Topography of the Active Site of the Saccharomyces cerevisiae Plasmalemmal Dicarboxylate Transporter Studied Using Lipophilic Derivatives of Its Substrates
D. A. Aliverdieva1*, D. V. Mamaev2, D. I. Bondarenko2, and K. F. Sholtz2
1Caspian Institute of Biological Resources, Dagestan Research Center, Russian Academy of Sciences, ul. Gadgieva 45, 367025 Makhachkala, Russia; E-mail: dinara_inbi@mail.ru; dinara0195@mail.ru2Bach Institute of Biochemistry, Russian Academy of Sciences, Leninsky pr. 33, 119071 Moscow, Russia; E-mail: dmamaev_inbi@mail.ru
* To whom correspondence should be addressed.
Received October 9, 2006
2-Alkylmalonates and O-acyl-L-malates have been found to competitively inhibit the dicarboxylate transporter of Saccharomyces cerevisiae cells, and the substrate derivatives chosen did not penetrate across the plasmalemma under the experiment conditions. Probing of the active site of this transporter has revealed a large lipophilic area stretching between the 0.72 to 2.5 nm from the substrate-binding site. Itaconate inhibited the transport fivefold more effectively than L-malate. This suggests the existence of a hydrophobic region immediately near the dicarboxylate-binding site (to 0.72 nm). The yeast plasmalemmal transporter was different from the rat liver mitochondrial dicarboxylate transporter. An area with variable lipophilicity adjoining the substrate-binding site has been revealed in the latter by a similar method. This area is mainly hydrophobic at distances up to 1.76 nm from the binding site and is separated by a hydrophilic region from 0.38 to 0.88 nm. Fumarate but not maleate competitively inhibited succinate transport into the S. cerevisiae cells. It is suggested that the plasmalemmal transporter binds the substrate in the trans-conformation. The prospects of the proposed approach for scanning lipophilic profiles of channels of different transporters are discussed.
KEY WORDS: dicarboxylate transporter of Saccharomyces cerevisiae plasma membrane, succinate, transport competitive inhibitors, fumarate, itaconate, 2-alkylmalonates, O-acyl-L-malates, lipophilic profile of the transporter channelDOI: 10.1134/S0006297907030030
Abbreviations: DeltaDeltaG) change in the isobaric-isothermic potential; FCCP) carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone; Rn) distribution coefficient of a compound with n carbon atoms in the aliphatic chain of the substituent; R) universal gas constant; SF-4867) 3,5-di-tert-butyl-4-oxybenzylidenemalononitrile; ve) rate of endogenous respiration.
Many C4-dicarboxylate transporters have been rather well
studied. These transporters are found in various biological membranes.
The transporters are present in the inner mitochondrial membrane [1], in the membrane of bacteria (in particular,
serovar typhimurium [2], Treponema
pallidum [3], Bradyrhizobium japonicum
[4]), and in the plasma membranes of higher
(e.g. rabbit kidney [5]) and lower eukaryotes
(Saccharomyces cerevisiae [6],
Schizosaccharomyces pombe [7],
Kluyveromyces lactis [8]). Molecules of
plasma membrane transporters are twofold larger than molecules of
mitochondrial transporters [9].
Na+/dicarboxylate symporters of higher eukaryotes contain
~600 amino acid residues [5, 9], whereas H+/dicarboxylate symporters of
yeast contain ~440 residues [7, 8, 10] (Sch. pombe
contains 438 amino acid residues [7]), which is
nearer to the size of bacterial transporters (about 400 residues [2-4]). Mitochondrial transporters
have six hydrophobic transmembrane alpha-helical segments [1, 11], whereas the plasma
membrane transporters have 11 [5, 12] or 12 such segments [7, 8]. In primary structure, these hydrophobic areas are
separated by hydrophilic regions exposed into solution [1, 13]. In the case of adenylate
transporter, all six segments form a channel [11],
and it is reasonable to suggest that mitochondrial transporters of
C4-dicarboxylates may have the same structure [14]. The alpha-helical transmembrane regions
of the plasma membrane transporter molecules encircle the channel by
nearly two layers [15, 16].
It is known that four of 12 segments determine the Km
value for succinate of the rabbit liver Na+/dicarboxylate
symporter [12]. Only four of 12 segments form the
inner surface of the human glucose transporter channel [15] and of many other plasma membrane transporters
[16]. Hydrophobic segments of nearly all known
C4-dicarboxylate transporters have unit polar amino acid
residues, but it is unclear whether they are exposed into the channel.
The tertiary structure has been established for some transporters of
hydrophilic substrates. The channels of these transporters are shown to
have both a hydrophilic and hydrophobic inner surface (similarly to the
glucose transporter [15] and the potassium channel
of bacteria [17], respectively). Three-dimensional
structures of dicarboxylate transporters are still unstudied. For such
transporters the probing of the channel near the substrate-binding site
using inhibitors (amphiphilic derivatives of these hydrophilic
substrates) is very informative [18].
We studied earlier the topography of the active site channel of the rat liver mitochondrial dicarboxylate transporter [18]. The studies were performed using competitive inhibitors, 2-monoalkylmalonates. Changes in the inhibition constants of these compounds (DeltaKi = Ki(n) - Ki(n-1)) on lengthening by one methylene link characterized the degree of lipophilicity in the region of the terminal methyl group binding [19]. It has been established that near the substrate-binding site and at the channel exit a small and large lipophilic area, respectively, is located, with a clearly expressed polar region between them. The sizes of these regions were 0.38, no less than 0.88, and 0.50 nm, respectively. Consequently, the outer semi-channel was of no less than 1.76 nm in length. Therefore, it was suggested that the substrate-binding site of the active site should be located in the middle of the membrane [19], because the thickness of its hydrophobic matrix was taken as 4.0 nm. Such symmetry is likely to characterize mitochondrial transporters, antiporters.
In the present work, O-acyl-L-malates and 2-alkylmalonates were used for probing of the active site of the S. cerevisiae plasma membrane dicarboxylate transporter. The activity of this transporter did not depend on the plasma membrane uncoupling, manifested itself in the presence of sodium ions (at pH 5.5), and nearly disappeared in mono-potassium medium [6].
Under certain conditions, amphiphilic inhibitors used by us can be inducers of permeability [20]. The primary structure has been established for H+/dicarboxylate symporters of K. lactis [8] and Sch. pombe [7], but they can display an additional, noncompetitive to substrate, sensitivity to amphiphiles when those behave as protonophores. This brought us to choose a less studied object.
On S. cerevisiae cells and preparations of rat liver mitochondria conditions were chosen to determine the rate of succinate transport into the cells [6] and mitochondria [18], using the corresponding endogenous coupled systems of substrate oxidation. This allowed us to avoid artifacts associated with use of radiolabeled substrates-metabolites on intact cells [21, 22] and changes in the Km associated with the reconstruction of transporters into liposomes, which was observed in the case of tricarboxylate [23] and adenylate [24] transporters.
MATERIALS AND METHODS
The S. cerevisiae strain Y-503 (Collection of the State Venture GNII Genetika, Russia) obtained at the Caspian Institute of Biological Resources, Dagestan Research Center, Russian Academy of Sciences, was used [25]. The tetraploid strain Y-503 has enhanced respiratory activity compared with S. cerevisiae haploid and diploid strains [26]. The cells of this strain grown in medium with a low content of glucose have considerably higher number of mitochondria [27]. Moreover, under these conditions of growth, the mitochondrial succinate/fumarate antiporter was not induced [28], and this allowed us to use fumarate as a competitive inhibitor of the transport across the plasmalemma. The yeast cells were grown for 10 h. Specific features of the growth, preparation, and measurement of respiration have been described [6]. Sodium succinate was added upon the stabilization of the cell endogenous respiration rate, and its value (ve) was subtracted from the total rate of oxidation [6, 29]. Rat liver mitochondria were isolated by a modification of Weinbach's method [30]. The rat liver mitochondrial protein was determined by Goa's method [31].
The rate of sodium succinate oxidation by the cells was determined at 30°C in 50 mM potassium phosphate buffer (pH 5.5) under stationary conditions. Under these conditions, the succinate transporter activity was slightly decreased, but this allowed us to retain limiting conditions of “succinate oxidase” transport across the plasmalemma for the yeast “preparation” during 12-14 h of preincubation [6]. Moreover, the presence of potassium in the incubation medium promotes maintaining of constant pH value in the cytoplasm of the respiring S. cerevisiae cells [32]. However, phosphate does not affect the affinity of succinate to the transporter [6]. In specially mentioned cases, 50 mM sodium phosphate buffer (pH 5.5) was used. The respiration of rat liver mitochondria was measured at 25°C in medium which contained 125 mM sucrose, 20 mM Tris-HCl (pH 7.2), 2 mM EDTA, 20 mM KCl, 2.5 mM MgCl2, and 10 mM KH2PO4 [18]. Rotenone (1 µM) was added immediately before the addition of the mitochondria, and the concentration of the protonophore 3,5-di-tert-butyl-4-oxybenzylidenemalononitrile (SF-4867) [33] was changed in dependence on the mitochondria concentration, taking into account its distribution coefficient [30]. Yeast respiration was measured without the protonophore [6].
Oxygen concentration was determined amperometrically in a thermostatted cell with closed electrodes [34]. Because the observable parameters of inhibition (I50) of the most hydrophobic compounds depended on the concentration of mitochondria or cells (B), the true I50 was determined by extrapolation to zero concentration of organelles in {I50; B} coordinates, as proposed by Heirwegh [35, 36]. This approach was used for higher 2-alkylmalonates, beginning from 2-dodecylmalonate, and higher O-acyl-L-malates, beginning from O-myristoylmalate. A linear type of dependence was the reason for us to use this approach [18, 36]. For low-molecular-weight itaconate and fumarate, which (possibly) penetrate across the plasmalemma, the Ki value was determined by varying the concentration of succinate (according to Lineweaver-Burk) and of the inhibitor itself (according to Dixon). The coincidence of the constants indicated the absence of the effector side influence on the components of the S. cerevisiae endogenous coupled system used to measure the concentration of the transported succinate in the cytoplasm.
The interaction of L-malate and malonate derivatives with the plasma membrane dicarboxylate transporter was studied under conditions when the rate of succinate transport across the plasmalemma limited the rate of its oxidation by the cells. Similarly, the interaction with the rat liver mitochondrial dicarboxylate transporter was studied when the oxidation rate was limited by the transport of this substrate into the matrix of the organelles. Just such conditions emerged upon the aerobic preincubation of the cells for 10-22 h at 0°C (10 mM potassium phosphate buffer, pH 5.5) [6] and preincubation from 0 to 15 h at 0°C (4 mM Tris-HCl, pH 7.4, 250 mM sucrose) of a preparation of tightly coupled mitochondria [18]. During the calculations, a slight influence of the inhibitors on the rate of endogenous respiration of the cells was taken into account [6]. The distribution coefficients of O-acyl-L-malates (Rn) in the system of octanol/water (10 mM phosphate buffer, pH 6.5 and 7.2) were determined by measuring the inhibitor concentrations in the aqueous phase containing Methylene Blue [37]. The distance from the plane formed by the lines connecting oxygen atoms in the carboxyl groups to the last carbon atom in the substituent of the malate and malonate derivatives in conformation with the minimum free energy was calculated using the Chemoffice MM2 program. This value was taken as the length of the molecular probe.
Reagents. L-Malic acid (Sigma, USA), BSA (Calbiochem, USA), Tris and rotenone (Serva, Germany), yeast extract (Difco Laboratories, USA), protonophores SF-4867 (Sumimoto Chem. Co., Japan) and FCCP (carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone) (Aldrich, USA), sodium malonate (ICN, USA), and domestic preparations of KCl, KH2PO4, and KOH of special purity were used. MgCl2, EDTA, sucrose, succinic, itaconic, maleic, and fumaric acids, and sodium succinate and pyruvate were recrystallized twice. 2-Alkylmalonates and O-acyl-L-malates were synthesized in our laboratory [18]. Water-insoluble reagents were dissolved in dimethylsulfoxide.
RESULTS
The rate of succinate transport into the S. cerevisiae cell was measured using an endogenous coupled system of succinate oxidation (further called cellular “succinate oxidase”). This system consists of the plasma membrane transporter, the yeast cell mitochondrial succinate oxidase. The rate of succinate transport in the rat liver intact mitochondria was also measured using an endogenous coupled system of succinate oxidation (further called mitochondrial “succinate oxidase”) consisting of the mitochondrial dicarboxylate transporter, succinate dehydrogenase, and “ubiquinol oxidase”. Under the conditions chosen, the rate was limited by the plasma membrane transporter in the S. cerevisiae cells [6] and by the mitochondrial transporter in the mitochondria [18].
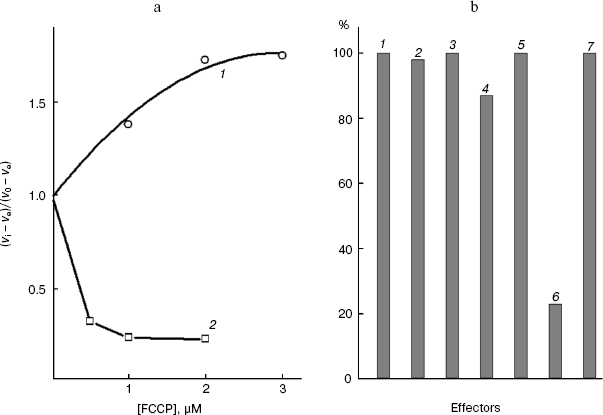
Figure 1a shows that low concentrations of the protonophore FCCP suppressed the activity of “pyruvate oxidase”, which under the chosen conditions is limited by the monocarboxylate transporter of the S. cerevisiae cell plasmalemma [6]. High concentrations of the protonophore activated “glucose oxidase”. The products of glucose metabolism are more intensively oxidized upon the uncoupling of the mitochondrial inner membrane of the yeast cells [6, 29], and the glucose transport across the S. cerevisiae plasmalemma is insensitive to the protonophore. O-Palmitoyl-L-malate (an amphiphilic inhibitor of the dicarboxylate transporter [6, 36, 37]) unable to penetrate into the cell at pH 5.5 does not inhibit “pyruvate oxidase” (Fig. 1b, the seventh effector), while the latter was effectively suppressed by the protonophore penetrating into the cell (Fig. 1b, the sixth effector). Thus, the lack of inhibition of the pyruvate symporter of the plasmalemma correlated with its impermeability for the amphiphilic dicarboxylic acid. Effectors 2-5 in Fig. 1b (the extreme members of the series of malonate and L-malate derivatives with a variable aliphatic substituent) had virtually no effect on “pyruvate oxidase” of these cells. These active inhibitors of mitochondrial “succinate oxidase” [18, 36] seemed not to penetrate into the cell, to mitochondria. Permeabilization of the rat liver mitochondrial inner membrane does not lead to inhibition of succinate dehydrogenase and “ubiquinol oxidase” by these derivatives of the rat liver dicarboxylate transporter substrates [18]. The inhibitors are promising for studies on the dicarboxylate transport, because they either fail to penetrate into the rat liver mitochondrial matrix, or do not inhibit the activities of the coupled system components [38].
Typical aliphatic derivatives of the cell plasmalemmal transporter substrates, O-palmitoyl-L-malate and 2-undecylmalonate, increased the Km of the yeast cellular “succinate oxidase” (Fig. 2a), not affecting the maximal rate of the process. The competitive type of action of these inhibitors indicates that both groups of compounds (for most of substances data not shown) interact with the same substrate-binding site in the active site of the transporter. Similarly, typical aliphatic derivatives of the rat mitochondrial transporter substrates, O-stearoyl-L-malate and 2-octylmalonate, increase the Km of the mitochondrial “succinate oxidase” (Fig. 2b), not affecting the maximal rate of the process (for the other substances data not shown). The competition relative to the same substrate suggests that O-acyl-L-malates and 2-alkylmalonates interact with the same substrate-binding site in the active site of each transporter under study.Fig. 1. a) Dependence of the relative oxidation rate of 10 mM glucose (1) and 20 mM sodium pyruvate (2) on FCCP concentration. S. cerevisiae cells (1.7 mg wet weight per ml) were aerobically preincubated at 0°C for 2-4 h. The substrate was added after the addition of the protonophore. In the presence of FCCP, pyruvate, and glucose the rates stabilized, respectively, 3, 4, and 2 min after the addition of the effector (time dependences not shown). b) The “pyruvate oxidase” activity with 20 mM pyruvate in the absence of the effector (1) and in the presence of 8 mM 2-pentylmalonate (2), 4 µM 2-pentadecylmalonate (3), 600 µM O-lauroyl-L-malate (4), 40 µM O-stearoyl-L-malate (5), 1 µM FCCP (6), and O-palmitoyl-L-malate (7). The S. cerevisiae cells (1.7 mg wet weight per ml) were aerobically preincubated at 0°C for 2-4 h.
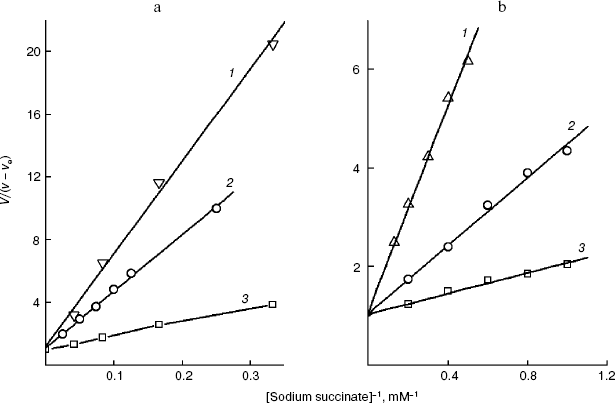
The linear dependence of inhibition in Dixon's coordinates indicates that the transport across the plasma membrane is still limiting the endogenous coupled system [29] in the presence of the effector and that the inhibitor binds with the transporter at the same site. Such dependences were obtained for every derivative of the S. cerevisiae plasmalemmal dicarboxylate transporter substrates. Figures 3a and 4a present the dependences for the extreme members of the series of 2-alkylmalonates: 2-pentylmalonate and 2-pentadecylmalonate, and in Figs. 3b and 4b such dependences are given for L-malate and O-stearoyl-L-malate. Similar dependences for the rat liver intact mitochondrial transporter were obtained by us earlier [19, 20]. These findings in total allowed us to compare the topography of the active sites (the substrate-binding site + the channel) of two transporters using the results of probing using two groups of substrate derivatives.Fig. 2. a) Dependence of the oxidation rate on succinate concentration in Lineweaver-Burk coordinates in the presence of 80 µM O-stearoyl-L-malate (1), 50 µM 2-undecylmalonate (2), and without the inhibitor (3). The S. cerevisiae cells (5 mg wet weight per ml) were aerobically preincubated at 0°C for 18 h. b) The dependence of the oxidation rate on the succinate concentration in Lineweaver-Burk coordinates in the presence of 8 µM O-stearoyl-L-malate (1), 0.1 mM 2-octylmalonate (2), and without the inhibitor (3). Rat liver mitochondria (0.5 mg protein per ml).
Fig. 3. Dependence of the oxidation rate of 8 mM succinate on the concentration of 2-pentylmalonate (a) and the dependence of the oxidation rate of 20 mM sodium succinate on the concentration of potassium L-malate in Dixon coordinates (b). The S. cerevisiae cells (10 mg wet weight per ml) were aerobically preincubated at 0°C for 18 h. The oxidation rate stabilized 10 min after the addition of 2-pentylmalonate.
Not all compounds were suitable for probing the S. cerevisiae plasmalemmal transporter. For 2-methyl- and 2-propylmalonate, the dependence in Dixon coordinates was biphasic. A relatively low activity of the cellular “succinate oxidase” compared to that of the mitochondrial “succinate oxidase” prevented obtaining the inhibition parameters for 2-heptadecylmalonate. The I50 values at the subzero cell concentration required to calculate the true I50 by extrapolation could be determined with wide scattering. Higher O-acyl-L-malates were promising as the longer probes than 2-alkylmalonates with the same number of carbon atoms in the substituent. Poor solubility (low constant of micelle production) prevented study of the effect on the yeast cellular transporter of the derivatives from O-butyroyl-L-malate to O-capryloyl-L-malate in the range of concentrations which we were interested in.Fig. 4. Dependence of the oxidation rate of 8 mM succinate on the concentration of 2-pentadecylmalonate (a) and the dependence of the oxidation rate of 24 mM sodium succinate on the concentration of O-stearoyl-L-malate in Dixon coordinates (b). The S. cerevisiae cells (10 mg wet weight per ml) were aerobically preincubated at 0°C for 18 h.
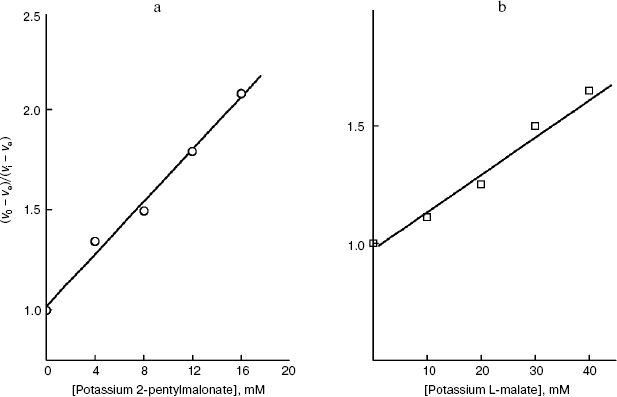
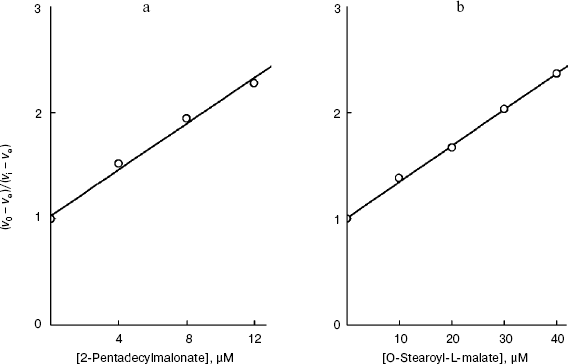
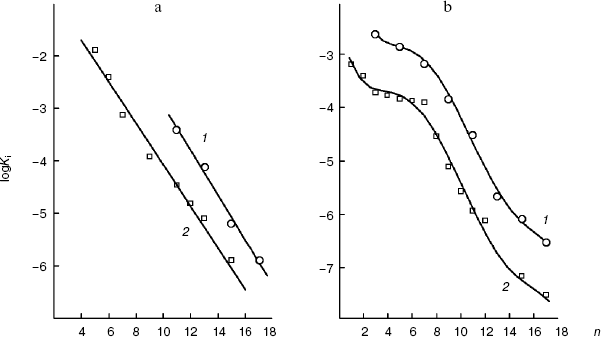
As shown in Fig. 5a, in the case of the S. cerevisiae cellular “succinate oxidase”, the Ki dependences on the number n of carbon atoms in the aliphatic chain are linear and parallel for O-acyl-L-malates and 2-alkylmalonates. The Ki value of the O-acyl-L-malate effect on the cellular dicarboxylate transporter is nearly an order higher than the Ki value of the effect of 2-alkylmalonates with the aliphatic chain of same length (DeltalogKi = 0.92). The dependences follow linear equations (logKi = -0.427n + 1.323 and log Ki = -0.396n - 0.115, respectively). With an increase in the hydrophobicity of the compound, its affinity for the active site increases.
In the case of the mitochondrial “succinate oxidase”, the Ki dependences on n for O-acyl-L-malates and 2-alkylmalonates are also very similar (Fig. 5b). Both curves have a plateau for n values from 4 to 8 (the binding with the polar zone) and the region of Ki decrease for n values from 8 to 15 (the binding with the “large lipophilic area”). It seems that similar areas of the O-acyl-malate and 2-monoalkylmalonate aliphatic chains are bound by the same parts of the transporter's channel. In total, O-acyl-L-malates display higher affinity for the mitochondrial transporter than for the plasma membrane transporter. We have earlier shown that the affinities of these transporters for L-malate differ by approximately one and a half orders of magnitude (0.5 mM [18] and 17 mM [29], respectively). This difference seems to be determined by the affinity of the inhibitor “heads” for the substrate-binding site. However, the tangent of the slope angle of the 2-alkylmalonate and O-acyl-L-malate dependences on the “large lipophilic area” of the mitochondrial transporter active site (logKi = -0.38n + 1.41) and tangents of the slope angles of the plasmalemmal transporter dependences were similar (-0.396 and -0.38, respectively).Fig. 5. a) Dependence of the inhibition constant (Ki) of the S. cerevisiae cellular “succinate oxidase” by O-acyl-L-malates (1) and 2-alkylmalonates (2) on the number (n) of carbon atoms in their aliphatic chains. b) The dependence of the inhibition constant (Ki) of the rat liver mitochondrial “succinate oxidase” by O-acyl-L-malates (1) and 2-alkylmalonates (2) on the number (n) of carbon atoms in their aliphatic chains.
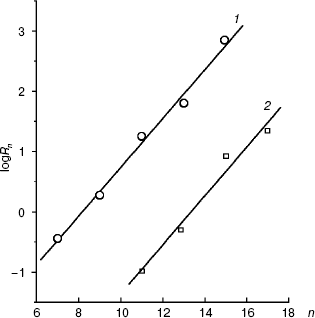
Figure 6 presents the dependences of the distribution coefficients (Rn) of O-acyl-L-malates in the system of octanol/water determined under the same conditions as the affinities of the plasma membrane transporter (pH 5.5) and mitochondrial transporter (pH 7.2). The dependences are described by linear equations: logRn = 0.416n - 3.46 and logRn = 0.406n - 5.42, respectively. The influence of pH on the logRn value for the zero member of the series seems to be associated with differences in the malate residue ionization. Modules of the coefficient values at n in the equations for the dependences logKi (n) and logRn (n) are similar. Consequently, the energy of interaction of the methyl link of the inhibitor aliphatic chain (DeltaDeltaG = RTDeltalogKi) with amino acid residues “lining” the channel in the active site of the transporters is due only to lipophilic interactions (DeltaDeltaG = RTDeltalogRn). Concurrently, this indicates that steric obstacles did not distort the conformation of the aliphatic “tail” of the inhibitor molecule in the state with the energy minimum, because steric obstacles to the binding would lower the interaction energy. Such a molecular probe (a set of inhibitors with different length) can be used as a “ruler”. Note that O-acyl-L-malate “ruler” is longer by the length of the ester bond.
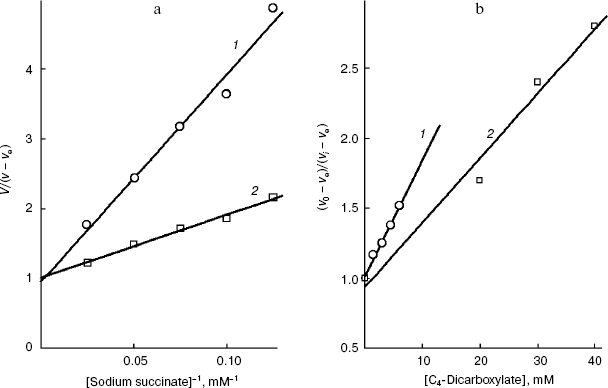
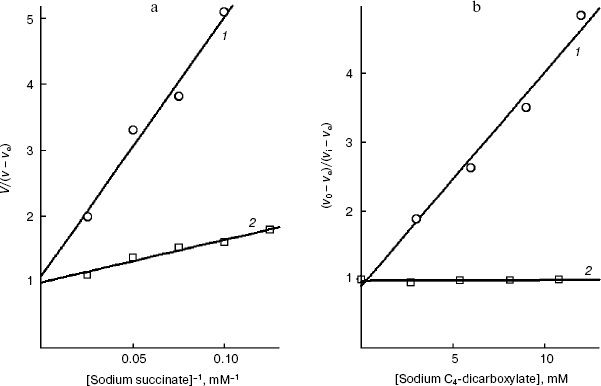
We have at our disposal two “rulers” (2-alkylmalonates and O-acyl-L-malates) for measuring the length of the lipophilic region of the yeast transporter channel, and it is desirable to elucidate whether they are “applied” to the reference point of the active site by the dicarboxylate head or by the “fatty tail” beginning. The polar area, which is present in the mitochondrial transporter profile (Fig. 5b), suggests the second variant. The ester group of O-acyl-L-malates seems to have no contact with the lipophilic zone of the active site [18]. We have studied the effect on the S. cerevisiae respiration of itaconate and L-malate, which possess, respectively, the hydrophobic methylene and hydrophilic hydroxyl group in the second position of the substrate molecule. Both substances failed to activate the endogenous respiration (L-malate because of its saturating concentration in the cytoplasm [6, 29]). Data presented in Fig. 7b show that the more hydrophobic competitive inhibitor (Fig. 7a) of the cellular “succinate oxidase” has the higher affinity (respectively, Ki corresponds to 4.15 ± 0.35 and 17.5 ± 1.1 mM). Thus, it seems that in the plasmalemmal transporter, the zone of the active center, which is adjacent to the binding sites of carboxyls of substrate, does not interact with the polar ester group of O-acyl-L-malates. Therefore, it is reasonable to expect that 2-alkylmalonates would have an order higher affinity than O-acyl-L-malates with the same length of the aliphatic chain, and just this is obvious.Fig. 6. Dependence of the distribution coefficient (Rn) in the octanol-water system on the number (n) of carbon atoms in the aliphatic chain of O-acyl-L-malates. Experiment conditions: 10 mM potassium phosphate buffer, pH 5.5 (1) and pH 7.2 (2).
It is known that the rat liver mitochondrial dicarboxylate transporter can bind maleate but not fumarate [1, 39]. Fumarate is shown (Fig. 8b) to effectively inhibit the transport of succinate across the yeast plasmalemma (Ki = 1.0 ± 0.1 mM) and competitively with succinate (Fig. 8a). Dicarboxylates seem to bind with the substrate-binding site of the trans-conformation of this S. cerevisiae transporter. The configuration influence on the inhibition efficiency is consistent with our earlier data on the pH dependence of the affinity, which has shown that succinate is transported as a dianion [6].Fig. 7. a) Dependence of the oxidation rate on the succinate concentration in Lineweaver-Burk coordinates in the presence of 10 mM sodium itaconate (1) and without the inhibitor (2) in 50 mM sodium phosphate buffer. The S. cerevisiae cells (5 mg wet weight per ml) were aerobically preincubated at 0°C for 18 h. b) The dependence of the oxidation rate of 10 mM sodium succinate on the concentration of sodium itaconate (1) and potassium L-malate (2) in Dixon coordinates. The S. cerevisiae cells (10 mg wet weight per ml) were aerobically preincubated at 0°C for 18 h.
Fig. 8. a) Dependence of oxidation rate on the succinate concentration in Lineweaver-Burk coordinates in the presence of 10 mM sodium fumarate (1) and without the inhibitor (2) in 50 mM sodium phosphate buffer. The S. cerevisiae cells (5 mg wet weight per ml) were aerobically preincubated at 0°C for 20 h. b) The dependence of the oxidation rate of 10 mM sodium succinate on the concentration of sodium fumarate (1) and sodium maleate (2) in Dixon coordinates. The S. cerevisiae cells (10 mg wet weight per ml) were aerobically preincubated at 0°C for 20 h.
DISCUSSION
The effects of O-acyl-L-malates and 2-alkylmalonates on the rate of succinate oxidation by S. cerevisiae cells have been studied. Because this rate is limited by the cell plasma membrane transporter [29], the monophasic dependences in Dixon coordinates (Figs. 3a, 4a, and 5a) indicate that compounds of both groups interact with the same protein, the dicarboxylate transporter. Comparison of the results of the active site probing using these competitive inhibitors (Fig. 2a) allowed us to investigate specific features of the substrate-binding site and its environment. Similar findings [18, 19] allowed us to probe the rat liver mitochondrial dicarboxylate antiporter.
Based on the data presented in Fig. 5a, it was suggested that the plasma membrane transporter channel should be hydrophobic along nearly its whole length and have no polar region at the distance of 0.47-0.97 nm from the malonate-binding site. The data shown in Fig. 5b indicate that the mitochondrial dicarboxylate channel has a polar region in this area and manifests variable lipophilicity. Thus, the substrate (C4-dicarboxylate) hydrophilicity fails to inevitably determine the shape of the lipophilic profile of the transporter active site. These findings can be compared with data on other mitochondrial antiporters of hydrophilic substrates. Note that variable lipophilicity has been shown to characterize the exposed into the channel surface of the third transmembrane segment of the S. cerevisiae mitochondrial citrate/malate antiporter [40]. The structure of this transporter resembles the tertiary structure of the adenylate transporter [14], which also has the substrate-binding site exposed into the channel [11]. All these data are consistent with the results of scanning of the lipophilic profile of the channel (more exactly, of the outer semi-channel) of the rat liver mitochondrial dicarboxylate transporter presented in Fig. 5b. However, the scanning of the transmembrane segment surface using successive substitutions of the amino acid residues by cysteine and the following study of its availability for hydrophilic SH-agents prevents the positioning of the polar residues on the third segment of the citrate/malate antiporter [40] with respect to the malate-binding site on the fourth segment [41]. Mitochondrial transporters are significantly different from the plasma membrane transporters in size, number of transmembrane segments, and functioning mechanisms; only the channel presence in the structure is their common feature. It is reasonable to suggest that this channel is the only place in the transporter's structure where the aliphatic chain of the competitive inhibitor can have a conformation with the minimum energy, according to the slope angles of the logarithm Ki dependences on the length of this chain. Thus, the applicability of the proposed approach has been shown for scanning the lipophilic profile of the channel in both groups of transporters.
In the mitochondrial citrate [42] and alpha-ketoglutarate [43] transporters, point substitution of the invariant arginine residues in the depth of the fourth transmembrane segment resulted in irreversible loss of activity. Both residues (Arg181 and Arg189) of the citrate transporter are exposed into the channel [11, 41]. However, the rat liver mitochondrial dicarboxylate transporter binds maleate [39]. Thus, as differentiated from the S. cerevisiae plasmalemmal dicarboxylate transporter (Fig. 8b), arginyls of all three transporters carrying L-malate in the mitochondria bind this substrate in the cis-position [39, 42, 43].
According to many data, conservative arginyls can act as cations binding carboxyls in the active site of dicarboxylate transporters. The neuron plasmalemmal Na+/glutamate symporter contains the conservative Arg479 residue responsible for specificity to glutamate and aspartate in the eighth transmembrane segment [44]. Four transmembrane segments near the C-end of the Na+/dicarboxylate rabbit symporter are responsible for succinate binding [12, 45], and segment VII of them includes the conservative Arg349, which is also present in the transporters of the human, rat, rabbit, and X. laevis kidneys [46]. Substitution of Arg349 by isoleucine by a point mutation causes a nearly complete loss of activity (although the inactive protein is present in the membrane). However, the substitution by the lysine residue has virtually no effect on the transporter activity, but increases sixfold its Km to succinate [47].
Some bacterial permeases and transporters, which carry succinate and malate in the yeast plasma membrane, are shown to contain conservative arginine residues inside the transmembrane lipophilic alpha-helical segments. The Dcu C transporter of S. typhimurium has Arg74 in the third segment and Arg209 in the sixth segment [2]. The dct M transporter of T. pallidum contains Arg143 in the fourth segment [3]. The dct P transporter of B. japonicum has Arg258 in the sixth segment [4]. The dicarboxylate transporter of K. lactis has Arg127 in the second segment and Arg174 in the fourth segment [8]. The transporter of Sch. pombe contains Arg219 in the fourth segment and Arg264 in the seventh segment [7].
The well studied plasma membrane transporters with known tertiary structure [15, 16, 48] are shown to have a single substrate-binding site (the selectivity determining region) per molecule. Based on the presence of unit arginine residues in the transmembrane segments, all transporters of C4-dicarboxylates are supposed to have a single substrate-binding site exposed into the channel. In molecules of the plasma membrane transporters with known three-dimensional structure the lipophilic segments calculated by hydropathic profiles do not begin in the same plane of the molecule and are not perpendicular to the membrane plane [16], as it is usually designated in schemes [4, 8, 16]. Therefore, it is impossible to predict the position of the substrate-binding site relative to the origin or end of the channel based on analysis of the arginyl locations in the primary structure. A distortion of the linear lipophilic profile of the channel (an emergence of the plateau, as in Fig. 5b) can be a result of a point mutation (substitution of a hydrophobic amino acid residue by a hydrophilic one) in the transporter and, thus, can be informative. Such an approach is promising for positioning this point on the transmembrane segment surface, which is exposed into the channel, relative to arginyls of the substrate-binding site. Although transmembrane segments of the rat liver mitochondrial dicarboxylate transporter contain unit serine residues [49], the plateau on the lipophilic profile is clearly expressed (Fig. 5b). However, citrate transporters of yeast mitochondria with point substitution of a unit residue in the third transmembrane segment are known to retain the functional activity of the unmodified transporter upon reconstruction into liposomes [41]. Thus, the probing by the substrate amphiphilic substrate of mutants with the locally modified channel of the transporter seems very promising.
It should be noted that results of transporter channel scanning by a labor-consuming “cysteine mutagenesis” under native conditions can supplement crystallographic data obtained at high resolution [15, 16]. If the crystallographic data contradicted the results of cation-induced modulation of the volt-dependent potassium channel conductivity [50, 51], the authors supposed that the structure should be distorted during the lipid-free crystallization. Therefore, development of new approaches for “scanning” of molecules under native conditions is promising. The development of easy approaches for studies on the three-dimensional structure will reduce a gap between numerous transporters with the established primary structure [9, 16] and a relatively low number of transporters with the known tertiary structure.
If the malate and malonate heads of the inhibitor of the S. cerevisiae plasmalemmal transporter are bound in the same site, that is indirectly confirmed by our data on the comparative efficiency of inhibition by itaconate and L-malate, the plasma membrane channel remains lipophilic at the distance of 2.5 nm from the substrate-binding site, which is equal to the length of O-stearoyl-L-malate. It seems that the channel is hydrophobic also at the greater distance from this site. The dependence presented in Fig. 5a has no tendency for deflection from the linearity at the greater values of n (Fig. 5b). The length of the hydrophobic part of the lipophilic profile was estimated as the difference between the lengths of O-stearoyl-L-malate (2.52 nm) and 2-pentylmalonate (0.75 nm), i.e. the longest and the shortest molecular probes. This difference was 1.74 nm. The lipophilic channel of the S. cerevisiae plasmalemmal dicarboxylate transporter resembles the channel of the potassium transporter of bacteria [17] and is different from the hydrophilic channel of the plasma membrane glucose transporter of human cells [15]. The length of the hydrophobic region of the potassium channel calculated by its three-dimensional structure is 2.4 nm [17]. At present, the dicarboxylate transporter under our study seems to be the only one for which the lipophilicity of the inner surface has been characterized.
The authors are grateful to S. A. Bulat for confirmation of this strain being a member of the S. cerevisiae taxon and also to S. V. Benevolensky and D. G. Kozlov for establishing of the Y-503 strain ploidy.
This work was supported by the Russian Foundation for Basic Research (project No. 04-04-49670).
REFERENCES
1.Palmieri, L., Runswick, M. J., Fiermonte, G.,
Walker, J. E., and Palmieri, F. (2000) J. Bioenerg. Biomembr.,
32, 67-77.
2.McClelland, M., Sanderson, K. E., Spieth, J.,
Clifton, S. W., Latreille, P., Courtney, L., Porwollik, S., Ali, J.,
Dante, M., Du, F., Hou, S., et al. (2001) Nature, 413,
852-856.
3.Fraser, C. M., Norris, S. J., Weinstock, G. M.,
White, O., Sutton, G. G., Dodson, R., Gwinn, M., Hickey, E. K.,
Clayton, R., et al. (1998) Science, 281, 375-388.
4.Kaneko, T., Nakamura, Y., Sato, S., Minamisawa, K.,
Uchiumi, T., Sasamoto, S., Watanabe, A., Idesawa, K., Iriguchi, M., et
al. (2002) DNA Res., 9, 189-197.
5.Pajor, A. M. (1995) J. Biol. Chem.,
270, 5779-5785.
6.Aliverdieva, D. A., Mamaev, D. V., Bondarenko, D.
I., and Sholtz, K. F. (2006) Biochemistry (Moscow), 71,
1161-1169.
7.Grobler, J., Bauer, R. E., Subden, H. J. J., and
van Vuuren, F. (1995) Yeast, 11, 1485-1491.
8.Lodi, T., Fontanesi, F., Ferrero, I., and Donnini,
C. (2004) Gene, 339, 111-119.
9.Chung, Y. J., Krueger, C., Metzgar, D., and Saier,
M. H., Jr. (2001) J. Bacteriol., 183, 1012-1021.
10.Queiros, O., Casal, M., Althoff, S.,
Moradas-Ferreira, P., and Leao, C. (1998) Yeast, 14,
401-407.
11.Pebay-Peyroula, E., Dahout-Gonzalez, C., Kahn,
R., Trezequet, V., Lauquin, G. J.-M., and Brandolin, G. (2003)
Nature, 426, 39-44.
12.Pajor, A. M., Sun, N., Bai, L. Q., Markovich, D.,
and Sule, P. (1998) Biochim. Biophys. Acta, 1370,
98-106.
13.Sholtz, K. F. (1994) Usp. Biol. Khim.,
34, 167-187.
14.Walters, D. E., and Kaplan, R. S. (2004)
Biophys. J., 87, 907-911.
15.Zuniga, F. A., Shi, G., Haller, J. F., Rubashkin,
A., Flynn, D. R., Iserovich, P., and Fischbarg, J. (