Cloning and Characterization of Zebra Fish SPATA4 Gene and Analysis of Its Gonad Specific Expression
Shangfeng Liu1,2, Bowen Liu1, Shan He1, Ying Zhao1, and Zhao Wang1,2*
1Department of Biological Sciences and Biotechnology, Tsinghua University, Beijing 100084, People's Republic of China; fax: +86-10-62772240; E-mail: liusf@mail.tsinghua.edu.cn; lbw@mails.tsinghua.edu.cn; hes@mails.tsinghua.edu.cn; y-zhao02@mails.tsinghua.edu.cn2Medical School, Tsinghua University, Beijing 100084, China; E-mail: zwang@tsinghua.edu.cn
* To whom correspondence should be addressed.
Received November 23, 2004
The spermatogenesis associated 4 gene (SPATA4, previously named TSARG2) was first cloned in human tissues and was reported to be a candidate spermatocyte apoptosis-related gene that is expressed specifically in testis. Analysis of SPATA4 expression and regulation in zebra fish may provide insight into the understanding of the complicated process of gonadogenesis. In this study, we cloned and characterized the SPATA4 gene from zebra fish (Danio rerio), which is homologous to human and mouse SPATA4. Zebra fish SPATA4 consists of six exons separated by five introns, as all SPATA4 genes in vertebrates. A promoter region was predicted using homologous blast and cloned for further study, and possible transcription factors were analyzed in this region. The putative protein encoded by this gene was analyzed using bioinformatics methods. Multi-tissue RT-PCR results demonstrated that the zebra fish SPATA4 gene is expressed specifically in testis and slightly in ovary. Analysis of the SPATA4 sequence and its spatial expression pattern indicate that this gene is highly conserved and may play an important role in the process of zebra fish gonadogenesis.
KEY WORDS: bioinformatics, gonad-specific expression, SPATA4, zebra fish, testis/ovary
The apoptosis of gonadic cells is a complicated process involving multi-gene interactions [1, 2] whose study may lead to further understanding of the apoptosis mechanism and the biological processes of gonad cells and clarify physiological and pathological processes of fertility [3]. Certain specific gene activities and proteins involved in this apoptosis process include Bcl-2 family, p53, and Fas [4]. It has been reported that 24 ESTs of the SPATA4 gene (previously named TSARG2) were first cloned from mouse testis spermatogenic cells by creating a mouse cryptorchidism model and making use of suppression subtractive hybridization [5]. Subsequently, the full-length cDNA sequence of the SPATA4 gene was cloned from a mouse testis cDNA library starting with the EST BE644542 and human SPATA4 gene was cloned using nested PCR and draft human genome searching [6]. The full length cDNA of the human SPATA4 gene was then cloned and deduced as a candidate oncogene [7]. The primary functional study indicated that the SPATA4 gene is highly conserved in various vertebrates including human, chimpanzee, mouse, and rat (unpublished).
The zebra fish (Danio rerio), a powerful vertebrate model organism, is becoming widely used to study vertebrate biology and human diseases because it is relatively easy to access a large number of mutations affecting a wide range of developmental pathways and physiological systems [8-10]. The study of genes that are specifically expressed in zebra fish gonad tissues is at a primitive stage [11], with only a few gonad-specific genes being identified. It has been reported that Sox30 is involved in the differentiation of male germ cells and locates in normal testes tissues [12]. Another gene, ziwi, is expressed exclusively in the zebra fish gonad [13]. Furthermore, two testicular cell lines of zebra fish with distinct functions have been established to support the development of male germ cells [14]. The more in-depth study of gonad specific genes in zebra fish would be highly important and revealing to understand the complex process of gonadogenesis.
In this study, we cloned and characterized the zebra fish SPATA4 gene using bioinformatics methods and analyzed its chromosomal localization and the putative protein. To primarily study the distribution of SPATA4 in zebra fish, multi-tissue RT-PCR was applied to determine its tissue-specific expression in various organ tissues from adult zebra fish. The results suggested that the zebra fish SPATA4 gene is highly conserved in vertebrates and specifically expressed in adult zebra fish gonad.
MATERIALS AND METHODS
Silicon cloning and analysis of zebra fish SPATA4. We cloned and analyzed the full length cDNA sequence and putative amino acid sequence of zebra fish SPATA4 gene (GeneBank accession No. AY651920) using bioinformatics methods. First the full length cDNA sequence of the human SPATA4 gene (GeneBank accession No. AY040204) was obtained directly from Genebank database at the National Center for Biotechnology Information (NCBI), which was used to blast the zebra fish EST database and found EST CN017877 and BI673645. A 942-bp fragment was obtained from the two ESTs and RT-PCR was performed to clone the full length cDNA of this gene. A PCR kit (Sangon, China) was used according to the standard protocol to clone and validate the open reading frame (ORF) of the SPATA4 gene in the zebra fish genome DNA. For PCR amplification, primers designed and synthesized based on the zebra fish genomic DNA sequence of SPATA4 were ZF1: 5´-CGG AAT TCA AAT GGC CTA TTC ACT GTC GCC-3´ and ZF2: 5´-CGG TCG ACC TTG TTG GGA AGG CAG GCA C-3´ according to the 942-bp fragment. The primers for beta-actin were designed as [15]: forward 5´-GTC CCT GTA CGC CTC TGG TCG-3´ and reverse 5´-GCC GGA CTC ATC GTA CTC CTG-3´ as positive control. PCR was carried out in 10 µl volumes containing 1 U of Taq DNA polymerase, 2 mM MgCl2, 250 µM of each dNTP, 0.4 µM of each primer, and 1 µl cDNA template from the reverse transcription reaction. Following an initial denaturation step at 95°C for 1 min 30 sec, the reaction was subjected to 35 cycles of amplification at 94°C for 40 sec, 58°C for 30 sec, 72°C for 40 sec, and a final extension for 5 min. A 2-µl sample of each reaction was size-fractioned by 2% (w/v) agarose gel electrophoresis and stained with ethidium bromide and photographed under UV light using Imagemaster VDS (Amersham Pharmacia Biotech, USA). This PCR fragment was cloned into pUCm-T vectors and sequenced.
Chromosomal localization of zebra fish SPATA4 was obtained by blasting the zebra fish genomic DNA with the cDNA sequence of SPATA4 using Ensemble Zebra Fish Genome Browser (http://www.ensembl.org/Danio_rerio/) and the adjacent EST marker fj36c01.×1 (GeneBank accession No. AW077435) was found. By blasting fj36c01.×1 in the Tuebingen Map of the Zebra Fish Genome, the positions of the gene loci were intercalated on the radiation hybrid map of zebra fish chromosome linkage group based on the Goodfellow T51 radiation hybrid panel [16-18] (http://wwwmap.tuebingen.mpg.de/).
Deduction and analysis of putative protein SPATA4. The molecular mass and isoelectric point of the putative protein SPATA4 from zebra fish were predicted using the Compute pI/Mw tool at the ExPASy molecular biology WWW server of the Swiss Institute of Bioinformatics (http://www.expasy.ch/). SignalP was used to search possible signal peptides [19] and the transmembrane regions, and orientation of zebra fish SPATA4 was detected using the Tmpred program (http://www.ch.embnet.org/software/TMPRED_form.html/). Its subcellular localization was indicated by PSORT [20, 21]. Motif searches were performed with PROSITE programs at the ExPASy server [22] (http://cn.expasy.org/prosite).
Zebra fish multi-tissue preparation. Zebra fish were obtained from the Laboratory of Developmental Biology and cultured in filtered and aerated water at 28.5°C and maintained on a 24-h cycle of 14-h light and 10-h darkness. Fish breeding was conducted according to established protocols [23]. Eight organs of wild-type zebra fish were dissected under the microscope as follows: brain (whole), eyes (whole), skin (without scales), ovary (whole), intestine (total), heart (whole), skeletal muscle, and testis (total). The tissues were transferred to 1.5 ml Eppendorf tubes and frozen in liquid nitrogen immediately and stored frozen at -70°C.
RNA extraction and RT-PCR. RT-PCR was conducted to reveal the tissue-specific expression of the SPATA4 gene in adult zebra fish. Total RNA was extracted from the tissues described above using an RNA Isolation Kit (Gentra, USA) according to the protocols recommended by the supplier. One milligram of total RNAs from each sample was used as the template for the synthesis of first strand cDNA by reverse transcriptase using an MMLV First Strand cDNA Synthesis Kit (Sangon). Primers were designed and synthesized as ZF3: 5´-GTG AAG CAA AAC ATC AGT CTG G-3´ and ZF4: 5´-CGG TCG ACC GAC AAG CTG CTT GAC TGG TGT TC-3´. The condition and protocol of PCR amplification was the same as described previously to detect the expression of SPATA4 gene in each sample.
Cloning and computer-aided analysis of the SPATA4 promoter region. We aligned the upstream sequence of zebra fish genomic DNA with the human and mouse promoter region using Clustal W at the European Bioinformatics Institute server to deduce the promoter of zebra fish SPATA4. PromoterInspector was used to predict the typical promoter of zebra fish SPATA4 at the Genomatix server (http://www.genomatix.de/). To clone the promoter region of this gene in zebra fish, primers were designed and synthesized as: pZF1, 5´-GCG GTA CCG GAT GGA TGA TGC ACA GTG CTT G-3´, and pZF2, 5´-AAG CTT AAA TGA AAT ATA TGA ACT AAT TAC CG-3´. Zebra fish genomic DNA was extracted with a DNA Extract Kit (Gentra) and used as templates, and PCR conditions and protocols were the same as described previously. The upstream sequence of the first exon of zebra fish SPATA4 genomic DNA was examined to search for known regulatory elements using MatInspector (http://www.genomatix.de/cgi-bin/matinspector/matinspector.pl).
RESULTS
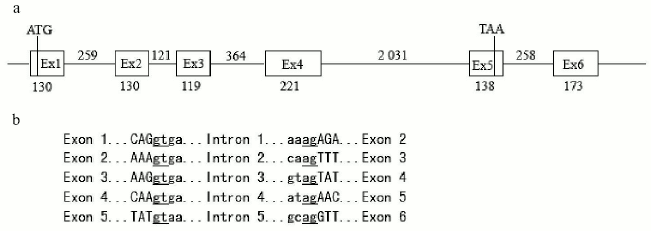
Cloning and analysis of the zebra fish SPATA4 gene. Primers were designed according to the 942 bp sequence mentioned previously and zebra fish cDNA was used in PCR. The PCR fragment was cloned into pUCm-T vectors and sequenced. The complete cDNA sequence of zebra fish SPATA4 is 942 bp in length containing six exons with the start codon located in the first exon and the stop codon in the fifth exon. The full length cDNA sequence of zebra fish SPATA4 and the amino acid sequence of the putative protein are shown in Fig. 1. The exon-intron junctions were determined by comparing zebra fish SPATA4 cDNA sequence with the genomic DNA sequence, which obeys the AG/GT rule (Fig. 2). A potential polyadenylation signal (AATAAA) was detected at nucleotide position 857.
By searching the full length cDNA of the SPATA4 gene in the zebra fish genome using Ensemble Genome Browser (http://www.ensembl.org/), zebra fish SPATA4 was mapped on chromosome 14 from 49,939,363 to 49,943,306 bp, and six exons were localized within this region in accordance with the prediction (Fig. 2). The closest EST marker fj36c01.×1 was used to locate the SPATA4 gene in the zebra fish genome. By blasting the Tuebingen Map of the Zebra fish Genome, the position of the gene loci of zebra fish SPATA4 was intercalated on the heat shock mapping (HS mapping) based on the Goodfellow T51 radiation hybrid panel [24], which was between the two simple sequence-length polymorphism markers (SSLP) z22107 to z21080 with a distance of 2.3 centimorgans from each other (Fig. 3).Fig. 1. cDNA sequence and predicted protein sequence of zebra fish SPATA4 gene. Polyadenylation signal is underlined; stop codon is indicated by an asterisk (*).
Fig. 2. Structure of genomic DNA sequence of zebra fish SPATA4 gene. a) Genomic DNA sequences from the database were aligned with cDNA sequence of SPATA4 to predict the intron-exon junctions. The start codon ATG is in the first exon and stop codon TAA in the fifth exon. The numbers indicate the length of each exon or intron (bp). b) Exon-intron junction sequences. Uppercase letters correspond to the exon sequences while lowercase letters correspond to intron sequences. Splice donor and acceptor sites are underlined. The junctions obey the AG/GT rule.
Analysis and deduction of the putative protein of SPATA4. Analysis of the zebra fish SPATA4 gene sequence using ORF finder at NCBI revealed an open reading frame of 675 bp encoding a putative protein of 224 amino acids with a molecular mass of 25,697 daltons and isoelectric point of 9.44. No signal peptide was predicted in the amino acid sequence of zebra fish SPATA4 using SignalP version 3.0, and no transmembrane domain could be detected using the TMpred program. The cellular location prediction using PSORT indicated that zebra fish SPATA4 is most probable a nuclear protein (43.5%) with low possibility to locate in cytoplasm (26.1%) and mitochondria (21.7%). Four motifs were identified using PROSITE within the amino acid sequence of zebra fish SPATA4 as follows: protein kinase C phosphorylation site, N-glycosylation site, N-myristoylation site, and casein kinase II phosphorylation site.Fig. 3. Chromosomal localization of the zebra fish SPATA4 gene. a) The blast result using Ensemble Zebra Fish Genome Browser (http://pre.ensembl.org/Danio_rerio/). A scale bar illustrates the physical map coordinates this particular region falls into. DNA contigs shown in alternating dark and light gray color depict the individual contig sequences that form the genome sequence assembly. Where no dark gray contig is shown, this indicates a gap in the assembly. The positions of Sequence Tagged Sites (STS) markers are indicated in italic directly under the contig map ideogram. Stippled boxes represent the zebra fish SPATA4 gene. b) The localization of SPATA4 is represented on the radiation hybrid map of zebra fish chromosome LG14 (LG for linkage group) [16]. Position of zebra fish SPATA4 on the chromosome is designated by an arrow. SSLPs are shown on the genetic map (left) created by the lab of Mark Fishman [27] with the distances between them indicated (in centimorgans). Labels on the radiation hybrid map (right) are all markers linked to at least one other marker with a LOD score of six or more. Gaps indicate two-point LOD scores of less than six. SSLPs are shown in standard font. Published genes and ESTs are underlined by wavy line. Other STSs including anonymous markers and BAC ends are underlined by straight solid line. Mutations are underlined by dashed line.
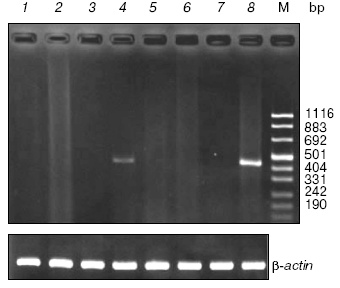
Spatial expression of the SPATA4 gene in adult zebra fish. RT-PCR was performed with mRNA extracted from eight organs of adult zebra fish to determine the tissue-specific expression pattern of the SPATA4 gene, which appeared as a band of 435 bp in 2% agarose gel. Abundant amplification product of this gene was detected using zebra fish SPATA4 specific primers and cDNA templates from testis tissues and slight amount of PCR products were observed when cDNA from ovary was used as template. No PCR products were amplified with cDNA derived from other tissues from zebra fish (Fig. 4).
Cloning and characterization of the SPATA4 promoter region. No typical promoter could be identified using bioinformatics methods or homologous alignments at the 5´ flanking region of zebra fish SPATA4. However, we have cloned the upstream region of the first exon of zebra fish SPATA4 genomic DNA, which was supposed to consist of the promoter region of this gene (Fig. 5). Possible transcription factors were predicted in the upstream region sequence of zebra fish SPATA4 genomic DNA using MatInspector at Genomatix server (http://www.genomatix.de/), such as prostate-specific homeodomain protein NKX3.1, GATA-binding factor 1 to 3, TTF1, MEF2, Gsh-1, cAMP-responsive element binding protein 1, Sox-5, etc.Fig. 4. Expression of the SPATA4 gene in eight organ tissues. Total RNA was extracted from adult zebra fish tissues for multi-tissue RT-PCR using the SPATA4 specific primers. From left to right are: 1) brain; 2) eye; 3) skin; 4) ovary; 5) intestine; 6) heart; 7) skeleton muscle; 8) testis; M) pUC 8 marker ladder. The expected 435 bp band was observed in zebra fish testis and ovary (upper panel). The same samples were assayed for beta-actin mRNA expression as a positive control (lower panel).
Fig. 5. The 5´-flanking region of SPATA4 in zebra fish. Primer sequence is underlined; exon 1 is shadowed; start codon is framed.
DISCUSSION
The SPATA4 gene was first cloned as an apoptosis-related testis-specific gene in mouse and human and was hypothesized to be related to spermatogenesis. To further study the characteristics and distribution of the SPATA4 gene, the DNA sequence was cloned and deduced putative protein was analyzed from adult zebra fish, which is a widely used and powerful model organism for studying vertebrates.
The sequence of the SPATA4 gene is highly conserved in human, mouse, rat, chimpanzee, and other vertebrates, all consisting six exons and five interval introns (unpublished). The subcellular localization and the property of the putative protein were analyzed indicating that SPATA4 is with the highest possibility a nuclear protein. According to the previous study, SPATA4 was speculated to be a spermatocyte apoptosis-related gene and candidate oncogene, indicating that the putative protein SPATA4 was potentially involved in the transcription signal pathway during gonadogenesis. The results of multi-tissue RT-PCR exhibited a spatial-specific expression pattern. The zebra fish SPATA4 gene is expressed specifically in adult zebra fish testis and a slight amount of expression was observed in ovary. We also cloned the sequence upstream of the first exon of zebra fish genomic DNA, which was considered to consist of the zebra fish SPATA4 promoter, and further study of its function will be performed.
The zebra fish is considered to be a gonochoristic, undifferentiated species, which means that both sexes pass through an ovary-like stage before differentiating into the phenotypic sex. In males, this includes a type of juvenile hermaphroditism. The males pass through a phase as females, before the final development of male gonads [25]. SPATA4 transcript was detected exclusively in human and mouse testis [6, 7, 26], but in zebra fish, there was a trace expression in ovary. This result may relate to the development process (especially sex control and sex reversal under natural conditions) of zebra fish. We speculated that this gene played an important role in growth regulation of germ cells in zebra fish. However, the mechanism and the significance of the spatial expression of SPATA4 in zebra fish and other vertebrates remain to be studied.
The promoter region of the zebra fish SPATA4 gene was obtained by PCR assay. Mutation of the human SPATA4 gene was not found in 122 cases of azoospermia, severe oligzoospermia, and cryptorchidism by PCR-SSCP, suggesting that this gene might carry out its function to up-regulate or down-regulate through promoter methylation [7]. The cloning of this promoter region will provide a useful tool for understanding the regulation of SPATA4 and will contribute new insight into its role in zebra fish testis and ovary.
This work was supported by research grants of the Tsinghua-Yue-Yuen Medical Sciences Fund (grant THYY20040008) and the China Postdoctoral Science Foundation (grant 2004035052). We thank the Laboratory of Developmental Biology, Tsinghua University (Beijing) for providing zebra fish samples.
REFERENCES
1.Eddy, E. M. (2002) Recent Progr. Horm. Res.,
57, 103-128.
2.Vanderhyden, B. (2002) Front. Biosci.,
7, 2006-2022.
3.Nakanishi, Y. (1995) Yakugaku Zasshi.,
115, 420-430.
4.Sasagawa, I., Yazawa, H., Suzuki, Y., and Nakada,
T. (2001) Arch. Androl., 47, 211-216.
5.Jiang, H., Li, L. Y., and Lu, G. X. (2001) Sheng
Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai), 33,
421-425.
6.Liu, S. F., Li, L. Y., Fu, J. J., Liu, G., Xing, X.
W., and Lu, G. X. (2002) Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue
Bao (Shanghai), 34, 378-382.
7.Liu, S. F., Lu, G. X., Liu, G., Xing, X. W., Li, L.
Y., and Wang, Z. (2004) Biochem. Biophys. Res. Commun.,
319, 32-40.
8.Driever, W., Solnica-Krezel, L., Schier, A. F.,
Neuhauss, S. C., Malicki, J., Stemple, D. L., Stainier, D. Y.,
Zwartkruis, F., Abdelilah, S., Rangini, Z., Belak, J., and Boggs, C.
(1996) Development, 123, 37-46.
9.Haffter, P., Granato, M., Brand, M.,
Mullins, M. C., Hammerschmidt, M., Kane, D. A., Odenthal, J., van
Eeden, F. J., Jiang, Y. J., Heisenberg, C. P., Kelsh, R. N.,
Furutani-Seiki, M., Vogelsang, E., Beuchle, D., Schach, U.,
Fabian, C., and Nusslein-Volhard, C. (1996) Development,
123, 1-36.
10.Dooley, K., and Zon, L. I. (2000) Curr. Opin.
Genet. Dev., 10, 252-256.
11.Braat, A. K., Speksnijder, J. E., and Zivkovic,
D. (1999) Int. J. Dev. Biol., 43, 745-760.
12.Osaki, E., Nishina, Y., Inazawa, J., Copeland, N.
G., Gilbert, D. J., Jenkins, N. A., Ohsugi, M., Tezuka, T.,
Yoshida, M., and Semba, K. (1999) Nucleic Acids Res.,
27, 2503-2510.
13.Tan, C. H., Lee, T. C., Weeraratne, S. D.,
Korzh, V., Lim, T. M., and Gong, Z. (2002) Mech. Dev.,
119, Suppl. 1, 221-224.
14.Kurita, K., and Sakai, N. (2004) Mol. Reprod.
Dev., 67, 430-438.
15.Chen, W. Y., John, J. A., Lin, C. H., and Chang,
C. Y. (2002) Biochem. Biophys. Res. Commun., 291,
798-805.
16.Geisler, R., Rauch, G.-J., Baier, H., van Bebber,
F., Bro, L., Dekens, M. P. S., Finger, K., Fricke, C., Gates, M. A.,
Geiger, H., Geiger-Rudolph, S., Gilmour, D., Glaser, S., Gnügge,
L., Habeck, H., Hingst, K., Holley, S., Keenan, J., Kirn, A., Knaut,
H., Lashkari, D., Maderspacher, F., Martyn, U., Neuhauss, S., Neumann,
C., Nicolson, T., Pelegri, F., Ray, R., Rick, J. M., Roehl, H., Roeser,
T., Schauerte, H. E., Schier, A. F., Schönberger, U.,
Schönthaler, H.-B., Schulte-Merker, S., Seydler, C., Talbot, W.
S., Weiler, C., Nüsslein-Volhard, C., and Haffter, P. (1999)
Nat. Genet., 23, 86-89.
17.Kelly, P. D., Chu, F., Woods, I. G.,
Ngo-Hazelett, P., Cardozo, T., Huang, H., Kimm, F., Liao, L., Yan, Y.
L., Zhou, Y., Johnson, S. L., Abagyan, R., Schier, A. F., Postlethwait,
J. H., and Talbot, W. S. (2000) Genome Res., 10,
558-567.
18.Woods, I. G., Kelly, P. D., Chu, F.,
Ngo-Hazelett, P., Yan, Y. L., Huang, H., Postlethwait, J. H., and
Talbot, W. S. (2000) Genome Res., 10, 1903-1914.
19.Nielsen, H., Engelbrecht, J., Brunak, S., and von
Heijne, G. (1997) Protein Eng., 10, 1-6.
20.Nakai, K., and Horton, P. (1999) Trends
Biochem. Sci., 24, 34-36.
21.Emanuelsson, O., Nielsen, H., Brunak, S., and von
Heijne, G. (2000) J. Mol. Biol., 300, 1005-1016.
22.Sigrist, C. J., Cerutti, L., Hulo, N., Gattiker,
A., Falquet, L., Pagni, M., Bairoch, A., and Bucher, P. (2002)
Brief Bioinform., 3, 265-274.
23.Westerfield, M. (1995) The Zebra fish Book: a
Guide for the Laboratory Use of Zebra fish (Danio rerio),
University of Oregon Press, Eugene, USA.
24.Kwok, C., Korn, R. M., Davis, M. E., Burt, D. W.,
Critcher, R., McCarthy, L., Paw, B. H., Zon, L. I., Goodfellow, P. N.,
and Schmitt, K. (1998) Nucleic Acids Res., 26,
3562-3566.
25.Bull, J. J. (1983) Evolution of Sex
Determining Mechanisms, Benjamin/Cummings, Menlo Park, CA.
26.Liu, S. F., Li, L. Y., Mo, Y. Q., Fu, J. J., Liu,
G., Xing, X. W., and Lu, G. X. (2003) Yi Chuan Xue Bao,
30, 943-948.
27.Knapik, E. W., Goodman, A., Ekker, M., et al.
(1998) Nat. Genet., 18, 338-343.