Characterization and Stability Investigation of Cu,Zn-Superoxide Dismutase Covalently Modified by Low Molecular Weight Heparin
H. W. Zhang, F. S. Wang*, W. Shao, X. L. Zheng, J. Z. Qi, J. C. Cao, and T. M. Zhang
Institute of Biochemical and Biotechnological Drugs, School of Pharmacy, Shandong University, Jinan 250012, China; fax: 0086-531-8380288; E-mail: fswang@sdu.edu.cn* To whom correspondence should be addressed.
Received February 18, 2005; Revision received March 21, 2005
Cu,Zn-superoxide dismutase (SOD) was chemically modified with low molecular weight heparin (LMWH). To characterize the conjugate, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and native polyacrylamide gel electrophoresis (native PAGE) with protein staining and polysaccharide staining were employed. The stabilities of the modified enzyme to heat, acid, alkali, and trypsin treatment were also investigated. SDS-PAGE of the conjugate presented two major bands, and native PAGE of the conjugate showed similar banding position with protein staining and polysaccharide staining, which was different from that of the unmodified SOD and LMWH/SOD mixture. Moreover, the conjugate migrated faster with increasing extent of the modification. Enhanced heat stability, acid resistance, alkali resistance, and anti-trypsin stability of the modified enzyme were observed compared with those of the unmodified enzyme. Results of the study suggest that covalent linkage in LMWH-SOD can be effectively characterized by electrophoretic techniques and the chemical modification of SOD with LMWH can enhance the stabilities of the enzyme. In addition, native PAGE with protein staining can be used to evaluate the extent of the modification.
KEY WORDS: Cu,Zn-superoxide dismutase, low molecular weight heparin, chemical modification, covalent conjugate, electrophoresis, stabilityDOI: 10.1134/S0006297906130165
Abbreviations: SOD) Cu,Zn-superoxide dismutase; LMWH) low molecular weight heparin; SDS-PAGE) sodium dodecyl sulfate polyacrylamide gel electrophoresis; native PAGE) native polyacrylamide gel electrophoresis.
Because of shortcomings of proteins used as medicines, such as short
half-life, antigenicity, and instability, increasing attention has been
given to chemical modification of proteins. Among the modifying agents,
polysaccharides are widely used [1, 2]. The polysaccharides conjugated to proteins could
form a masking layer on the surface of the proteins and exert
protective effects for the protein molecules. Cu,Zn-superoxide
dismutase (SOD), a free radical scavenger, has the ability to protect
cells or tissues from oxidative damage. As a promising candidate for
many clinical practices, attempts have been made in recent years to
improve the pharmacokinetic properties and reduce the immunogenicity of
the native enzyme [3, 4]. Among
the approaches used, PEGylation was a successful strategy, and
polyethylene glycol-superoxide dismutase (PEG-SOD) conjugate is
commercially available. The successful development of PEG-SOD conjugate
has invoked a new round of exploration of chemical modification of SOD
with other novel modifying agents aiming to convey targeting or produce
more satisfactory bioactivities [5]. Low molecular
weight heparin (LMWH), a mucopolysaccharide, has antithrombin and
anti-inflammatory activities and shows growing clinical applications
[6, 7]. Therefore, with the
anticipation that the combination of the two bioactive molecules, LMWH
and SOD, could provide improved properties, such as long half-life, low
immunogenicity, increased stability, and enhanced anti-inflammatory
activity, we used LMWH to chemically modify SOD.
In this paper we report on the characterization of LMWH-SOD conjugate using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and native PAGE with protein staining and polysaccharide staining and on some of the stabilities of the modified enzyme including heat stability, acid and alkali resistance, and anti-trypsin stability.
MATERIALS AND METHODS
Chemicals. Cu,Zn-SOD (Yanghe Biotechnological Co. Ltd., China), LMWH (4-5 kD, Dongcheng Biochemical Co. Ltd., China), trypsin (Yuanju Biotechnological Co. Ltd., China), and low molecular weight protein SDS markers (Shanghai Institute of Biochemistry, Chinese Academy of Sciences) were used. Other chemicals and reagents were of analytical grade.
Enzymatic activity assay. SOD activity was assayed according to the improved pyrogallol autooxidation method [8].
Preparation of LMWH-SOD conjugate. SOD was dissolved in 0.3 M sodium carbonate buffer (pH 9.5), the activated LMWH prepared by periodate oxidation [9] was added, and the reaction solution was agitated slowly in dark at 4°C for 48 h. During the reaction process, aliquots of the solution were pipetted out at 0, 12, 24, and 48 h for electrophoretic analysis. The reaction was terminated by adding glycine. Subsequently, the solution was subjected to a DEAE-Sepharose FF (Pharmacia, Sweden) column (2.6 × 40 cm) chromatography and the eluent possessing enzymatic activity was collected, desalted, concentrated, and lyophilized.
Considering the possible non-covalent interactions between SOD and LMWH under the designed modification conditions, the same amount of SOD and LMWH were mixed together in 0.3 M sodium carbonate buffer (pH 9.5) and the mixture was also slowly stirred in dark at 4°C for 48 h. Samples were taken at different time points for electrophoresis.
Electrophoresis. Electrophoresis was performed using the method described by Li et al. [10]. For SDS-PAGE, a discontinuous system was employed; the total monomer concentration of the stacking gel was 3% (w/v) and the separation gel was 15% (w/v). Dimensions of the gel immersed in running buffer (0.1% SDS, 0.05 M Tris/0.384 M glycine buffer, pH 8.3) were 8 × 8 × 0.1 cm. Samples in loading buffer (1% SDS, 1% mercaptoethanol, 20% glycerol, 0.02% bromophenol blue, 0.01 M Tris-HCl, pH 8.0) were incubated at 100°C for 5 min before electrophoresis and enzymatic activity analysis, whereas some samples of the LMWH-SOD conjugate were incubated at 50°C for 5 min in order to investigate the effect of the temperature. Electrophoresis was performed at room temperature for approximately 2 h, and the system was programmed to a two-step mode with applying constant current 10 mA in stacking gel and 20 mA in the separation gel. Gels were stained with Coomassie Blue R-250 (Fluka, Germany) overnight and were then washed in destaining solution for 4 h.
For native PAGE in a discontinuous system, the total monomer concentration of the stacking gel was 3.75% and the separation gel was 10%. The electrophoretic program was set as for SDS-PAGE except that the running buffer was 0.05 M Tris/0.384 M glycine buffer (pH 8.3) and the loading buffer was 40% sucrose solution containing 0.1% bromophenol blue. Gels were stained with 0.05% Coomassie Blue R-250 solution containing 20% sulfosalicylic acid for 1 h and were then washed in destaining solutions for 2 h, or were stained with Alcian Blue for 1 h and then washed with 2% acetic acid for 2 h.
Acid and alkali treatment of LMWH-SOD conjugate. The LMWH-SOD conjugate and unmodified SOD were dissolved in a 0.2 M Na2HPO4/0.1 M citric acid buffer (pH 5.2) or in 0.1 M Na2CO3/NaHCO3 buffer (pH 10.8) and then incubated at 25°N for 6 h.
Heat and trypsin treatment of LMWH-SOD conjugate and unmodified SOD. The LMWH-SOD conjugate or unmodified SOD were dissolved in 0.2 M phosphate buffer (pH 7.0) and then incubated at 80°N for 1 h. The LMWH-SOD conjugate and unmodified SOD were dissolved in 2 ml of 1 mg/ml trypsin solution buffered at pH 7.84 with 0.2 M Na2HPO4/0.2 M NaH2PO4 buffer and incubated at 25°N for 1 h.
RESULTS
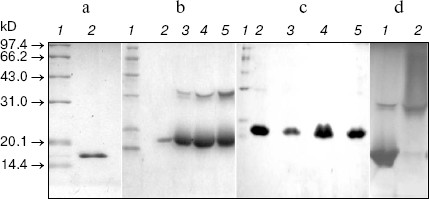
SDS-PAGE of LMWH-SOD conjugate and LMWH/SOD mixture. The molecular weight of SOD was characterized by SDS-PAGE, and a single band was shown at the position of 16 kD (Fig. 1a). LMWH-SOD conjugate presented heterogeneity on SDS-PAGE, and protein bands near 30 kD showed increasing density with the extending of modification reaction time (Fig. 1b). Moreover, the residual enzymatic activity of the LMWH-SOD conjugate after the treatment at 100°C for 5 min in the loading buffer increased with the extending of the reaction time. In contrast, the samples of the LMWH/SOD mixture incubated at 4°N for different time presented the same migration rate and homogeneity as unmodified SOD (Fig. 1c). In addition, Fig. 1d demonstrates that LMWH-SOD conjugate incubated at 50°C for 5 min in the loading buffer exhibited a continuous distribution over a wide molecular weight range above 32 kD.
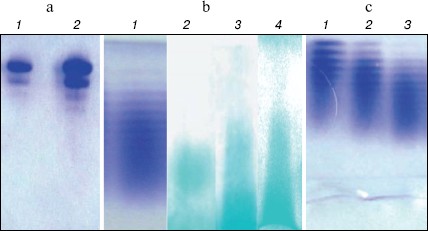
Native PAGE of LMWH-SOD conjugate and LMWH/SOD mixture. The unmodified SOD exhibited almost the same banding patterns on native PAGE compared with the LMWH/SOD mixture incubated at 4°N for 48 h (Fig. 2a, see color insert, p. 5), suggesting that non-covalent interactions did not occur between LMWH and SOD. However, the protein bands of LMWH-SOD conjugate presented significantly wider distribution along the electrophoretic lane, and polysaccharide staining with Alcian Blue also exhibited the differences of LMWH-SOD conjugate, LMWH/SOD mixture, and LMWH (Fig. 2b, see color insert, p. 5). The protein bands and polysaccharide bands of LMWH-SOD conjugate showed similar patterns for the most part, while the polysaccharide bands of the LMWH/SOD mixture were similar to that of LMWH but the banding distribution was wider than that of the LMWH-SOD conjugate. In addition, LMWH-SOD conjugates obtained by modifying for 12, 24, and 48 h formed “decreasing ladder” protein bands on the native PAGE (Fig. 2c, see color insert, p. 5).Fig. 1. SDS-PAGE analysis of unmodified SOD, LMWH-SOD conjugate, and LMWH/SOD mixture. a: 1) Markers; 2) unmodified SOD. b) LMWH-SOD conjugates of different modification time: 1) markers; 2-5) samples modified for 0, 12, 24, and 48 h. c) LMWH/SOD mixture: 1) markers; 2-5) samples incubated for 0, 12, 24, and 48 h. d) LMWH-SOD conjugate incubated at different temperature for 5 min in the loading buffer: 1) 100°C; 2) 50°C.
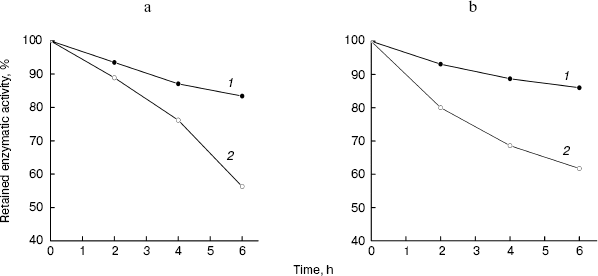
Stability of LMWH-SOD conjugate under acidic and basic conditions. For LMWH-SOD conjugate and unmodified SOD, both acid and alkali treatments resulted in the loss of the enzymatic activity, but LMWH-SOD conjugate always showed superior endurance than unmodified SOD towards the treatments. Compared with enzymatic activity at zero time, the residual activity of LMWH-SOD conjugate after 6 h was 83% in the acid treatment and 86% in alkali treatment, while that of unmodified SOD was 56 and 62%, respectively. The time-courses of the enzymatic activity are shown in Fig. 3a (acid treatment) and Fig. 3b (alkali treatment).Fig. 2. Native PAGE analysis of unmodified SOD, LMWH-SOD conjugate, and LMWH/SOD mixture. a) Protein staining: 1) unmodified SOD; 2) LMWH/SOD mixture incubated at 4°C for 48 h. b) Protein staining or polysaccharide staining: 1) LMWH-SOD conjugate stained with Coomassie Blue R-250; 2-4) Alcian Blue staining of LMWH-SOD conjugate, LMWH/SOD mixture, and LMWH. c) Protein staining of LMWH-SOD conjugates of different modification time: 1-3) samples modified for 12 (1), 24 (2), and 48 h (3).
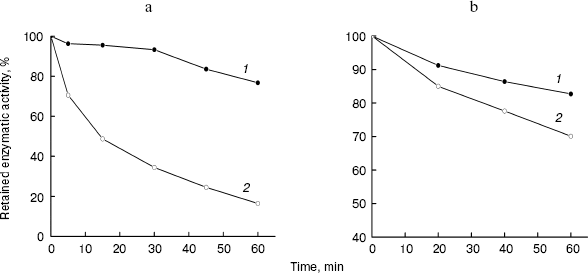
Heat stability and anti-trypsin stability of LMWH-SOD conjugate. The residual activities of LMWH-SOD conjugate and unmodified SOD after incubation at 80°N for 1 h, compared with enzymatic activity at 0 h, were 77 and 16.5%, respectively, suggesting that the modified SOD is more endurable than native SOD towards heat treatment. The time-courses of the enzymatic activity are shown in Fig. 4a.Fig. 3. Effects of acid (pH 5.2) (a) and alkali treatment (pH 9.5) (b) at 25°C on the enzymatic activity of LMWH-SOD conjugate (1) and unmodified SOD (2).
Anti-proteolytic stability can provide biomolecules more stability in vivo and hence prolong their half-life, which is helpful to enhance the bioavailability of the biomolecules. In this study, trypsin was selected as the reference enzyme to investigate the proteolytic susceptibility of LMWH-SOD conjugate. The time-courses of activity of LMWH-SOD conjugate and unmodified SOD incubated with trypsin are shown in Fig. 4b. Although the two curves present a similar trend in the course of the incubation, enhanced anti-trypsin stability was observed in LMWH-SOD conjugate with the residual enzymatic activity of 83% after 1 h incubation with trypsin compared with 70% for the unmodified SOD.Fig. 4. Effects of heat (80°C) (a) and trypsin treatment (at 25°C) (b) for 60 min on the enzymatic activity of LMWH-SOD conjugate (1) and unmodified SOD (2).
DISCUSSION
It has been well established that non-covalent interactions can occur between proteins and polysaccharides in a complex manner [11, 12]. In the preparation of LMWH-SOD conjugate, high pH value and high ionic strength were adopted to minimize the non-covalent interactions and the effects of these measures could be satisfactorily evaluated by electrophoresis.
As illustrated in Fig. 1b, SDS-PAGE of LMWH-SOD conjugate gave two major bands corresponding to 16 and 32 kD molecular weight. In contrast, the LMWH/SOD mixture showed the same banding pattern as unmodified SOD, with one band at 16 kD (Figs. 1c and 1a). According to the molecular structure of SOD, the protein band of 16 kD was the subunit of SOD and 32 kD was the intact SOD molecule. Moreover, enzymatic activity assay revealed that LMWH-SOD conjugate pretreated with mercaptoethanol for 5 min at 100°N before electrophoresis still retained enzymatic activity, but the unmodified SOD and LMWH/SOD mixture lost their enzymatic activity totally. Based on these results, a conclusion could be drawn that the covalent linkage between LMWH and SOD stabilized dimeric SOD, and the stabilizing effect increased with increasing the extent of the modification. In addition, pretreatment at different temperatures resulted in different electrophoretic behavior of LMWH-SOD conjugate (Fig. 1d). After pretreatment at 50°N, the modified SOD showed heterogeneity with widely banding distribution above 32 kD on the gel, whereas after pretreatment at 100°N only two bands remained. It can be speculated that covalent attachment of the heterogeneous LMWH to SOD produces the heterogeneity, and incubation at 100°N for a period of time can break the covalent bonds but is not enough to completely depolymerize the SOD dimer as it can do to the native enzyme in the same pretreatment period.
Although the electrophoretic behavior of the LMWH/SOD mixture on SDS-PAGE was different from that of LMWH-SOD conjugate, the non-covalent interactions were not visualized on the gel. Native PAGE of unmodified SOD with protein staining exhibited two major bands which were believed to belong to SOD isoforms with different isoelectric points because both bands showed enzymatic activity in SOD activity assay by the nitroblue tetrazolium reduction method [13] (data not shown). The LMWH-SOD conjugate, in spite of its increased molecular weight, migrated faster on the native gel due to enhanced negative charge because of the covalent attachment of LMWH. Moreover, a similar banding pattern of the LMWH/SOD mixture and LMWH with polysaccharide staining suggests that under the conditions used the non-covalent interactions are weak, which is considered as beneficial information for chemical modification of SOD with LMWH. Under non-denaturing PAGE conditions, charge was the major parameter to affect electrophoretic behavior. With increased binding of LMWH, the LMWH-SOD conjugates migrated faster (Fig. 2c) and hence evaluation of the combination extent was feasible with PAGE.
Improved stabilities of LMWH-SOD conjugate to heat, acid, alkali, and trypsin treatment were observed. The results are consistent with previous reports [14, 15]. Enhanced stabilities are believed to be due to the following possible reasons [16]: decreased conformational freedom of the enzyme and hence reduced possibility to contact unfavorable regents, formation of a shielding layer on the enzyme surface, and electrostatic repulsion of inhibitors or hydrolases by charges carried by some modifying regents. In the present study, enhancement of heat stability was most significant, especially in the absence of mercaptoethanol that breaks disulfide bonds in proteins. Cysteine residues at position 55 and 144 can form a disulfide bond within a single subunit of SOD, and this disulfide bond is important for the oligomerization and enzymatic activity [17]. Therefore, breaking the disulfide can cause loss of enzyme integrity and activity. Although LMWH-SOD conjugate also exhibited a somewhat increased stability under electrophoresis pretreatment conditions (i.e., in the presence of mercaptoethanol), the modified SOD was more durable to heat treatment in the absence of mercaptoethanol. However, a more detailed study by other techniques of the conformation-activity relationship of the modified SOD is needed.
In conclusion, data from our study demonstrate that the covalent linkage of SOD and LMWH can be monitored by electrophoretic techniques and the extent of combination can also be evaluated. Moreover, after modification, enhanced stabilities of SOD are obtained towards acid, alkali, heat, and trypsin.
This work was supported by a grant from the National Natural Science Foundation of China (No. 30270492).
REFERENCES
1.Fujita, T., Furutsu, H., Nishikawa, M., Takakura,
Y., Sezaki, H., and Hashida, M. (1992) Biochem. Biophys. Res.
Commun., 189, 191-196.
2.Maksimenko, A. V., Tischenko, E. G., and Golubykh,
V. L. (1999) Cardiovasc. Drugs Ther., 13, 479-484.
3.Nakaoka, R., Tabata, Y., Yamaoka, T., and Ikada, Y.
(1997) J. Control. Release, 46, 253-261.
4.Sonmez, K., Demirogullari, B., Turkyilmaz, Z.,
Karabulut, B. S., Kale, N., and Basaklar, A. C. (2001) Res. Commun.
Mol. Pathol. Pharmacol., 110, 97-106.
5.Muzykantov, V. R. (2001) J. Control.
Release, 71, 1-21.
6.Hodl, R., and Klein, W. (2003) J. Clin. Pharm.
Ther., 28, 371-378.
7.Ginzburg, E., Cohn, S. M., Lopez, J., Jackowski,
J., Brown, M., and Hameed, S. M. (2003) Br. J. Surg., 90,
1338-1344.
8.Deng, B. Y., Yuan, Q. S., and Li, W. J. (1991)
Progr. Biochem. Biophys., 18, 163 (in Chinese).
9.Tam, S. C., Blumenstein, J., and Wong, J. T. (1976)
Proc. Natl. Acad. Sci. USA, 73, 2128-2131.
10.Li, J. W., Xiao, N. G., Yu, R. Y., Yuan, M. X.,
Chen, L. R., Chen, Y. H., and Chen, L. T. (eds.) (1994) Experimental
Principles and Methods of Biochemistry [in Chinese], Peking
University Publishers, Beijing, pp. 189-194, 216-223.
11.Galazka, V. B., Smith, D., Ledward, D. A., and
Dickinson, E. (1999) Food Hydrocolloids, 13, 81-88.
12.Fugedi, P. (2003) Mini. Rev. Med. Chem.,
3, 659-667.
13.Beauchamp, C., and Fridovich, I. (1971)
Analyt. Biochem., 44, 276-287.
14.Beckman, J. S., Minor, R. L., Jr., White, C. W.,
Repine, J. E., Rosen, G. M., and Freeman, B. A. (1988) J. Biol.
Chem., 263, 6884-6892.
15.Veronese, F. M., Caliceti, P., Schiavon, O., and
Sergi, M. (2002) Adv. Drug Deliv. Rev., 54, 587-606.
16.Roberts, M. J., Bentley, M. D., and Harris, J. M.
(2002) Adv. Drug Deliv. Rev., 54, 459-476.
17.Yuan, Q. S. (ed.) (2001) Modern Enzymology
[in Chinese], East China University of Science and Technology
Publishers, Shanghai, pp. 127-128, 304.