sign of what I call the urea syndrome because of the shock that the scientific community had two hundred years ago after learning that urea can be synthesized by a chemist from inorganic materials. It was assumed that organic chemicals could only be produced by a vital force present in living organisms. Perhaps, when we describe signal transduction pathways properly, we would realize that their similarity to the radio is not superficial. In fact, engineers already see deep similarities between the systems they design and live organisms [2].
Another argument is that we know too little to analyze cells in the way engineers analyze their systems. But, the question is whether we would be able to understand what we need to learn if we do not use a formal description. The biochemists would measure rates and concentrations to understand how biochemical processes work. A discrepancy between the measured and calculated values would indicate a missing link and lead to the discovery of a new enzyme, and a better understanding of the subject of investigation. Do we know what to measure to understand a signal transduction pathway? Are we even convinced that we need to measure something? As Sydney Brenner noted, it seems that biochemistry disappeared in the same year as communism [3]. I think that a formal description would make the need to measure system's parameters obvious and would help to understand what these parameters are.
An argument that is usually raised privately is why to bother with all these formal languages if one can make a living by continuing with purely experimental research that took years to learn. There are at least two reasons. One is that formal approaches would make our research more meaningful, more productive and might indeed lead to miracle drugs. A more immediate reason is that formal approaches may become a basic part of biology sooner than we, experimental biologists, expect. This transition may be as rapid as that from slides to PowerPoint presentations, a change that forced some graphics designers to learn how to use a computer and put others out of work.
Of course, a plea for a formal approach in biology is not new. The general systems theory, developed by Ludwig von Bertalanffy because of his fascination with the complexity of live organisms, was formulated 60 years ago, as well as his concept of organisms as physical systems [4]. Bertalanffy's fundamental studies have been followed by several attempts to approach cells as systems, the latest of which, system biology, has been rapidly developing into an active field [5-11]. Available computer power and advances in analysis of complex systems raise hope that this time the system approach will change from an esoteric tool that is considered useless by many experimental biologists, to a basic and indispensable approach of biology.
The question is how to facilitate this change, which is not exactly welcomed by many experimental biologists, to put it mildly [12]. Learning computer programming was greatly facilitated by BASIC, a language that was not very useful to solve complex problems, but was very efficient in making one comfortable with using a computer language and demonstrating its analytical power. Similarly, a simple language that experimental scientists can use to introduce themselves to formal descriptions of biological processes would be very helpful in overcoming a fear of long-forgotten mathematical symbols. Several such languages have been suggested [13, 14] but they are not quantitative, which limits their value. Others are designed with modeling in mind but are too new to judge as to whether they are user-friendly [15]. However, I hope that it is only a question of time before a user-friendly and flexible formal language will be taught to biology students, as it is taught to engineers, as a basic requirement for their future studies. My advice to experimental biologists is to be prepared.
REFERENCES
1.Gall, J. G. (1996) Views of the Cell. A
Pictorial History, The American Society of Cell Biology,
Bethesda.
2.Csete, M. E., and Doyle, J. C. (2002)
Science, 295, 1664-1669.
3.Brenner, S. (1995) Curr. Biol., 5,
332.
4.Von Bertalanffy, L. (1969) General System
Theory, Revised Edn., George Braziller, New York.
5.Bhalla, U. S., and Iyengar, R. (1999)
Science, 283, 381-387.
6.Bhalla, U. S., Ram, P. T., and Iyengar, R. (2002)
Science, 297, 1018-1023.
7.Bray, D. (1995) Nature, 376,
307-312.
8.Davidson, E. H., Rast, J. P., Oliveri, P., Ransick,
A., Calestani, C., Yuh, C. H., Minokawa, T., Amore, G., Hinman, V.,
Arenas-Mena, C., et al. (2002) Dev. Biol., 246,
162-190.
9.Guet, C. C., Elowitz, M. B., Hsing, W., and
Leibler, S. (2002) Science, 296, 1466-1470.
10.Hartwell, L. H., Hopfield, J. J., Leibler, S.,
and Murray, A. W. (1999) Nature, 402, C47-52.
11.Schoeberl, B., Eichler-Jonsson, C., Gilles, E.
D., and Muller, G. (2002) Nat. Biotechnol., 20,
370-375.
12.Bray, D. (2001) Nature, 412,
863.
13.Kohn, K. W. (1999) Mol. Biol. Cell,
10, 2703-2734.
14.Pirson, I., Fortemaison, N., Jacobs, C., Dremier,
S., Dumont, J. E., and Maenhaut, C. (2000) Trends Cell. Biol.,
10, 404-408.
15.Maimon, R., and Browning, S. (2001)
Diagrammatic Notation and Computational Structure of Gene
Networks, Proc. 2nd Int. Conf. Systems Biology.
http://www.icsb-2001.org/Papers/21_Maimon_Paper.pdf