|
DNA-[Adenine] Methylation in Lower EukaryotesS. HattmanDepartment of Biology, University of Rochester, Rochester, NY 14627-0211, USA; fax: 585-275-2070; E-mail: modDNA@mail.rochester.edu
|
Received October 4, 2004
DNA methylation in lower eukaryotes, in contrast to vertebrates, can involve modification of adenine to N6-methyladenine (m6A). While DNA-[cytosine] methylation in higher eukaryotes has been implicated in many important cellular processes, the function(s) of DNA-[adenine] methylation in lower eukaryotes remains unknown. I have chosen to study the ciliate Tetrahymena thermophila as a model system, since this organism is known to contain m6A, but not m5C, in its macronuclear DNA. A BLAST analysis revealed an open reading frame (ORF) that appears to encode for the Tetrahymena DNA-[adenine] methyltransferase (MTase), based on the presence of motifs characteristic of the enzymes in prokaryotes. Possible biological roles for DNA-[adenine] methylation in Tetrahymena are discussed. Experiments to test these hypotheses have begun with the cloning of the gene. Orthologous ORFs are also present in three species of the malarial parasite Plasmodium. They are compared to one another and to the putative Tetrahymena DNA-[adenine] MTase. The gene from the human parasite P. falciparum has been cloned.
KEY WORDS: DNA methylation, DNA methyltransferase, lower eukaryotes, motifs, open reading frames, Plasmodium, Tetrahymena
This mini-review does not present an exhaustive account of the literature on DNA methylation in lower eukaryotes. Rather, after a general overview, I will address two systems that are of special interest to my laboratory. These are works in progress and their continuation is subject to the vagaries of the grant funding process. Should that process prove unfavorable, then this article may well describe what might have been.
DNA MODIFICATION IN PRO- AND EUKARYOTES
Post-replicative enzymatic modifications of DNA are epigenetic changes that add another level of information to the cell genome. These modifications are usually in the form of the methylated bases: N6-methyladenine (m6A), N4-methylcytosine (m4C), or C5-methylcytosine (m5C). Whereas m4C is confined to prokaryotes, m5C and m6A are found in prokaryotes and eukaryotes (see below). The in vivo production of methylated bases is mediated by adenine- and cytosine-specific DNA methyltransferases (MTases). These enzymes catalyze the transfer of a methyl group from donor S-adenosyl-L-methionine (AdoMet) to the C5 ring-carbon of cytosine (Cyt) or to the exocyclic amino group of adenine (Ade) (N6) or Cyt (N4). The reaction products are methylated DNA and S-adenosyl-L-homocysteine (AdoHcy). The DNA-[Cyt-C5] MTases are much better understood mechanistically than the DNA-[amino] MTases. For the former, not only has the chemical mechanism of catalysis been elucidated, but also three-dimensional structures of binary and ternary complexes of some MTases with their substrates have been solved by X-ray crystallography [1-4]. A most surprising and exciting result was that the Cyt residue to be methylated is “flipped out” of the DNA helix [3, 4]. An intermediate stable binary complex is formed by a covalent bond between the flipped Cyt-C6 (which activates the C5 atom) and a cysteine residue within the enzyme active site. Such an intermediate has not been observed with the [Ade-N6]- or [Cyt-N4]-amino MTases, which appear to transfer the methyl moiety directly to the exocyclic amino group. For members of the latter group, TaqI, PvuII, DpnM, and RsrI [5-8], 3-D structures have been reported; but only ternary complexes of TaqI [9] and T4Dam [10, 10a] have been co-crystallized with DNA (a small synthetic duplex) and AdoHcy.
In addition to the three methylated bases, hyper-modified bases may also be found in DNA; these may partly or completely replace one of the four common bases. Depending on the specific case, modification may occur at the level of nucleotide pool metabolism, or as a post-replicative event. For a time hyper-modified bases were thought to be confined to the prokaryotic world (primarily bacteriophages), but that changed with the discovery of 5-hydroxymethyluracil (5-hmUra) in certain dinoflagellates [11]. Subsequently, hyper-modified bases have been documented in several other lower eukaryotes (see [12] for a review, and references therein). Examples of hyper-modified bases include: 5-hydroxymethylcytosine (with or without covalently attached glucosyl residues) in the Escherichia coli T-even phages [13, 14]; 5-hmUra in Bacillus subtilis phages; 5-hydroxycytosine in Rhizobium phage RL38J1 [15], which is also hexosylated; alpha-glutamylthymine in B. subtilis phage SP10; N6-(1-acetamido)adenine in phage Mu [16]; 2-aminoadenine in Shigella phage S-2L; and alpha-putrescinylthymine in Pseudomonas phage phiW14. In addition to dinoflagellates, 5-hmUra is in the DNA of kinetoplastid protozoa (e.g., Trypanosoma brucei) [17] and the green alga Euglena gracilis [18]. The function(s) of hyper-modified bases has not been fully elucidated, but in the cases of T-even phages and phage Mu, it protects the viral DNA against host-controlled endonucleolytic degradation and, thereby, extends their host ranges [19-21].
In prokaryotes, DNA methylation affects such diverse phenomena as determination of DNA-host specificity [22], strand selection specificity during DNA-mismatch repair [23], control of chromosome replication and segregation [24], positive regulation of phage Mu mom gene transcription [25], pilus phase-variation [26], and multiple gene expression in E. coli [27, 28]. An essential role in virulence was discovered in a variety of bacterial pathogens [29], where dam- mutants, defective for the Dam DNA MTase activity, were found to be avirulent; such strains were effective in producing live vaccine against murine typhoid fever [30].
In higher eukaryotes DNA methylation has been implicated in the regulation of gene expression [31-33], mammalian X-chromosome inactivation [34], parental imprinting [35], mutation, and human disease, such as the ICF, Rett, and Fragile-X syndromes [36], and cancer [37]. Deamination of m5C produces thymine (Thy), while deamination of Cyt produces uracil (Ura). Although Ura is removed from DNA by a uracil glycosylase, Thy generated from m5C is not removed efficiently and is mutagenic because it produces a G/T mismatch. Since deamination of Ade and m6Ade yield the same product, hypoxanthine, adenine-methylation is not a potentially mutagenic event.
In addition to their biological importance, the wide distribution of DNA MTases has made them ideal for evolutionary studies of sequence-specific DNA-binding proteins. Direct analysis of methylation sites from oligonucleotide sequencing [38, 39] proved superior to the indirect method of protection against cleavage by known restriction nucleases. The first comparative analysis of conserved amino acid sequence motifs in DNA MTases was reported [40] for three prokaryotic Dam enzymes, all of which methylate the palindromic tetranucleotide sequence, GATC [38, 41]. One conserved motif, DPPY, was noted. This is a sub-set of (D/N/S) PP (Y/F), which is now known to be part of the so-called motif IV, found in virtually every prokaryotic DNA-[amino] MTase [42-44]. As more MTase genes were cloned and sequenced, amino acid sequence alignments revealed additional conserved motifs, of which I-VIII and X were the most common (I and IV being the most highly conserved). Motifs X and I-III have been assigned to the AdoMet-binding domain, and motifs IV-VIII to the active site sub-domain. The least conserved of all the domains, the target recognition domain (TRD) interacts with substrate DNA. DNA-[N6-Ade and N4-Cyt amino] MTases have been classified into six groups (alpha through zeta, summarized in Fig. 1) based on the relative order of their motifs (X-I-II-III vs. IV-VIII and the TRD) [42, 44]; however, at the time of the original proposal [42], there were no known members of the delta,_epsilon,_or zeta class. In contrast to the extensive information available on the prokaryote enzymes, no eukaryotic DNA-[adenine] MTase has been characterized.
Fig. 1. Order of motifs of different classes of DNA-[amino] MTases (adapted from [42] and [44]). Groups are classified according to relative order of motifs: the AdoMet binding region X-I-II-III, the catalytic region IV-VIII, and DNA target recognition domain (TRD) (highlighted in light gray). Note that alpha, delta. epsilon_and beta, gamma, zeta) form two circularly permuted sets.
DNA METHYLATION IN Tetrahymena
Characteristic of ciliated protozoa, Tetrahymena thermophila contains two differentiated nuclei: (i) a germinal (diploid, 2C) micronucleus responsible for maintaining genetic continuity through sexual cycles and (ii) a somatic (polycopy, 45C) macronucleus responsible for gene expression during vegetative growth [45]. Ciliates are the only major group of protists to have evolved separate germ line and somatic nuclei. Procedures are available for the isolation of large quantities of nuclei from vegetative or conjugating cells [46], for mass transformation [47], and for gene replacements in either the vegetative or germ-line nucleus [48-50].
Thirty years ago the first example of DNA-adenine methylation in a eukaryotes was reported in the ciliate Tetrahymena [51], where the polycopy, transcriptionally active macronuclear (MAC) DNA was shown to contain ca. 0.8% of its adenine residues in the form of m6A. Surprisingly, methylation was not detectable in the transcriptionally-inactive diploid, micronuclear (MIC) DNA. Subsequently, m6A has been found in the nuclear DNA of other ciliates [52] including Paramecium aurelia [53], Oxytricha fallax [54], and Stylonichia mytilius [55], as well as several other lower eukaryotes including the green algae Chlorella spp. [56] and Chlamydomonas reinhardtii [57], and the dinoflagellate Peridinium triquetrum [11]. Mitochondrial DNA of a higher plant has been reported to contain m6A [58]. MAC DNA from the ciliates Colpoda inflata [59] and Blepharisma japonicum [60] has been reported to contain m5C, while the ciliate Stylonichia lemnae, had no detectable m5C in either MAC or MIC DNA; however, transposon-like sequences were methylated during the MAC differentiation process following conjugation [61]. Thus, the nature of lower eukaryote DNA methylation pattern exhibits a wide range of variation, as observed in prokaryotes.
Although Tetrahymena MAC DNA contains m6A, there was no m5C detected in either MAC or MIC DNA [62]; this was the first reported example of a eukaryote lacking DNA-cytosine methylation. Chromatin structure influences DNA methylation since m6A was found preferentially in internucleosomal linker regions [63]. Taking advantage of this property, it was demonstrated that nucleosomes were “phased”, i.e., they remained specifically positioned and did not randomize over the DNA during vegetative growth [64]. A positive role for DNA methylation in gene regulation in Tetrahymena was suggested by the fact that MAC DNA is transcriptionally active and methylated, whereas MIC DNA is transcriptionally inactive and unmethylated. This is an issue that requires further investigation.
A DNA-[adenine] MTase activity was partially purified from Tetrahymena macronuclei and used to methylate Tetrahymena and micrococcal DNAs in vitro [65]. Since Tetrahymena macronuclear DNA served as a good methyl acceptor, this indicated that DNA methylation in vivo must be incomplete. Subsequent analysis of labeled DNA dinucleotides (generated by enzymatic digestion) showed that the nearest neighbors of m6A were 5´ N (any base) and 3´ T or C, indicating that methylation occurs in a subset of the sequence 5´ N-A-Y 3´ (where Y = T or C). However, nearest neighbor analysis of cell DNA methyl-labeled in vivo during growth in the presence of [3H-methyl]-methionine revealed that the 3´ nearest neighbor was primarily T. (This difference has not been resolved to date.) However, since 5´ N-m6A-T 3´ was produced in both cases, this suggested that AT is part of the Tetrahymena DNA MTase recognition site. If all AT sequences were to be methylated in vivo, then 36% of the Ade residues should be m6A, not the observed 0.8%. In this regard, in vivo the palindromic sequence GATC, a subset of NAT, is essentially unmethylated [66-68]. Therefore, the MTase probably requires more sequence information than N-A-Y, for example in a sequence like GGTNACC, which is the site methylated by the bacterial enzyme M.EcaI [69]. In such a site, although the 5´ nearest neighbor to the target A is degenerate, the m6A content will be low since a GC-rich, heptameric recognition site will occur very infrequently in the AT-rich DNA of Tetrahymena. Furthermore, since DNA methylation is predominantly in the linker regions of nucleosomes [63], some structural feature(s) of chromatin and/or chromatin-bound proteins inhibit complete in vivo methylation. Thus, NAC sites that are methylatable in vitro with naked DNA might not be accessible in chromatin in vivo.
Since Tetrahymena is DNA-[adenine] methylation proficient, a BLAST search of the genome database was conducted to look for an open reading frame (ORF) capable of encoding a DNA-[adenine] MTase. It should be noted that UAA and UAG, read as stop codons in most organisms, are read as glutamine (Q) in ciliates [70, 71], so only UGA serves as a translational stop codon. A candidate DNA-[adenine] MTase ORF was identified, containing 391 residues with 27 internal in-frame UAA and UAG codons coding for Q. In Table 1, the Tetrahymena motifs I and IV are compared with several representative prokaryotic sequences. It is clear that the putative T. thermophila MTase (designated M. TthP) motifs have a high degree of homology to those of the prokaryote enzymes.
Table 1. Conserved motif I and IV sequences
in prokaryotic DNA-[adenine-N6] MTases*
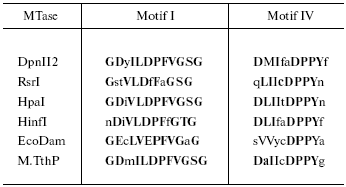
*Capital letters indicate similar functional groups; bold letters
indicate consensus residues. The first four MTases belong to the
beta class [42] in which motif IV is
proximal to the N-terminal end. In the alpha class of MTases
(e.g., EcoDam), motif I is proximal to the N-terminal end, as it
is for M.TthP.
In addition to the DNA MTases, there are other AdoMet-dependent enzymes; viz., MTases that act on RNAs, proteins, lipids, polysaccharides, or small molecules. Physical characterization of about a dozen of these proteins showed a high degree of structural similarity [72], although this conservation was not seen at the amino acid sequence level. However, analysis of a larger set of non-DNA MTases revealed some conserved motifs [73]. While one of these bears some similarity to motif I, the signature motif IV sequence (D/N) PP (Y/F/W) was lacking. A search of the yeast genome with these motifs turned up 26 potential new MTases [74], but no putative DNA MTase; however, as will be discussed later, Saccharomyces cerevisiae and Schizosaccharomyces pombe do contain complete DNA-[adenine] MTase-like ORFs. The HemK family of protein MTases has a motif (LIVMAC)-(LIVFYWA)-x-(D/N)-P-P-(Y/F/W) that has some similarity to motif IV [75]. Coincidentally, the T. thermophila translation release factor eRF1 has elements of both motif IV (DPPFG) and motif I, but these sequences are not found in other eRFs [76]. Amino acid sequences in known RNA MTases have some similarity with DNA MTases; however, none contained the characteristic DPPY [77]. Therefore, while AdoMet-dependent non-DNA MTases share elements of similarity with some motifs in DNA-[adenine] MTases, they are generally distinguishable from each other. Nevertheless, proof that the cloned Tetrahymena ORF encodes a DNA-[adenine] MTase requires experimental verification. To this end, PCR primers were designed and used to amplify a 2.2 kb region containing the ORF, as well as 5´ and 3´ flanking regions. This amplified fragment has been cloned into the pGEM-T vector (unpublished) for the purpose of generating constructs to produce MAC and MIC gene knockouts (experiments are currently in progress). If the cloned ORF actually encodes the Tetrahymena DNA MTase, then we can also investigate whether DNA-[adenine] methylation has an essential function(s) in cell growth and development.
WHAT ROLE(S) MIGHT DNA-[ADENINE] METHYLATION PLAY IN THE LIFE
CYCLE OF Tetrahymena
We know that MAC DNA is methylated and transcriptionally active, while MIC DNA is unmethylated and transcriptionally inactive. This difference suggests that DNA methylation might play a positive regulatory role in transcription during vegetative cell growth. In addition, DNA methylation might also be a player in the chromosome processing events that occur following conjugation [78]. After cell pairing, a series of MIC meiotic divisions is followed by reciprocal exchanges and fusion of the haploid gametic nuclei to produce a diploid fertilization nucleus in each of the exconjugants. This undergoes two mitotic divisions giving rise to four equivalent product nuclei; these develop into two new MICs and two new MACs, which then segregate into the daughter cells upon refeeding and resumption of division. In the meantime, the original parental MAC is degraded and eliminated. In the nascent developing MAC, termed the anlage, after one to two rounds of DNA replication (DNA content > 4 C to 8 C), there occur a series of programmed stage- and site-specific chromosomal alterations. In brief, the former MIC chromosomes are fragmented in a specific pattern, producing roughly 200 sub-chromosomal fragments varying in length from 100-1000 kb [79, 80]. MAC gene segments are either protected by telomere addition or joined. In the transition from anlage to mature MAC, the internal deletions result in an overall loss of about 15% of the MIC DNA sequence complexity [81, 82]. Moreover, developing anlage DNA undergoes de novo methylation.
The chromosome fragmentation described above occurs through hundreds of sites through specific cleavages within a conserved 15 nt long chromosome breakage sequence, Cbs. The Cbs consensus sequence 5´ AAAGAGGTTGGTTTA 3´/5´ TAAACCAACCTCTTT 3´ has been shown to be necessary and sufficient to specify a breakage site [83]. The Cbs sequence is no longer found in the mature MAC DNA of vegetatively growing cells. MAC-destined sequences are excised, followed by eventual elimination of internal unique and repetitive MIC sequences [81, 84].
How might DNA methylation be important for chromosome processing during Tetrahymena anlage development? One of the Cbs complementary strands, 5´ TAAACCAACCTCTTTT 3´, contains two AAC sequences, where A denotes a potential methylation target, based on the partial characterization of the Tetrahymena DNA MTase sequence specificity [65]. Thus, during anlage development, Cbs sequences could undergo de novo DNA methylation and become targets for nicking/cleavage and further processing events. The occurrence of DNA methylation-dependent cleavage is not without precedent. For example, the S. pneumoniae restriction endonuclease, DpnI, cuts DNA at palindromic GATC sites only when they are adenine-methylated on both strands [85]; other methylation-dependent endonucleases have been reported as well [86-88]. In contrast, since MIC DNA lacks methylation, its Cbs sequences remain refractory to these events. In studies on methylation of several specific GATC sites in ribosomal RNA genes [66] or in the vicinity of the H4-I and 73-kD heat shock protein genes, respectively [67], it was found that the onset of DNA methylation correlated with the time of anlage DNA synthesis and rearrangement. However, GATC methylation represents only a minor fraction of all DNA methylation, and there is still nothing known about methylation of non-GATC sites at early growth stages.
Fan and Yao [89] prepared a series of plasmid constructs, each containing a single base-substitution at one of the 15 conserved positions within a cloned Cbs. Each construct was separately microinjected into developing macronuclei and its fate tracked with respect to Cbs function, as monitored by the occurrence of specific cleavage. With respect to variants in the two AAC sites, the following results were observed: (i) substitution of either of the two 5´ A residues (to T or G) resulted in partial function; (ii) substitution of either of the two internal A residues (to G) abolished function; (iii) substitution of either of the two 3´ C residues (to G, A or T) abolished function. These results showed a good correlation between the loss of Cbs function (absence of the appropriate cleavage) and loss of potential (N-A-C) methylation sites. It should be mentioned that some substitutions at other sites also abolished Cbs function. This is not unexpected if a sequence-specific nuclease/glycosylase were involved in the process; that is, methylation may be necessary but not sufficient for cleavage. Thus, it would be extremely interesting to examine Cbs processing in the developing anlage under DNA-[adenine] MTase gene knockout conditions.
DNA METHYLATION IN THE HUMAN MALARIA PARASITE Plasmodium
falciparum?
The genome of P. falciparum is relatively small, comprising about (2.5-3.0)*107 bp. It is distributed over 14 chromosomes with lengths of (6-36)*105 bp [90]. Nuclear DNA has an unusually high AT content of about 81% [91]; however, a computer analysis of a 36 kb segment of known DNA nucleotide sequence showed that the coding regions were 69% AT and flanked by 86% AT regions [92]. In 1982, it was reported that P. falciparum is devoid of any detectable DNA methylation, since HPLC analysis failed to reveal either m6A or m5C [90]. Almost ten years later, the same group used methylation-specific restriction nucleases to probe Plasmodium DNA for methylated bases within several specific nucleotide sequences [92]. The authors failed to detect m6A in GATC sequences, but they suggested that there might be partial methylation (m5C) at one MspI (CCGG) site within the DHFR-TS gene. The apparent lack of modified bases may have discouraged any further interest in studying Plasmodium DNA methylation. Nevertheless, we became interested in the possibility that Plasmodium spp. might be DNA methylation-proficient. This was prompted by several considerations, one of which was based on evolutionary grounds. Apicomplexans (such as Plasmodium spp.) are members of the same phylogenetic group (the clade Alveolata) as ciliates and dinoflagellates. Since some ciliates and dinoflagellates contain m6A in their nuclear DNA, it seemed reasonable that the same might be true of Plasmodium. Secondly, in silico analyses of the available sequence data bases unexpectedly identified potential ORFs that appear to code for DNA-[adenine] MTases in seven eukaryotes [93] including Leishmania major, S. cerevisiae, S. pombe, Arabidopsis thaliana, Drosophila melanogaster, Caenorhabditis elegans, and Homo sapiens, none of which previously have been found to contain detectable levels of m6A. These ORFs contained amino acid sequence motifs characteristic of well-known prokaryote DNA-[adenine] MTases [38]. If a functional enzyme is present in these organisms, then one has to ask whether they actually contain m6A in their DNA, albeit at levels that have hitherto escaped detection.
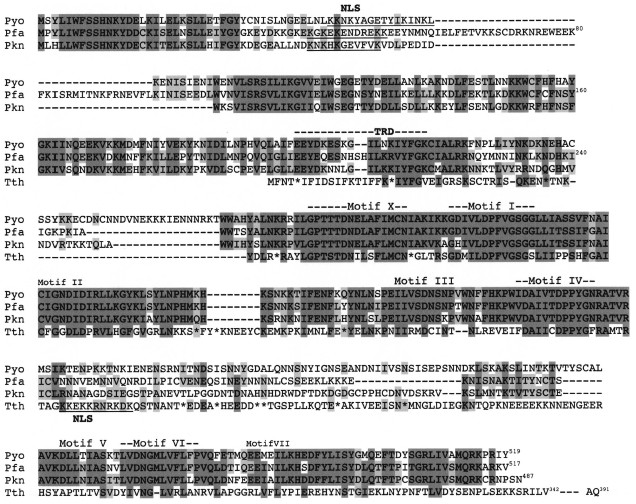
Encouraged by the above observations, we carried out a BLAST search to look for a potential DNA-[adenine] MTase gene in the genomes of Plasmodium spp. Indeed, ORFs were identified in the malarial parasite for humans (P. falciparum), rodents (P. yoelii), and simians (P. knowlesi) (unpublished results). The three orthologous ORFs encode proteins of 517 (Pfa), 519 (Pyo), and 487 (Pkn) residues, and they all appear to lack introns. The three sequences are compared to each other and to the putative Tetrahymena DNA-[adenine] MTase (Fig. 2). The lysine-rich clusters in the NH2-terminal regions of the Plasmodium proteins appear to be good nuclear localization signals (NLS). In contrast, the putative Tetrahymena NLS is located internally, downstream from motif IV. It is not clear to which class the Plasmodium MTase would belong since the location of the TRD is not known (see Fig. 1). However, if the TRD is located in the N-terminal region (as suggested in Fig. 2), then these enzymes would be members of the zeta class of DNA-[adenine] MTases.
In Table 2 (taken in part from [93]), the sequences of the most highly conserved motifs I and IV are compared with those observed in other eukaryote sequences [94]. It must be asked whether any of these putative eukaryote DNA-[adenine] MTase ORFs actually encodes a catalytically active enzyme. Since the largest of the three H. sapiens encoded proteins lacks motif X and is less than 200 residues long [93], these genes are likely to be (truncated) pseudogenes. While the two yeast proteins appear to have the requisite motifs, m6A has not been detected in yeast DNA (less than one m6A per 2000 Ade residues [21]). It should be noted that the mosquito Anopheles gambiae contains an ORF that encodes a protein exhibiting a high degree of homology to the putative S. cerevisiae MTase (unpublished observation). It is possible that these organisms have a DNA-[adenine] MTase pseudogene, since deletion of the S. cerevisiae ORF does not affect cell viability. In this regard, in S. pombe the pmt1 gene encodes an inactive form of DNA-[cytosine-C5] MTase, psiM.SpoI, which can be restored to catalytic activity by deletion of a single amino acid residue [95]. Why S. pombe should have retained this gene through evolution remains to be seen; perhaps it functions purely through its ability to bind DNA and/or interact with other protein(s).Fig. 2. Comparison of deduced amino acid sequences of putative DNA-[adenine] MTases encoded by three Plasmodium spp. and Tetrahymena. Sequences were aligned using BLAST and edited manually. Pyo, Plasmodium yoelii; Pfa, Plasmodium falciparum; Pkn, Plasmodium knowlesi; Tth, Tetrahymena thermophila. Asterisks in the Tth sequence correspond to Q residues encoded by UAA or UAG. Residues with a high degree of chemical similarity or identity are denoted in dark gray when shared by at least three of the enzymes and in pale gray when shared by two enzymes. Putative nuclear localization signals (NLS) are underlined. The approximate extents of the various motifs are denoted by the dashes. Motif VIII is not indicated since it is generally poorly conserved among MTases. The approximate target recognition domain (TRD) is based on several considerations: homology among the three Plasmodium species, and partial homology to a fairly well conserved sequence in the TRD of the Dam MTase family; viz., VPFG(K/R). The other motifs show variable identity/similarity to prokaryote DNA-[adenine] MTase classes as follows: X (gamma); I (beta); II (beta, gamma); III (alpha, beta, or gamma); IV (alpha, beta, or gamma); V (alpha or gamma); VI (alpha or gamma); VII (alpha).
Table 2. Comparison of motifs I and IV of
putative DNA-[adenine] MTases in various eukaryotes and two
representative prokaryotes*

*Capital letters indicate similar functional groups; bold
letters indicate consensus residues (taken in part from [93]).
Do the Plasmodium spp. ORFs actually encode DNA-[adenine] MTases and, if so, are the proteins enzymatically active? To address this, we analyzed enzymatic digests of P. falciparum (red cell stage) DNA by HPLC for the presence of dm6A (mononucleotide); however, we could not unequivocally detect its presence (S. L. Schlagman and S. Hattman, unpublished observations). We have cloned the putative Pfa MTase gene and then sub-cloned it into different expression vectors under regulatable promoters (N. Young, S. L. Schlagman, and S. Hattman, unpublished). These constructs were separately transferred into E. coli and yeast cells. Cultures were subjected to conditions that would induce transcription of the cloned gene, and cellular DNA was isolated and analyzed for the presence of dm6A. In no case did we observe evidence of DNA-[adenine] methylation (unpublished observations). These negative results are inconclusive, however, since some Plasmodium-specific post-translational modification and/or other processing event may be required for enzymatic activity. Moreover, if the target site occurs infrequently in yeast and E. coli, then methylation would have gone undetected because the m6A content would be too low. Finally, it is possible that Plasmodium DNA methylation occurs in a cell-cycle regulated manner, such as has been observed in several prokaryotic species [96, 97], or as a developmental stage-specific event that does not include the red cell stage. Clearly, the outcome of the Tetrahymena gene-knockout experiment is of great importance because of its homology to the Plasmodium MTase ORFs. Thus, evidence for the Tetrahymena gene encoding a DNA-[adenine] MTase would lend support for a similar function of the Plasmodium ORFs.
Considering that DNA-[cytosine] methylation in plant and mammalian genomes is known to affect an array of important biological processes, it is surprising how little is known about DNA-[adenine] methylation in lower eukaryotes. Not only might it be involved in regulating gene expression, but it might be essential for chromosome processing during ciliate development. One may ask whether DNA-[adenine] methylation in lower eukaryotes has been retained through evolution because it is a player in important cellular functions. If so, then a reinvestigation into its presence and possible biological role(s) in the life cycle of P. falciparum is long overdue, especially considering that malaria continues to affect millions of people worldwide. Since potential DNA-[adenine] MTase ORFs have been reported in a variety of lower and higher eukaryotes, one has to consider the possibility that m6A is present in the DNA of these organisms, albeit at levels too low to detect by current methodologies. Since the detection limit is ca. one m6A per 3000 Ade residues, thousands of m6A residues per genome could be present and still escape detection. In this regard, for a long time it was believed that D. melanogaster was devoid of m5C [98], but this was only recently proved to be incorrect [99, 100]. Indeed, it is unfortunate that there is no chemistry currently available that can distinguish m6A from A, as in the bisulfate sequencing method, which distinguishes between m5C and C.
This article is dedicated to my old and dear friend, Prof. Boris Vanyushin. For more than 30 years, we have both watched (and participated) with wonder and pleasure as the field of DNA methylation has burgeoned into an important discipline.
This work was supported by a US Public Health Service grant from the Fogarty International Center (R03 TW05755).
I thank Dr. Dan Goldberg (Washington University, St. Louis, MO) for supplying P. falciparum DNA for HPLC analysis.
REFERENCES
1.Cheng, X., Kumar, S., Posfai, J., Pflugrath, J. W.,
and Roberts, R. J. (1993) Cell, 74, 299-307.
2.Cheng, X., Kumar, S., Klimasauskas, S., and
Roberts, R. J. (1993) Cold Spring Harbor Symp. Quant. Biol.,
58, 331-338.
3.Klimasauskas, S., Kumar, S., Roberts, R. J., and
Cheng, X. (1994) Cell, 76, 357-369.
4.Reinisch, K. M., Chen, L., Verdine, G. L., and
Lipscomb, N. (1995) Cell, 82, 143-153.
5.Labahn, J., Granzin, J., Schluckebier, G.,
Robinson, D. P., Jack, W. E., Schildkraut, I., and Saenger, W. (1994)
Proc. Natl. Acad. Sci. USA, 91, 10957-10961.
6.Gong, W., O'Gara, M., Blumenthal, R. M., and Cheng,
X. (1997) Nucleic Acids Res., 25, 2702-2715.
7.Tran, P. H., Korszun, Z. R., Cerritelli, S.,
Springhorn, S. S., and Lacks, S. A. (1998) Structure, 6,
1563-1575.
8.Scavetta, R. D., Thomas, C. B., Walsh, M. A.,
Szegedi, S. S., Joachimiak, A., Gumport, R. L., and Churchill, M. E.
(2000) Nucleic Acids Res., 28, 3950-3961.
9.Goedecke, K., Pignot, M., Goody, R. S., Scheidig,
A. J., and Weinhold, E. (2001) Nature Struct. Biol., 8,
121-125.
10.Yang, Z., Horton, J. R., Zhou, L., Zhang, X.,
Dong, A., Zhang, X., Schlagman, S. L., Kossykh, V., Hattman, S., and
Cheng, X. (2003) Nature Struct. Biol., 10, 849-855.
10a. Horton, J. R., Liebert, K., Hattman, S., Jeltsch, A., and Cheng, X.
(2005) Cell, 121, 349-361.
11.Rae, P. M. (1976) Science, 194,
1062-1064.
12.Gommers-Ampt, J. H., and Borst, P. (1995)
FASEB J., 9, 1034-1042.
13.Wyatt, G. R., and Cohen, S. S. (1952) Nature
(London), 170, 1072-1073.
14.Lehman, I. R., and Pratt, E. A. (1960) J.
Biol. Chem., 235, 3254-3259.
15.Hsu, F. F., Crain, P. F., Swinton, D. L.,
Hattman, S., and McCloskey, J. A. (1988) Adv. Mass Spectr.,
11B, 1340-1341.
16.Swinton, D., Hattman, S., Crain, P. F., Cheng,
C.-S., Smith, D. L., and McCloskey, J. A. (1983) Proc. Natl. Acad.
Sci. USA, 80, 7400-7404.
17.Borst, P., and van Leeuwen, F. (1997) Mol.
Biochem. Parasitol., 90, 1-8.
18.Dooijes, D., Chaves, I., Kleft, R., Dirks-Mulder,
A., Martin, W., and Borst, P. (2000) Nucleic Acids Res.,
28, 3017-3021.
19.Hattman, S., and Fukasawa, T. (1963) Proc.
Natl. Acad. Sci. USA, 50, 297-300.
20.Toussaint, A. (1976) Virology, 70,
17-27.
21.Kahmann, R. (1982) Cold Spring Harb. Symp.
Quant. Biol., 47, 639-646.
22.Wilson, G. G., and Murray, N. E. (1991) Annu.
Rev. Genet., 25, 585-627.
23.Modrich, P. (1987) Annu. Rev. Biochem.,
56, 435-466.
24.Messer, W., and Noyer-Weidner, M. (1988)
Cell, 54, 735-737.
25.Hattman, S. (1982) Proc. Natl. Acad. Sci.
USA, 79, 5581-5521.
26.Van de Woude, M. W., Braaten, B. A., and Low, D.
A. (1992) Molec. Microbiol., 6, 2429-2435.
27.Marinus, M. G. (1984) in DNA Methylation.
Biochemistry and Biological Significance (Razin, A., Cedar, H., and
Riggs, A. D., eds.) Springer-Verlag, New York, pp. 81-109.
28.Barras, F., and Marinus, M. G. (1989) Trends
Genet., 5, 139-143.
29.Mahan, J., and Low, D. A. (2001) ASM News,
67, 356-361.
30.Heithoff, D. M., Sinsheimer, R. L., Low, D. A.,
and Mahan, M. J. (1999) Science, 284, 967-970.
31.Cedar, H. (1988) Cell, 53, 3-4.
32.Razin, A., and Cedar, H. (1991) Microbiol.
Rev., 55, 451-458.
33.Doerfler, W. (1983) Annu. Rev. Biochem.,
52, 93-124.
34.Riggs, A. D. (1975) Cytogenet. Cell
Genet., 14, 9-25.
35.Barlow, D. P. (1995) Science, 270,
1610-1613.
36.Robertson, K. D., and Wolffe, A. P. (2000)
Nature Rev. Gen., 1, 11-19.
37.Laird, P. W., and Jaenisch, R. (1996) Annu.
Rev. Genet., 30, 441-464.
38.Hattman, S., Brooks, J. E., and Masurekar, M.
(1978) J. Mol. Biol., 126, 367-380.
39.Hattman, S., van Ormondt, H., and deWaard, A.
(1978) J. Mol. Biol., 119, 361-376.
40.Hattman, S., Wilkinson, J., Swinton, D.,
Schlagman, S., Macdonald, P. M., and Mosig, G. (1985) J.
Bacteriol., 164, 932-937.
41.Lacks, S., and Greenberg, B. (1977) J. Mol.
Biol., 114, 153-168.
42.Malone, T., Blumenthal, R. M., and Cheng, X.
(1995) J. Mol. Biol., 253, 618-632.
43.Chandrasegaran, S., and Smith, H. O. (1987) in
Structure and Expression, Vol. 1: From Proteins to
Ribosomes (Sarma, R. H., and Sarma, M. H., eds.) Adenine Press,
Schenectady, NY, pp. 149-156.
44.Bujnicki, J. M. (2002) BMC Evol. Biol.,
2, 3-13.
45.Madireddi, M. T., Smothers, J. F., and Allis, C.
D. (1995) Devel. Biol., 6, 305-315.
46.Gaertig, J., and Gorovsky, M. A. (1992) Proc.
Natl. Acad. Sci. USA, 89, 9196-9200.
47.Gaertig, J., Gu, L., Hai, B., and Gorovsky, M. A.
(1994) Nucleic Acids Res., 22, 5391-5398.
48.Gaertig, J., and Gorovsky, M. A. (1995) Meth.
Cell Biol., 47, 559-569.
49.Cassidy-Hanley, D., Lee, J., Bowen, J., VerPlank,
L. A., Gaertig, J., Gorovsky, M. A., and Bruns, P. J. (1997)
Genetics, 146, 135-147.
50.Bruns, P. J., and Cassidy-Hanley, D. (2000)
Meth. Cell Biol., 62, 501-513.
51.Gorovsky, M. A., Hattman, S., and Pleger, G. L.
(1973) J. Cell Biol., 56, 697-701.
52.Gutiérrez, J. C., Callejas, S., Borniquel,
S., and Martin-Gonzalez, A. (2000) Int. Microbiol., 3,
139-146.
53.Cummings, D. J., Tait, A., and Goddard, J. M.
(1974) Biochim. Biophys. Acta, 374, 1-11.
54.Rae, P. M. M., and Spear, B. B. (1978) Proc.
Natl. Acad. Sci. USA, 75, 4992-4996.
55.Ammermann, D., and Steinbrück, G. (1981)
Eur. J. Cell Biol., 24, 154-156.
56.Van Etten, J. L., Schuster, A. M., Girton, L.,
Burbank, D. E., Swinton, D., and Hattman, S. (1985) Nucleic Acids
Res., 13, 3471-3478.
57.Hattman, S., Kenny, C., Berger, L., and Pratt, K.
(1978) J. Bacteriol., 135, 1156-1157.
58.Vanyushin, B. F., Alexandrushkin, N. I., and
Kirnos, M. D. (1988) FEBS Lett., 233, 397-399.
59.Palacios, G., Martin-Gonzalez, A., and
Gutiérrez, J. C. (1994) Cell Biol. Inter., 18,
223-228.
60.Salvini, M., Durante, M., Citti, L., and Nobili,
R. (1984) Experientia, 40, 1401-1403.
61.Juranek, S., Wieden, H.-J., and Lipps, H. J.
(2003) Nucleic Acids Res., 31, 1387-1391.
62.Pratt, K. (1981) Ph. D. dissertation,
University of Rochester, Rochester, NY.
63.Pratt, K., and Hattman, S. (1981) Mol. Cell.
Biol., 1, 600-608.
64.Pratt, K., and Hattman, S. (1983) J.
Protozool., 30, 592-598.
65.Bromberg, S., Pratt, K., and Hattman, S. (1982)
J. Bacteriol., 150, 993-996.
66.Blackburn, E. H., Pan, W.-C., and Johnson, C. C.
(1983) Nucleic Acids Res., 11, 5131-5145.
67.Harrison, B. S., Findly, R. C., and Karrer, K. M.
(1986) Mol. Cell. Biol., 6, 2364-2370.
68.Capowski, H. E., Wells, J. M., Harrison, G. S.,
and Karrer, K. M. (1989) Mol. Cell. Biol., 9,
2598-2605.
69.Brenner, V., Venetianer, P., and Kiss, A. (1992)
Nucleic Acids Res., 18, 354-359.
70.Horowitz, S., and Gorovsky, M. A. (1985) Proc.
Natl. Acad. Sci. USA, 82, 2452-2455.
71.Martindale, D. W. (1989) J. Protozool.,
36, 29-34.
72.Fauman, E. B., Blumenthal, R. M., and Cheng, X.
(1999) in S-Adenosylmethionine-Dependent Methyltransferases:
Structure and Function (Cheng, X., and Blumenthal, R. M., eds.)
World Scientific Publishing, pp. 1-38.
73.Kagan, R. M., and Clarke, S. (1994) Arch.
Biochem. Biophys., 310, 417-427.
74.Niewmierzycka, A., and Clarke, S. (1999) J.
Biol. Chem., 274, 814-824.
75.Nakahigashi, K., Kubo, N., Narita, S., Shimaoka,
T., Goto, S., Oshima, T., Mori, H., Maeda, M., Wada, C., and Inokuchi,
H. (2002) Proc. Natl. Acad. Sci. USA, 99, 1473-1478.
76.Karamyshev, A. L., Ito, K., and Nakamura, Y.
(1999) FEBS Lett., 457, 483-488.
77.Bujnicki, J. M., Feder, M., Radlinska, M., and
Blumenthal, R. M. (2002) J. Mol. Evol., 55, 431-444.
78.Bruns, P. J., and Brussard, T. B. (1974)
Genetics, 78, 831-841.
79.Yao, M.-C. (1989) in Mobile DNA (Berg, D.
E., and Howe, M. M., eds.) ASM Press, Washington, D. C., pp.
715-734.
80.Katzen, A. L., Lann, G. M., and Blackburn, E. H.
(1981) Cell, 24, 313-320.
81.Austerberry, C. F., Allis, C. D., and Yao, M.-C.
(1984) Proc. Natl. Acad. Sci. USA, 81, 7383-7387.
82.Yokoyama, R. W., and Yao, M.-C. (1982)
Chromosoma (Berl.), 85, 11-22.
83.Yao, M.-C., and Yao, C.-H. (1987) Cell,
48, 779-788.
84.Lauth, M. R., Spear, B. B., Heumann, J., and
Prescott, D. M. (1976) Cell, 7, 67-74.
85.De la Campa, A. G., Springhorn, S. S., Kale, K.,
and Lacks, S. A. (1988) J. Biol. Chem., 263,
14696-14702.
86.Sladek, T. L., Novak, J. A., and Maniloff, J.
(1986) J. Bacteriol., 165, 219-225.
87.Stewart, F. J., and Raleigh, E. A. (1998)
Biol. Chem., 379, 611-616.
88.Nelson, M., and McClelland, M. (1991) Nucleic
Acids Res., 19, 2045-2071.
89.Fan, Q., and Yao, M.-C. (2000) Nucleic Acids
Res., 28, 895-900.
90.Triglia, T., Willems, T. E., and Kemp, D. J.
(1992) Parasitol. Today, 8, 225-229.
91.Pollack, Y., Katzen, A. L., Spira, D. T., and
Golenser, J. (1982) Nucleic Acids Res., 10, 539-546.
92.Weber, J. L. (1987) Gene, 52,
103-109.
93.Shorning, B. Yu., and Vanyushin, B. (2001)
Biochemistry (Moscow), 66, 753-762.
94.Pollack, Y., Kogan, N., and Golenser, J. (1991)
Exp. Parasitol., 72, 339-334.
95.Pinarbasi, E., Elliott, J., and Hornby, D. A.
(1996) J. Mol. Biol., 257, 804-813.
96.Stephens, C., Reissenauer, A., Wright, R., and
Shapiro, L. (1996) Proc. Natl. Acad. Sci. USA, 93,
1210-1214.
97.Kossykh, V. G., and Lloyd, R. S. (2004) J.
Bacteriol., 186, 2061-2067.
98.Urieli-Shoval, S., Gruenbaum, Y., Sedat, J., and
Razin, A. (1982) FEBS Lett., 146, 148-152.
99.Gowher, H., Leismann, O., and Jeltsch, A. (2000)
EMBO J., 19, 6918-6923.
100.Lyko, F., Ramsahoye, B. H., and Jaenisch, R.
(2000) Nature, 408, 538-540.