Analysis of Interactions of DNA Polymerase beta and Reverse Transcriptases of Human Immunodeficiency and Mouse Leukemia Viruses with dNTP Analogs Containing a Modified Sugar Residue
N. A. Lebedeva1, T. A. Seredina2, V. N. Silnikov1, T. V. Abramova1, A. S. Levina1, S. N. Khodyreva1, N. I. Rechkunova1, and O. I. Lavrik1*
1Institute of Chemical Biology and Fundamental Medicine, Siberian Division of the Russian Academy of Sciences, pr. Lavrentieva 8, 630090 Novosibirsk, Russia; fax: (3832) 333-677; E-mail: lavrik@niboch.nsc.ru2Novosibirsk State University, ul. Pirogova 2, 630090 Novosibirsk, Russia
* To whom correspondence should be addressed.
Received August 25, 2004; Revision received September 29, 2004
Substrate properties of various morpholinonucleoside triphosphates in the reaction of DNA elongation catalyzed by DNA polymerase beta, reverse transcriptase of human immunodeficiency virus (HIV-1 RT), and reverse transcriptase of Moloney murine leukemia virus (M-MuLV RT) were compared. Morpholinonucleoside triphosphates were utilized by DNA polymerase beta and HIV-1 reverse transcriptase as substrates, which terminated further synthesis of DNA, but were virtually not utilized by M-MuLV reverse transcriptase. The kinetic parameters of morpholinoderivatives of cytosine (MorC) and uridine (MorU) were determined in the reaction of primer elongation catalyzed by DNA polymerase beta and HIV-1 reverse transcriptase. MorC was a more effective substrate of HIV-1 reverse transcriptase and significantly less effective substrate of DNA polymerase beta than MorU. The possible use of morpholinonucleoside triphosphates as selective inhibitors of HIV-1 reverse transcriptase is discussed.
KEY WORDS: viral reverse transcriptases, terminating substrates, DNA polymerase beta, selective inhibition of viral DNA polymerases
Reverse transcriptases, which stimulate rapid incorporation of viruses into eukaryotic cells, can be selectively suppressed with dNTP analogs containing a modified ribose residue. Such analogs terminate the reaction of DNA elongation due to introduction of the dNMP residue modified at the terminal 3´-hydroxyl of the growing DNA chain. In particular, this principle is used in creating preparations, which suppress the development of human immunodeficiency virus. These preparations include nucleotide analogs with modified ribose, such as azidothymidine and acyclovir [1]. It has been found that dNTP containing a modified ribose residue can be easily utilized by DNA polymerase beta, which is a key DNA polymerase of base excision repair (BER), and this results in inhibition of BER system and seems to promote the development of side effects [2, 3]. Studies on the interaction of DNA polymerases with new dNTP analogs are very interesting because these analogs are promising as terminators of DNA synthesis and, thus, effective inhibitors of viral polymerases. The inhibition of the DNA synthesis catalyzed by DNA polymerase beta by new analogs might not be too strong, and this would be of help in creating effective and less toxic inhibitors. A number of morpholinonucleoside triphosphates were studied in the present work as possible new inhibitors. Their substrate properties in the reaction of DNA elongation catalyzed by DNA polymerase beta and reverse transcriptases of human immunodeficiency and mouse leukemia viruses were compared. The purpose of this work was to find inhibitors effectively suppressing the DNA synthesis catalyzed by viral polymerases and less affecting the repair synthesis of DNA.
MATERIALS AND METHODS
Non-radioactive dNTP and T4 polynucleotide kinase from SibEnzyme (Russia), [gamma32-P]ATP from Biosan (Russia), and reagents for electrophoresis and buffer components from Sigma (USA) were used. Other reagents were of special purity or chemical purity qualification.
Recombinant proteins DNA polymerase beta and HIV-1 reverse transcriptase were isolated from Escherichia coli as described in [4, 5].
Reverse transcriptase of Moloney murine leukemia virus (M-MuLV) lacking the RNase H domain was kindly presented by Biosan company. Before use, this reverse transcriptase was additionally purified on a column with phosphocellulose P-11 to remove minor admixtures. The preparation of reverse transcriptase (15 mg, 3.5 mg/ml in buffer containing 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 5 mM dithiothreitol, 50% glycerol, and 0.1% NP-40) was fivefold diluted in buffer A (50 mM Tris-HCl, pH 8.0, 0.1% NP-40, 1 mM EDTA, 10 mM beta-mercaptoethanol) and placed onto a column with phosphocellulose (3 ml) equilibrated with 50 mM NaCl in buffer A. The column was washed with 10 ml of 0.1 M NaCl in buffer A. The protein was eluted with a linear concentration gradient of NaCl in buffer A (0.1-0.6 M; the total volume of the gradient was 40 ml). The purity of the enzyme was assessed by Laemmli electrophoresis in polyacrylamide gel [6]. Reverse transcriptase was eluted with 0.29 M NaCl. Fractions containing the homogenous enzyme were combined and dialyzed against buffer A and then against the storage buffer: 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 5 mM dithiothreitol, 50% glycerol, and 0.1% NP-40.
Morpholinonucleoside triphosphates were synthesized as described in [7].
Oligonucleotides used in this work were synthesized by Eurogentec (Belgium):
primer 5´-GGCGATTAAGTTGGG-3´.
templates 5´-GGAAGACCCTGACGTTACCCAACTTAATCGCC-3´,
5´-GGAAGACCCTGACGTTGCCCAACTTAATCGCC-3´.
The 5´-end of the oligonucleotide was radiolabeled as described in [8]. The labeled oligonucleotides were purified by electrophoresis as described in [9]. The primer and template were hybridized at equimolar ratio by heating at 95°C for 5 min and following slow cooling to room temperature.
Synthesis of DNA catalyzed by various DNA polymerases. The radiolabeled primer was elongated with DNA polymerase beta or with HIV-1 or M-MuLV reverse transcriptases in 10 µl of reaction mixture containing standard buffer components (50 mM Tris-HCl, pH 8.6 at 25°C, 50 mM NaCl, 5 mM MgCl2 or 1.5 mM MnCl2), 0.05 µM complex 5´-P32-labeled primer-template, 0.1 µM DNA polymerase (one of them), and 100 µM terminator or mixture of all terminators (100 µM each), or mixture of the natural dNTP (10 µM each). The mixtures were incubated for 45 min at 37°C. The reaction was stopped by addition of 5 µl of 0.1% Bromophenol Blue and 50 mM EDTA in 90% formamide. The samples were heated for 3 min at 95°C. The products were separated by electrophoresis in 20% polyacrylamide gel (acrylamide/bis-acrylamide 20 : 1) with 8 M urea. Electrophoresis was performed in 50 mM Tris-borate buffer (pH 8.3).
Determination of kinetic parameters. Kinetic parameters for the terminating substrates in the reaction catalyzed by DNA polymerase beta or HIV-1 reverse transcriptase were determined in 10 µl of the reaction mixture containing standard buffer components (50 mM Tris-HCl, pH 8.6 at 25°C, 50 mM NaCl, 5 mM MgCl2), 0.05 µM duplex 5´-32P-labeled primer-template, 0.1 µM DNA polymerase (one of them), and 0.05-10 µM of one of the terminators. The reaction samples were treated as in the previous case. The quantities of the initial and of the one step elongated primer were determined by scanning of the gels with a phosphorimager (Molecular FX; BioRad, USA). Values of kinetic parameters (Km, Vmax, kcat) were determined from the Michaelis-Menten equation based on experimental data using the Microcal Origin program (Microcal Software, USA).
RESULTS AND DISCUSSION
Analogs of dNTP containing the modified ribose residue, morpholinonucleoside triphosphates (terminating substrates), which were prepared as analogs of natural deoxyribonucleotides, are especially interesting due to inexpensive starting reagents and low toxicity [10, 11]. Such derivatives were first obtained many years ago, but they did not become widely used as terminators of DNA synthesis catalyzed by DNA polymerases [12]. It was earlier shown that the morpholinothymidine triphosphate derivative is a terminating substrate of the thermophilic Taq DNA polymerase and is specifically incorporated into positions complementary to adenine. This was associated with termination of the further synthesis of the chain, i.e., this analog seemed to be a suitable substrate for creating a DNA-sequencing set [13].
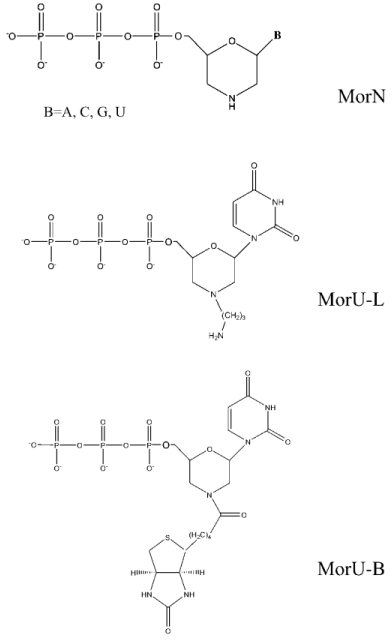
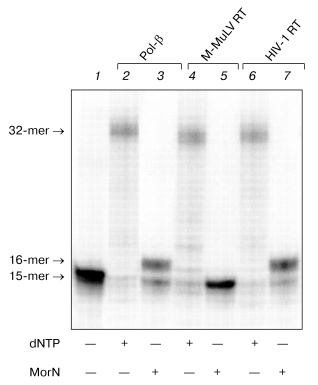
We have studied the substrate properties of some new dNTP analogs containing the morpholino residue instead of deoxyribose (MorA, MorC, MorG, and MorU) in the reaction of DNA primer elongation catalyzed by various DNA polymerases including reverse transcriptases of human immunodeficiency and mouse leukemia viruses (HIV-1 RT and M-MuLV RT, respectively). The structures of these new dNTP analogs are shown in Fig. 1. Figure 2 shows that in the presence of all four natural nucleoside-5´-triphosphates all used DNA polymerases generate a radiolabeled product, which corresponds in length to the product of the primer completion (lanes 2, 4, 6). However, in the presence of all four morpholinonucleoside triphosphates, the initial DNA primer is elongated by only one step in the case of DNA polymerase beta and HIV-1 reverse transcriptase (lanes 3, 7), which suggests termination of the reaction of the DNA chain elongation. With M-MuLV reverse transcriptase, no reaction of initial primer elongation was observed (lane 5). It should be noted that increase in the concentration of morpholinonucleoside triphosphates as well as changes in the experimental conditions did not result in the primer elongation. Thus, these dNTP analogs were not substrates of M-MuLV RT. However, they were competitive inhibitors of this enzyme (data not presented). Consequently, M-MuLV RT interacted with morpholinonucleoside triphosphates but did not utilize them as substrates for the reaction of elongation.
Fig. 1. Structural formulas of dNTP analogs.
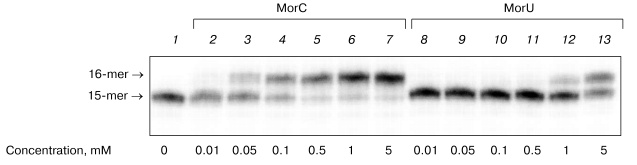
To determine the optimal conditions for the reaction of DNA elongation, the ability of HIV-1 reverse transcriptase for incorporating morpholinonucleoside residues (MorU and MorC) complementary to the template base depending on the enzyme and substrate concentrations and also on the reaction duration was studied. Figure 3 shows results of electrophoretic separation of products of the reaction catalyzed by HIV-1 RT in the presence of different concentrations of the terminators. One can see that the 32P-labeled primer in the presence of MorC was elongated with virtually every used concentration of the dNTP analog (from 0.05 to 5 mM). Under conditions chosen for the reaction, only decrease in the MorC concentration to 10 µM resulted in disappearance of the primer elongation products (Fig. 3, lane 2). In the presence of MorU, the primer elongation was observed only at high concentration of the substrate beginning from 1 mM (Fig. 3, lane 12). Note that increase in the HIV-1 RT concentration to 1 µM and in the reaction duration caused the appearance of products of the primer 3´-end elongation even at 10 µM substrate (MorC).Fig. 2. Electrophoretic separation of products of reactions catalyzed by DNA polymerase beta and M-MuLV and HIV-1 reverse transcriptases. Lane 1 corresponds to the primer-template complex. The reaction mixtures containing 0.05 µM duplex 32P-labeled primer-template, mixture of analogs (0.1 mM each), mixture of dNTPs (0.01 mM each), the enzymes Pol-beta, HIV-1 RT, or M-MuLV RT, and standard buffer components (see “Materials and Methods”) were incubated for 45 min at 37°C. Conditions of the separation: 20% polyacrylamide gel (acrylamide/bis-acrylamide 20 : 1) with 8 M urea.
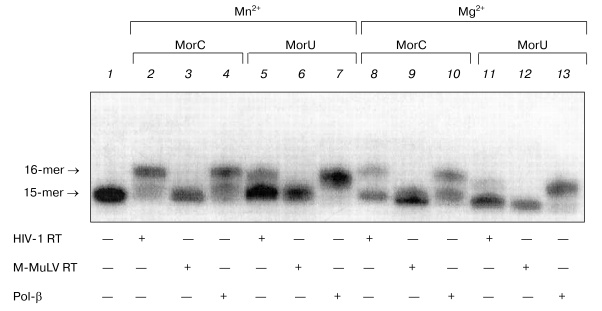
It is known that substitution of the bivalent magnesium cation by manganese is associated with increase in the number of errors in the DNA replication catalyzed by DNA polymerases from various sources [14-23]. Moreover, in the presence of manganese ions T7 DNA polymerase, Taq DNA polymerase, and DNA polymerase I of E. coli more effectively utilize dideoxyribonucleoside-5´-triphosphates as substrates [24, 25]. Therefore, it was interesting to compare the effect of manganese ions on the interaction of the terminating substrates with HIV-1 and M-MuLV reverse transcriptases to the effect on DNA polymerase beta. Figure 4 shows that the substitution of magnesium by manganese increases the efficiency of incorporation of dNTP analogs in the case of DNA polymerase beta and HIV-1 reverse transcriptase (Fig. 4, lanes 2, 4, 5, 7). In the presence of manganese ions, HIV-1 reverse transcriptase utilizes MorU even at the concentration of 0.5 mM (lane 5). However, in the case of M-MuLV RT residues of the terminating substrates were not incorporated into the 3´-end of the primer even in the presence of manganese ions (Fig. 4, lanes 3, 6).Fig. 3. Electrophoretic separation of products of reactions catalyzed by HIV-1 reverse transcriptases in the presence of different concentrations of MorC and MorU. Lane 1 corresponds to the primer-template complex. The reaction mixtures containing 32P-labeled primer-template, 0.1 µM HIV-1 RT, MorC, MorU, and buffer (see “Materials and Methods”) were incubated at 37°C for 45 min. Conditions of the separation: 20% polyacrylamide gel (acrylamide/bis-acrylamide 20 : 1) with 8 M urea.
To study in more detail the interaction of dNTP with DNA polymerases in the polymerase reaction, the kinetic parameters (Km and Vmax) of the elongation reaction were quantitatively determined for the terminating substrates in the presence of the primer-template complex. The Km and Vmax values were determined for MorC and MorU as substrates of the reaction of the primer elongation catalyzed by DNA polymerase beta and HIV-1 reverse transcriptase. Values of kinetic parameters Km and Vmax of the elongation reaction were determined in the presence of MgCl2. The Km and Vmax values are presented in the table.Fig. 4. Effect of Mn2+ on the substrate properties of MorC and MorU in reactions catalyzed by Pol-beta, HIV-1 RT, or M-MuLV RT. Lane 1 corresponds to the primer-template complex. The reaction mixtures containing 32P-labeled primer-template, MorC, MorU, Pol-beta, HIV-1 RT, M-MuLV RT, and buffer (see “Materials and Methods”) were incubated at 37°C for 45 min. Conditions of the separation: 20% polyacrylamide gel (acrylamide/bis-acrylamide 20 : 1) with 8 M urea.
Values of kinetic parameters Km and
Vmax for the terminating substrates MorC (numerator)
and MorU (denominator) in the reaction catalyzed by HIV-1 reverse
transcriptase as compared to DNA polymerase beta

On certain assumptions, the value of the Michaelis constant characterizes the affinity of the substrate for the enzyme. This value is useful for assessing tightness of the substrate-enzyme complex [26]. The table shows that in the case of HIV-1 RT, the Km value for MorU is an order higher than for MorC, whereas Vmax value is lower; thus, MorU is a less effective substrate for HIV-1 RT than MorC. However, MorC is utilized by Pol-beta worse than MorU, and this seems to provide its sparing effect on the repair synthesis of DNA.
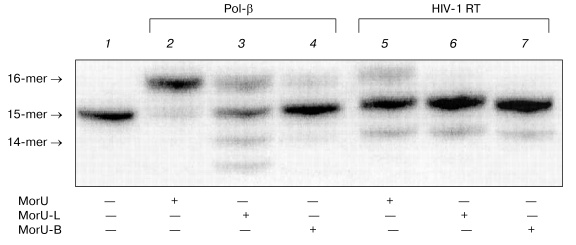
Then the sensitivity of DNA polymerases to modification of ribose was studied using analogs with different substitutes in the morpholino residue. Synthesis of these dNTP analogs will be described elsewhere. The structural formulas are presented in Fig. 1. Compounds were prepared which contained a hydrocarbon linker with NH2 group bound to the nitrogen atom in the morpholino residue (MorU-L); another analog contained the biotin residue (MorU-B). DNA polymerase beta effectively utilized the analog without additional modifications as a substrate (Fig. 5, lane 2), whereas the primer was 50% elongated in the case of the analog with the hydrocarbon linker MorU-L (Fig. 5, lane 3) and only 10% in the case of MorU-B (lane 4). In the case of HIV-1 RT, any modification of the morpholino residue eliminated the substrate properties of a dNTP analog in the reaction of primer elongation (lanes 6, 7).
Thus, based on the findings, we conclude that the most effective terminating substrate of HIV-1 reverse transcriptase is MorC, which seems promising as a specific inhibitor of this enzyme. Substrates with bulky substituents in the deoxyribose residue can be used only for the directed inhibition of the DNA synthesis catalyzed by Pol-beta. The directed inhibition of the activity of DNA polymerase beta is also interesting for practice because of the increase in the concentration of this enzyme observed in proliferating cells [27].Fig. 5. Substrate properties of analogs containing different modifications of the morpholino residue. Lane 1 corresponds to the primer-template complex. The reaction mixtures containing 32P-labeled primer-template, MorU, MorU-L, MorU-B, Pol-beta, HIV-1 RT, and buffer (see “Materials and Methods”) were incubated at 37°C for 45 min. Conditions of the separation: 20% polyacrylamide gel (acrylamide/bis-acrylamide 20 : 1) with 8 M urea.
Then we studied the resistance of DNA containing on the 3´-end residues terminating the further synthesis against the 3´-->5´ exonuclease activities concomitant with DNA polymerases. Such an analysis is essential for searching for the most effective and stable pathways for inhibition of the virus-induced DNA synthesis. In fact, the 3´-->5´ exonuclease cleavage of the 3´-end terminating residue can completely neutralize its inhibiting effect on synthesis of viral DNA.
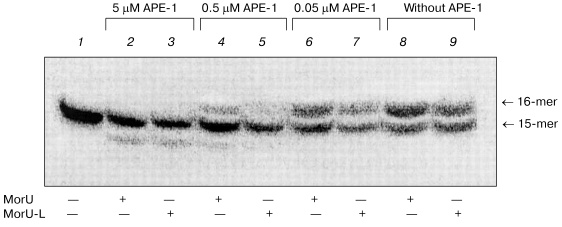
Apurinic/apyrimidinic endonuclease 1 (APE-1) is a protein interacting with intermediates of base excision repair. APE-1 displayed 3´-->5´ exonuclease activity [28], which manifested itself stronger in the case of misincorporation of the dNMP residue into the 3´-end of DNA [29]. DNA polymerase beta is known to interact with APE-1 during base excision repair [30]. Consequently, the 3´-->5´ exonuclease activity of APE-1 can contribute to accuracy of the repair synthesis, and this is very important because Pol-beta lacks the correcting 3´-->5´ exonuclease activity but is characterized by frequent errors in the replication of DNA. APE-1 can remove from the 3´-end of the primer residues of antiviral nucleoside analogs, such as 3´-azido-3´-deoxythymidine, 3´-dideoxythymidine, etc. [29]. Therefore, the ability of APE-1 to remove residues of the dNTP analogs under consideration was studied after their incorporation into the 3´-end of the primer catalyzed by DNA polymerase beta. The results are shown in Fig. 6. After elongation with DNA polymerase beta of the 32P-labeled primer in the presence of one of the dNTP analogs, the reaction mixtures were heated at 95°C for 5 min to inactivate Pol-beta and then cooled to continue the reaction of exonuclease cleavage. The efficiency of removing of the 3´-terminal residues of dNTP analogs was tested at varied concentrations of APE-1. The 3´-terminal residue of the dNTP analog containing the hydrocarbon linker with NH2 group bound to the nitrogen atom in the morpholino residue (MorU-L) was removed more efficiently than the residue MorU without modifications in the morpholino residue (Fig. 6, compare lanes 4 and 5, 6 and 7). At high concentration of APE-1 (5 µM), along with removing of residues of dNTP analogs, the initial primer was degraded (Fig. 6, lanes 2, 3), and this is in agreement with our earlier data [31]. Thus, it is concluded that morpholinonucleoside triphosphates terminate the DNA synthesis catalyzed by DNA polymerase beta, but the associated inhibition of the repair synthesis can be not very significant because APE-1 removes the terminating residue of the nucleotide due to its 3´-->5´ exonuclease activity. Due to effective cooperation with DNA polymerase beta [30, 32], APE-1 can provide a sparing effect of such inhibitors on the repair synthesis concurrently with the effective inhibition of the DNA synthesis catalyzed by HIV-1 reverse transcriptase.
This work was supported by the Russian Foundation for Basic Research (the project Nos. 02-04-48404, 04-04-48525).Fig. 6. 3´-->5´ exonuclease excision of modified dNMP residues by APE-1. Lane 1, primer-template complex. After inactivation of Pol-beta._APE-1 was added into reaction mixtures at the designated concentration.
REFERENCES
1.Ostrovskii, V. A. (1999) Soros Educational
J., 9, 44-51.
2.Copeland, W. C., Chen, M. S., and Wang, T. S.
(1992) J. Biol. Chem., 267, 21459-21464.
3.Garcia-Diaz, M., Bebenek, K., Sabariegos, R.,
Dominguez, O., Rodriguez, J., Kirchhoff, T., Garcia-Palomero, E.,
Picher, A. J., Juarez, R., Ruiz, J. F., Kunkel, T. A., and Blanco, L.
(2002) J. Biol. Chem., 277, 13184-13191.
4.Drachkova, I. A., Petruseva, I. O., Safronov, I.
V., Zakharenko, A. L., Shishkin, G. V., Lavrik, O. I., and Khodyreva,
S. N. (2001) Bioorg. Khim., 27, 179-204.
5.Martin-Hernandez, A. M., Domingo, E., and
Menendez-Arias, L. (1996) EMBO J., 15, 4434-4442.
6.Laemmli, U. K. (1970) Nature, 227,
680-685.
7.Abramova, T. V., Bakharev, P. A., Vasilyeva, S. V.,
and Silnikov, V. N. (2004) Tetrahedron Lett., 45,
4361-4364.
8.Yamshchikov, V. F. (1990) in Methods of
Molecular Genetics and Genetic Engineering (Salganik, R. I., ed.)
[in Russian], Nauka, Moscow, pp. 112-114.
9.Maniatis, T. (1989) Molecular Cloning: a
Laboratory Manual, Cold Spring Harbor University Press.
10.Summerton, J., and Weller, D. (1997) Antisense
Nucleic Acids Drug. Devel., 7, 187-195.
11.Summerton, J. (1999) Biochim. Biophys.
Acta, 1489, 141-158.
12.Erlanger, B. F., and Beiser, S. M. (1964)
Proc. Natl. Acad. Sci. USA, 52, 68-74.
13.Marciacq, F., Sauvaigo, S., Issartel, J.-P.,
Mouret, J.-F., and Molko, D. (1999) Tetrahedron Lett.,
40, 4673-4676.
14.Beckman, R. A., Mildvan, A. S., and Loeb, L. A.
(1985) Biochemistry, 27, 546-553.
15.Copeland, W. C., Lam, N. K., and Wang, T. S.
(1993) J. Biol. Chem., 268, 11041-11049.
16.El-Deiry, W. S., Donwey, K. M., and So, A.
G. (1984) Proc. Natl. Acad. Sci. USA, 81, 7378-7382.
17.El-Deiry, W. S., So, A. G., and Donwey, K. M.
(1988) Biochemistry, 24, 5810-5817.
18.Eger, B. T., and Benkovic, S. J. (1992)
Biochemistry, 31, 9227-9236.
19.Goodman, M. F., Keener, S., Guidotti, S. E., and
Branscomb, W. (1983) J. Biol. Chem., 258, 3496-3475.
20.Pelletier, H., Sawaya, M. R., Wolfle, W., Wilson,
S. H., and Kraut, J. (1996) Biochemistry, 35,
12762-12777.
21.Riccetti, M., and Buc, H. (1993) EMBO J.,
12, 387-396.
22.Seal, G., Sheaman, C. W., and Loeb, L. A. (1979)
J. Biol. Chem., 254, 5229-5237.
23.Sirover, M. A., and Loeb, L. A. (1977) J.
Biol. Chem., 252, 3605-3610.
24. Tabor, S., and Richardson, C. C. (1989) Proc. Natl. Acad.
Sci. USA, 86, 4076-4080.
25. Brandis, J. W., Edwards, S. G., and Johnson, K. A. (1996)
Biochemistry, 35, 2189-2200.
26.Rechkunova, N. I., Okhapkina, S. S., Anarbaev, R.
O., Lokhova, I. A., Degtyarev, S. Kh., and Lavrik, O. I. (2000)
Biochemistry (Moscow), 65, 609-614.
27.Servant, L., Bieth, A., Hayakawa, H., Cazaux, C.,
and Hoffmann, J. S. (2002) J. Mol. Biol., 315,
1039-1047.
28.Demple, B., Herman, T., and Chen, D. S. (1991)
Proc. Natl. Acad. Sci. USA, 88, 11450-11454.
29.Chou, K. M., and Cheng, Y. C. (2002)
Nature, 415, 655-659.
30.Bennett, R. A., Wilson III, D. M., Wong, D., and
Demple, B. (1997) Proc. Natl. Acad. Sci. USA, 94,
7166-7169.
31.Lebedeva, N. A., Khodyreva, S. N., Favre, A., and
Lavrik, O. I. (2003) Biochem. Biophys. Res. Commun., 300,
182-187.
32.Wong, D., and Demple, B. (2004) J. Biol.
Chem., 279, 25268-25275.