REVIEW: Energetics of Alkalophilic Representatives of the Genus Bacillus
M. S. Muntyan*, I. V. Popova, D. A. Bloch, E. V. Skripnikova, and V. S. Ustiyan
Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119992 Moscow, Russia; fax: (7-095) 939-3181; E-mail: muntyan@genebee.msu.su* To whom correspondence should be addressed.
Received October 11, 2004
Cytochrome and lipid composition of membranes is considered as the attributes required for adaptation of the alkalophiles to alkaline conditions. Respiratory chains of alkalophilic representatives of the genus Bacillus are discussed. Special attention is paid to the features of the Na+-cycle of these bacteria and to the features determining halo- and alkalotolerant phenotype, which have been reported due to recent achievements in genomics.
KEY WORDS: Bacillus pseudofirmus, Bacillus halodurans, halotolerant, alkalotolerant, alkalophile, sodium cycle, cytochrome, terminal oxidase, respiratory chain, fatty acids
The adaptation mechanisms of microorganisms to alkaline environment have been studied over the last three decades in many laboratories all over the world. In spite of considerable contribution of scientists to the study of alkalophily, the energy coupling principles are not yet clear in a variety of the alkalophilic bacterial representatives. Isolated in 1989, the alkalophilic bacterium Bacillus sp. FTU (recently renamed Bacillus halodurans FTU [1]) was one of the objects in our laboratory studies. In the same period similar investigations of other alkalophilic bacteria, Bacillus firmus RAB and Bacillus firmus OF4, were carried out under the direction of T. Krulwich in the USA. The past decade marked a full revision of numerous bacterial subdivisions due to the wide use of molecular biology methods including sequence analysis of 16S RNA. The revision touched upon the genus Bacillus as well. A new group of 11 alkalophilic representatives was proposed within the genus [2]. The two most intensively studied strains, B. firmus OF4 [3] and B. halodurans FTU [4], were re-identified at nearly the same time in Russia and in the USA. Both strains appeared to be representatives of the same new alkalophilic species Bacillus pseudofirmus and were renamed. The specific names OF4 and FTU were left in their new titles. Simultaneous tolerance to high NaCl concentrations up to 18% (3.1 M) and to high alkalinity (pH 10) is the feature of the species B. pseudofirmus. Based on these characteristics the species appears in the triplet of the most extremotolerant, new alkalophilic Bacillus species [2, 4] and is close to the recently identified extremely alkalophilic and halotolerant Oceanobacillus iheyensis isolated from deep-sea sediments (1050 m) [5].
A reversed DeltapH on the cytoplasmic membranes of the alkalophilic and alkalotolerant representatives of genus Bacillus was found as compared to neutrophilic bacteria, that is, the pH inside the cells is more acidic than outside the cells by 0.5-2 units under the alkaline environment. Calculations show [6, 7] that in spite of higher DeltaPsi the resulting DeltaµH+ is lower on the membrane of such bacteria as compared with the neutrophilic species. Obviously, the proton coupling mechanism of oxidative phosphorylation with respiration in the way proposed by P. Mitchell in the chemiosmotic theory is ineffective in the alkalophilic bacteria. Nevertheless, the growth yield of the alkalophiles under alkaline environment is comparable with that of the neutrophilic species. This implies the alkalophilic bacteria must possess alternative effective mechanisms of energy transduction and accumulation. The sodium cycle proposed by V. P. Skulachev [8] might be one of the possible findings. The sodium cycle conception was supported in studies of many gram-negative alkalophiles. Both primary Na+-potential generators (NADH-CoQ-reductases) and Na+-potential consumers (flagella, symporters) were found in the gram-negative bacteria [9]. The alkalophilic Bacillus energetics seems to present an alternative way of overcoming low deltaµH+, although part of the elaborated energetic mechanisms are common in all alkalophiles.
MEMBRANE CYTOCHROMES
One of the common features of alkalotolerant and alkalophilic bacteria is a multiple magnification of cytochrome content in membranes as compared with neutrophilic bacteria. In B. pseudofirmus FTU this magnitude increased threefold upon pH increase from 7 to 8.6 and reached 4.9 nmol/mg protein [10] at the expense of cytochromes b and in greater extent of induction of cytochromes c. In neutrophilic bacteria of genus Bacillus, this magnitude was of 0.76-1 nmol/mg protein [11, 12] and was comparable with the total cytochrome content in Escherichia coli. In alkalophilic Bacillus alkalophilus and B. firmus RAB, the total cytochrome content (5.5 nmol/mg protein) considerably exceeded the same value (1.4 nmol/mg protein) in those mutant strains capable of growth at neutral pH only [13]. An increase in the cytochrome content in these strains like in B. pseudofirmus FTU took place at the expense of the cytochrome b and increase in cytochrome c. In the facultatively alkalophilic strain Bacillus YN-2000 capable of growth at pH 7-10, the cytochrome content increased from 0.9 to 4.7 nmol/mg protein upon increasing the alkalinity of the medium over the specified pH interval [14]. It was shown that in alkalophilic Bacillus, cytochromes c were represented by acidic proteins of redox potential lower by 150 mV on average in contrast to soluble and membrane cytochromes of neutrophilic species. It was assumed that a multiple induction of low potential cytochromes c in membranes allowed alkalophiles to maintain larger DeltaPsi across the membranes [15].
In the membranes from B. pseudofirmus FTU, cytochromes a, b, and c were found among the heme-containing proteins [10]. Hemes O and D widely-spread in most gram-negative bacteria were not detected in those membranes [16]. Cytochrome bd induction was reported in B. pseudofirmus OF4 under growth on glucose at neutral pH or under growth on mineral medium at pH 10.5 up to the stationary phase of growth [17], in B. subtilis under growth on glucose [18], and in thermophilic species Bacillus stearothermophilus [19] and Bacillus coagulans under the simultaneous temperature increase and oxygen decrease in the growth medium [20]. Cytochrome o was found in membranes of some thermophilic species, B. stearothermophilus [21] and Bacillus PS3 [22]. Cytochrome o was not detected in membranes of other species, for example in B. subtilis, as well as in the complete sequence of those bacterial genomes. Only the CtaA gene deletion disturbing heme A biosynthesis in cells led to cytochrome o induction in B. subtilis [18]. The lack of cytochrome o in B. pseudofirmus FTU like in B. subtilis is possibly genetically determined. According to recent data on the complete genome sequencing, operons of cytochromes bd and/or bo3 are present in Bacillus halodurans C-125 [23], O. iheyensis [5], and Geobacillus stearothermophilus [24].
TERMINAL OXIDASES
In view of our negative data on the presence of heme O in B. pseudofirmus FTU membranes, it became apparent that cytochrome bo playing presumably a role of a terminal oxidase was not a part of the bacterial respiratory chain, and that another terminal oxidase represented by cytochromes b functioned in the membranes. Oxidases of cbb3- and bb3-type, on one hand, and of bo3-type, on the other, are characterized by similar optical spectra and it is difficult to draw a conclusion on the presence of any of them without a chromatographic analysis. We have conducted experiments that enabled to make a clear difference between the two groups of oxidases [25]. Starting from heme composition of B. pseudofirmus FTU membranes and isolated oxidases [16] and from the N-terminal sequence of oxidase subunits [1], we concluded that at least two enzyme complexes, cytochromes caa3 and cbb3, function in the terminal part of the bacterial respiratory chain.
Cytochrome-c-oxidase of the caa3-type is a member of the Sox M-subfamily [26] of the oxidase superfamily with a heme-copper binuclear center like the mitochondrial aa3-type oxidase. The caa3-type oxidase has been found generally in alkalophilic and thermophilic representatives of the genus Bacillus: Bacillus sp. PS3, B. stearothermophilus, B. firmus RAB, B. pseudofirmus OF4, and thermophilic bacteria. The neutrophilic and mesophilic species B. subtilis, Bacillus cereus, and Bacillus brevis synthesize this oxidase only during sporulation, whereas the quinol oxidase of the aa3-type is typical in usual conditions. In the opinion of some authors, the covalent bond of cytochrome c with the oxidase subunit II prevents cytochrome c from washing out of gram-positive bacterial envelopes [27]. Some other authors associated the caa3 supercomplex emergence with bacterial thermophily. However, none of the hypotheses explains the reasons for the emergence of such an enzyme. Interestingly, one more oxidase type with covalently bound cytochrome c, cytochrome aco3, was found in alkalophilic Bacillus cognii YN-2000 [28] and in thermophilic Bacillus sp. PS3 [29]. Cytochromes c in these oxidases, like other alkalophilic Bacillus cytochromes c, are characterized by redox potential much lower than in neutrophilic species. One can assume the occurrence in bacterial membranes of oxidases with covalently bound cytochrome c is an adaptive device for alkaline and high temperature environmental conditions. As shown for B. pseudofirmus OF4, even partial deficiency in the caa3-type oxidase by mutation made it impossible for the strain to live under alkaline conditions [30]. Proton pumping by the caa3-type oxidase was studied both in whole cells [22, 31] and in artificial membranes [32]. However, the H+/e- stoichiometry of this energy generator has never been studied thoroughly earlier. We have worked out a reconstitution method of the caa3-type oxidase into liposomes and showed the enzyme H+/e- coefficient is 1 and does not depend on pHout [33].
The two revealed and isolated terminal oxidases of B. pseudofirmus FTU were analyzed for reversible binding of carbon monoxide (CO). Interestingly, all the examined oxidases from several bacteria were classed into two groups: the first bound CO with tau1/2 = 25-30 msec and the other combined CO much more rapidly, with tau1/2 less than that in the first group by at least one order of magnitude. Oxidases of the aa3-, caa3-, bo3-, and ba3-type appeared to comprise the first group, and oxidases of the bd- and bb3-/cbb3-type were assigned to the second group [34-36]. As followed from our data as well as from the results of other laboratories, oxidases of the first group were proton pumps capable of proton translocation from inside to outside of the cell membrane [33, 37]. Concerning the second group of oxidases, the bd-type oxidase was found to have no ion translocation activity [37]. Data on the cbb3-type oxidase are controversial to date; there are different reports on the H+/e- stoichiometry coefficient for the enzyme: 0 [23], 0.2 [38], and 0.6-1 [39]. To study this subject more advanced reconstitution techniques need to be worked out. The common attribute of cytochrome oxidases of alkalophilic Bacillus is the higher content of the negatively charged amino acid residues in the protein structure compared to those of neutrophilic species. One can assume such characteristics are an adaptation of the bacteria to alkaline pH of the environment [40].
OTHER ELECTRON TRANSPORT CHAIN COMPONENTS
The analysis of the genome sequences testifies that a NADH-CoQ-reductase of type II (complex I) operates in the respiratory chain of genus Bacillus representatives. Such an enzyme of alkalophilic Bacillus YN-1 was isolated and characterized, and a gene encoding the enzyme was sequenced [41]. There has been no report on the presence of the type I H+-NADH-CoQ-reductase and Na+-NADH-CoQ-reductase (NQR) in Bacillus corroborated by genetic analysis so far, though such enzymatic activity was stated for alkalophilic Bacillus strains [40, 42]. Genes for succinate-menaquinone oxidoreductase (complex II) were identified in B. subtilis [43], alkalophilic B. cognii YN-2000, and facultative alkalophile B. pseudofirmus OF4 [15]. The enzyme complex was isolated and purified from these strains. A bc-complex (complex III) was isolated from thermophilic Bacillus PS3 [44]; its presence was noted also in B. subtilis [45] and in alkalophilic B. firmus RAB and B. pseudofirmus OF4 [46]. A bc-complex operon (qcr) was found in B. subtilis [47] and in thermophilic B. stearothermophilus K1041 [48].
LIPID COMPOSITION
The adaptation of bacteria to alkaline environments is not restricted to the amino acid composition difference of membrane proteins. The alkalophilic bacteria contain high concentration of squalene and anionic phospholipids, especially of cardiolipin [49], as compared to bacteria inhabiting neutral and low-saline niches. The studies of membrane vesicles from the strict and facultative alkalophiles indicate that membrane permeability decreased upon the squalene concentration increase and it increased upon diacylglycerol concentration increase. In the haloalkalophiles, unsaturated fatty acids predominate over saturated ones. In the membranes of gram-positive bacteria that include representatives of the genus Bacillus, branched fatty acids are biomarkers. However, unlike the alkalotolerant bacteria, both strictly alkalophilic and neutrophilic species are unable to survive the pH shift of the environment respectively in neutral or inversely in the alkaline region. Change in the lipid composition of the membranes is one of the specific adaptation mechanisms of tolerant microorganisms that make it possible for them to withstand sharp salinity and alkalinity decreases in the environment. Phosphatidylglycerol and cardiolipin content in the membranes of facultative alkalophile Bacillus spp. was shown to rise during cultivation at extremely alkaline pH in contrast with the same under neutral pH conditions [15]. The cardiolipin content in B. pseudofirmus OF4 membranes increases by a factor of 1.3 during the transition from neutral to alkaline pH in the growth medium and it is more than 5 times higher compared with the same value in neutrophilic B. subtilis [50]. Comparative study of liposomes from strictly alkalophilic B. firmus RAB and facultative alkalophilic B. pseudofirmus OF4 demonstrated that the membranes of the obligate alkalophiles characteristic of higher unsaturated and branched fatty acid content lost integrity at neutral pH. The facultative alkalophiles have lower passive conductivity at both neutral and alkaline pH as compared with strict alkalophiles. It was shown that cardiolipin and/or phosphatidylglycerol, glycolipid, and branched fatty acid content increased in the lipid composition of gram-positive bacteria on increase in medium salinity [51]. Adaptation to changed pH and salinity is followed in a few minutes upon the effect by a sharp and manifold increase in the rate of synthesis of the substituting lipid that continued during the next 1-2 h simultaneously with a full stop of bacterial growth [52]. Thereafter a return to the initial rates of lipid synthesis and growth recovery takes place. The pathways of energy supply in bacteria during adaptation conditions have been poorly studied. An application of the “free” respiration concept to the study of this problem substantially extends understanding of energetic mechanisms of tolerant bacteria adaptation [53]. One can assume that the concentration of fatty acids increased in cells during reconstruction of lipid composition in bacterial membranes at the expense of fatty acid biosynthesis activation, and fatty acids thereby effect the proton conductivity of the membranes. We have found that low concentrations of fatty acids had “mild” uncoupling effect on B. pseudofirmus FTU though they were ineffective in decreasing membrane potential in cells during the action of respiratory pumps [54]. The value of the “mild” uncoupling effect of natural uncoupler fatty acids found in bacteria might be in “free” respiration induction similar to the well studied effect in mitochondria.
FEATURES OF THE Na+-CYCLE
The genomic epoch has opened possibilities to identify a set of halo- and alkalotolerant bacterial phenotype characteristics by comparison of alkalophilic and neutrophilic bacterial genes. The complete genome sequence has become available for at least six representatives of the genus Bacillus: two halo- and alkalophilic species, B. halodurans C-125 [23], O. iheyensis [5], and four moderately halotolerant, neutrophilic species, B. subtilis [55], Bacillus anthracis [56], G. stearothermophilus [24], and B. cereus [57]. In 2002, the comparison of the genomes of the first three species and several gram-positive non-bacillus neutrophilic species was carried out [5]. Among the observed adaptation mechanisms, the following devices might be important for the alkalophily: 1) ABC-transporters of branched amino acids; 2) acidic polymers of the cell wall; 3) H+- and Na+-symporters of dicarboxylic acids; 4) Na+/H+-antiporter coded by the Mrp and Nha genes; 5) Na+-channel in Na+-dependent flagellar motor. Halotolerance might be provided by osmoprotector accumulation by means of the following systems: 1) K+- and glycine-betaine-transporters, and 2) Na+/proline/pantothenate-symporters. One can see that all Na+-cycle elements are used by gram-positive representatives of alkalophilic Bacillus species. The following consumers of Na+-potential in all alkalophilic Bacillus were brought out reliably: Na+-dependent flagellar motor, and transporters of sugars, dicarboxylic acids, and proline [15]. Among the well-known Na+-pumping structures, Na+/H+-antiporters encoded by the Mrp and Nha genes may provide a mechanism for the reduction of Na+ balance inside the cell and closing the Na+-cycle. Alkalophilic Bacillus bacteria thereby use a different variant of the Na+-cycle as compared with alkalophilic gram-negative bacteria. They seem to use any primary pump for the Na+-potential generation but they do apply a secondary Na+-pump. The scheme shows the Na+-cycle known for B. pseudofirmus at present.
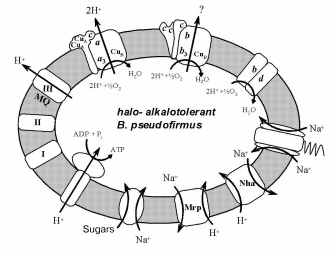
Electron-transport chain components and some membrane transporters using H+ and Na+ gradients on the membrane of halo- and alkalotolerant bacteria Bacillus pseudofirmus.
The experimental work was supported by the Russian Foundation for Basic Research (grant No. 02-04-49107) and the Ministry for Education and Science (grant “Leading Scientific Schools” No. 1710.2003.04).
REFERENCES
1.Grinkevich, V. A., Lysenko, A. M., Muntyan, M. S.,
Skripnikova, E. V., and Afrikyan, E. K. (1997) Biochemistry
(Moscow), 62, 718-724.
2.Nielsen, P., Fritze, D., and Priest, F. G. (1995)
Microbiology, 141, 1745-1761.
3.Takami, H., and Krulwich, T. A. (2000)
Extremophiles, 4, 19-22.
4.Muntyan, M. S., Tourova, T. P., Lysenko, A. M.,
Kolganova, T. V., Fritze, D., and Skulachev, V. P. (2002)
Extremophiles, 6, 195-199.
5.Takami, H., Takaki, Y., and Uchiyama, I. (2002)
Nucleic Acids Res., 30, 3927-3935.
6.Skulachev, V. P. (1994) Antonie Van
Leeuwenhoek, 65, 271-284.
7.Krulwich, T. A., Ito, M., and Guffanti, A. A.
(2001) Biochim. Biophys. Acta, 1505, 158-168.
8.Skulachev, V. P. (1991) Biosci. Rep.,
11, 387-444.
9.Hase, C. C., Fedorova, N. D., Galperin, M. Y., and
Dibrov, P. A. (2001) Microbiol. Mol. Biol. Rev., 65,
353-370.
10.Muntyan, M. S., and Skripnikova, E. V. (1993)
Biochemistry (Moscow), 58, 928-933.
11.Doi, R., and Halvorson, H. (1961) J.
Bacteriol., 81, 51-58.
12.Yamaguchi, T., Tamura, G., and Arima, K. (1966)
Biochim. Biophys. Acta, 124, 413-414.
13.Lewis, R. J., Belkina, S., and Krulwich, T. A.
(1980) Biochem. Biophys. Res. Commun., 95, 857-863.
14.Yumoto, I., Fukumori, Y., and Yamanaka, T. (1991)
J. Biochem., 110, 267-273.
15.Yumoto, I. (2002) J. Biosci. Bioeng.,
93, 342-353.
16.Muntyan, M. S., Ustiyan, V. S., Viryasov, M. B.,
and Skulachev, V. P. (1995) Biochem. Biophys. Res. Commun.,
207, 55-61.
17.Hicks, D. B., Plass, R. G., and Quirk, P. G.
(1991) J. Bacteriol., 173, 5010-5016.
18.Svensson, B., Lubben, M., and Hederstedt, L.
(1993) Mol. Microbiol., 10, 193-201.
19.Sakamoto, J., Koga, E., Mizuta, T., Sato, C.,
Noguchi, S., and Sone, N. (1999) Biochim. Biophys. Acta,
1411, 147-158.
20.Jones, M. V., Spencer, W. N., and Edwards, C.
(1984) J. Gen. Microbiol., 130, 95-101.
21.Nikaido, K., Noguchi, S., Sakamoto, J., and Sone,
N. (1998) Biochim. Biophys. Acta, 1397, 262-267.
22.Sone, N., and Fujiwara, Y. (1991) J.
Biochem., 110, 1016-1021.
23.Takami, H., Nakasome, K., Takai, Y., Maeno, G.,
Sasaki, R., Masui, N., Fujii, F., Hirama, C., Nakamura, Y., Ogasawara,
N., Kuhara, S., and Horikoshi, K. (2000) Nucleic Acids Res.,
28, 4317-4331.
24.Takami, H., Nishi, S., Lu, J., Shimamura, S., and
Takaki, Y. (2005) Extremophiles, in press.
25.Muntyan, M. S., Dinarieva, T. Y., Baev, M. V.,
and Netrusov, A. I. (2002) Arch. Biochem. Biophys., 398,
118-124.
26.Castresana, J., and Saraste, M. (1995) Trends
Biochem. Sci., 20, 443-448.
27.Ishizuka, M., Machida, K., Shimada, S., Mogi, A.,
Tsuchiya, T., Ohmori, T., Souma, Y., Gonda, M., and Sone, N. (1990)
J. Biochem., 108, 866-873.
28.Yumoto, I., Takahashi, S., Kitagawa, T.,
Fukumori, Y., and Yamanaka, T. (1993) J. Biochem., 114,
88-95.
29.Sone, N., Ogura, T., Noguchi, S., and Kitagawa,
T. (1994) Biochemistry, 33, 849-855.
30.Krulwich, T. A., Ito, M., Gilmour, R., Sturr, M.
G., Guffanti, A. A., and Hicks, D. B. (1996) Biochim. Biophys.
Acta, 1275, 21-26.
31.Yaginuma, A., Tsukita, S., Sakamoto, J., and
Sone, N. (1997) J. Biochem. (Tokyo), 122, 969-976.
32.Elferink, M. G. L., De Wit, J. G., Driessen, A.
J. M., and Konings, W. N. (1993) Eur. J. Biochem., 214,
917-925.
33.Muntyan, M. S., and Popova, I. V. (2000) EBEC
Short Reports, 11, 244.
34.Muntyan, M. S., Bloch, D. A., Drachev, L. A., and
Skulachev, V. P. (1993) FEBS Lett., 327, 347-350.
35.Muntyan, M. S., Bloch, D. A., Ustiyan, V. S., and
Drachev, L. A. (1993) FEBS Lett., 327, 351-354.
36.Muntyan, M. S., Ludwig, B., Zickermann, I., and
Starshinova, N. P. (1998) FEBS Lett., 429, 216-220.
37.Puustinen, A., Finel, M., Haltia, T., Gennis, R.
B., and Wikstrom, M. (1991) Biochemistry, 30,
3936-3942.
38.Arslan, E., Kannt, A., Thony-Meyer, L., and
Hennecke, H. (2000) FEBS Lett., 470, 7-10.
39.Toledo-Cuevas, M., Barquera, B., Gennis, R. B.,
Wikstrom, M., and Garcia-Horsman, J. A. (1998) Biochim. Biophys.
Acta, 1365, 421-434.
40.Hicks, D. B., and Krulwich, T. A. (1995)
Biochim. Biophys. Acta, 1229, 303-314.
41.Xu, X., Koyama, N., Cui, M., Yamagishi, A.,
Nosoh, Y., and Oshima, T. (1991) J. Biochem., 109,
678-683.
42.Kostyrko, V. A., Semeykina, A. L., Skulachev, V.
P., Smirnova, I. A., Vagina, M. L., and Verkhovskaya, M. L. (1991)
Eur. J. Biochem., 198, 527-534.
43.Von Wachenfeldt, C., and Hederstedt, L. (1992)
FEMS Microbiol. Lett., 100, 91-100.
44.Kutoh, E., and Sone, N. (1988) J. Biol.
Chem., 263, 9020-9026.
45.Yu, J., and Le Brun, N. E. (1998) J. Biol.
Chem., 273, 8860-8866.
46.Trumpower, B. (1990) Microbiol. Rev.,
54, 101-129.
47.Yu, J., Hederstedt, L., and Piggot, P. J.
(1995) J. Bacteriol., 177, 6751-6760.
48.Sone, N., Sawa, G., Sone, T., and Noguchi, S.
(1995) J. Biol. Chem., 270, 10612-10617.
49.Clejan, S., Krulwich, T. A., Mondrus, K. R., and
Seto-Young, D. (1986) J. Bacteriol., 168, 334-340.
50.Krulwich, T. A., Federbush, J. G., and Guffanti,
A. A. (1985) J. Bacteriol., 162, 768-772.
51.Bygraves, J. A., and Russell, N. J. (1988)
Food Microbiol., 5, 109-116.
52.Russell, N. J., Adams, R., Bygraves, J., and
Kogut, M. (1986) FEMS Microbiol. Lett., 39, 103-107.
53.Skulachev, V. P. (1962) Ratio between
Oxidation and Phosphorylation in Respiratory Chain [in Russian],
Academic Press of the USSR, Moscow.
54.Popova, I. V., Bodrova, M. E., Mokhova, E. N.,
and Muntyan, M. S. (2004) Biochemistry (Moscow), 69,
1165-1169.
55.Kunst, F., Ogasawara, N., Moszer, I., Albertini,
A. M., Alloni, G., Azevedo, V., Bertero, M. G., Bessieres, P., Bolotin,
A., Borchert, S., Borriss, R., Boursier, L., Brans, A., Braun, M.,
Brignell, S. C., Bron, S., Brouillet, S., Bruschi, C. V., Caldwell, B.,
Capuano, V., Carter, N. M., Choi, S.-K., Codani, J.-J., Connerton, I.
F., Cummings, N. J., Daniel, R. A., Denizot, F., Devine, K. M.,
Dusterhoft, A., Ehrlich, S. D., Emmerson, P. T., Entian, K.
D., Errington, J., Fabret, C., Ferrari, E., Foulger, D.,
Fritz, C., Fujita, M., Fujita, Y., Fuma, S., Galizzi,
A., Galleron, N., Ghim, S.-Y., Glaser, P., Goffeau, A.,
Golightly, E. J., Grandi, G., Guiseppi, G., Guy, B. J., Haga,
K., Haiech, J., Harwood, C. R., Henaut, A., Hilbert, H.,
Holsappel, S., Hosono, S., Hullo, M.-F., Itaya,
M., Jones, L., Joris, B., Karamata, D., Kasahara,
Y., Klaerr-Blanchard, M., Klein, C., Kobayashi,
Y., Koetter, P., Koningstein, G., Krogh, S., Kumano, M.,
Kurita, K., Lapidus, A., Lardinois, S., Lauber,
J., Lazarevic, V., Lee, S.-M., Levine, A., Liu,
H., Masuda, S., Mauel, C., Medigue, C., Medina, N., Mellado,
R. P., Mizuno, M., Moestl, D., Nakai, S., Noback, M., Noone,
D., O'Reilly, M., Ogawa, K., Ogiwara, A., Oudega, B., Park,
S.-H., Parro, V., Pohl, T. M., Portetelle, D., Porwollik, S.,
Prescott, A. M., Presecan, E., Pujic, P., Purnelle, B.,
Rapoport, G., Rey, M., Reynolds, S., Rieger,
M., Rivolta, C., Rocha, E., Roche, B., Rose,
M., Sadaie, Y., Sato, T., Scanlan, E., Schleich,
S., Schroeter, R., Scoffone, F., Sekiguchi, J.,
Sekowska, A., Seror, S. J., Serror, P., Shin, B.-S., Soldo, B.,
Sorokin, A., Tacconi, E., Takagi, T., Takahashi,
H., Takemaru, K., Takeuchi, M., Tamakoshi, A., Tanaka,
T., Terpstra, P., Tognoni, A., Tosato,
V., Uchiyama, S., Vandenbol, M., Vannier, F.,
Vassarotti, A., Viari, A., Wambutt, R., Wedler, E.,
Wedler, H., Weitzenegger, T., Winters, P., Wipat,
A., Yamamoto, H., Yamane, K., Yasumoto, K., Yata,
K., Yoshida, K., Yoshikawa, H.-F., Zumstein, E., Yoshikawa,
H., and Danchin, A. (1997) Nature, 390, 249-256.
56.Read, T. D., Peterson, S. N., Tourasse, N.,
Baillie, L. W., Paulsen, I. T., Nelson, K. E., Tettelin, H., Fouts, D.
E., Eisen, J. A., Gill, S. R., Holtzapple, E. K., Okstad, O. A.,
Helgason, E., Rilstone, J., Wu, M., Kolonay, J. F., Beanan, M. J.,
Dodson, R. J., Brinkac, L. M., Gwinn, M., DeBoy, R. T., Madpu, R.,
Daugherty, S. C., Durkin, A. S., Haft, D. H., Nelson, W. C., Peterson,
J. D., Pop, M., Khouri, H. M., Radune, D., Benton, J. L., Mahamoud, Y.,
Jiang, L., Hance, I. R., Weidman, J. F., Berry, K. J., Plaut, R. D.,
Wolf, A. M., Watkins, K. L., Nierman, W. C., Hazen, A., Cline, R.,
Redmond, C., Thwaite, J. E., White, O., Salzberg, S. L., Thomason, B.,
Friedlander, A. M., Koehler, T. M., Hanna, P. C., Kolsto, A.-B., and
Fraser, C. M. (2003) Nature, 423, 81-86.
57.Ivanova, N., Sorokin, A., Anderson, I., Galleron,
N., Candelon, B., Kapatral, V., Bhattacharyya, A., Reznik, G.,
Mikhailova, N., Lapidus, A., Chu, L., Mazur, M., Goltsman, E., Larsen,
N., D'Souza, M., Walunas, T., Grechkin, Y., Pusch, G., Haselkorn, N.,
Fonstein, M., Ehrlich, S. D., Overbeek, R., and Kyrpides, N. (2003)
Nature, 423, 87-93.