REVIEW: Nucleocytoplasmic Transport of Proteins
A. V. Sorokin, E. R. Kim, and L. P. Ovchinnikov*
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: p50@rambler.ru; ovchinn@vega.protres.ru* To whom correspondence should be addressed.
Received April 10, 2007
In eukaryotic cells, the movement of macromolecules between the nucleus and cytoplasm occurs through the nuclear pore complex (NPC)--a large protein complex spanning the nuclear envelope. The nuclear transport of proteins is usually mediated by a family of transport receptors known as karyopherins. Karyopherins bind to their cargoes via recognition of nuclear localization signal (NLS) for nuclear import or nuclear export signal (NES) for export to form a transport complex. Its transport through NPC is facilitated by transient interactions between the karyopherins and NPC components. The interactions of karyopherins with their cargoes are regulated by GTPase Ran. In the current review, we describe the NPC structure, NLS, and NES, as well as the model of classic Ran-dependent transport, with special emphasis on existing alternative mechanisms; we also propose a classification of the basic mechanisms of protein transport regulation.
KEY WORDS: nucleocytoplasmic transport, nuclear pore complex, nucleoporin, exportin, importin, karyopherin, nuclear export signal, nuclear localization signal, RanDOI: 10.1134/S0006297907130032
Abbreviations: CaM) Ca2+-binding protein calmodulin; CAS) cellular apoptosis susceptibility gene; CGC) central gated channel; CRS) cytoplasmic retention signal; ER) endoplasmic reticulum; INM) inner nuclear membrane; LBD) ligand-binding domain; NES) nuclear export signal; NLS) nuclear localization signal; cNLS) classical (basic) NLS; NPC) nuclear pore complex; NTF2) nuclear transport factor; ONM) outer nuclear membrane; RanBP) Ran-binding proteins; RRM) RNA recognition motifs; STAT proteins) signal transducers and activators of transcription; TM) transmembrane nucleoporins; WGA) wheat germ agglutinin.
In eukaryotic cells, the cytoplasm and the nucleus intercommunicate via
nuclear pore complexes in the nuclear membrane. The nuclear pore
complex (NPC) consisting of about 30 different proteins, nucleoporins
[1], forms a channel and regulates
nucleocytoplasmic transport of varying types of RNAs [2], membrane proteins (receptors) [3], and soluble proteins [4].
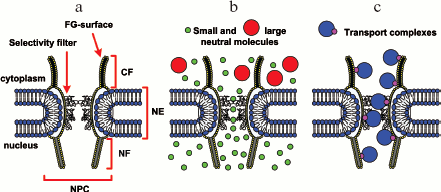
The nuclear pore complex is a great transporter that runs through a nuclear membrane. Ions, as well as small neutral proteins that do not bind to nucleoporins, run through the nuclear pore complex due to diffusion [5]. In this case, they pass via a tunnel 8 to 10 nm in diameter and nearly 45 nm in length [6, 7] (Fig. 1b). If molecules bind to nucleoporins, the diameter of the tunnel increases to 40 nm [8, 9], and the transport proceeds much faster (Fig. 1c) [10, 11]. The selective filter for small proteins represents a network of unfolded hydrophobic polypeptide nucleoporins lining the central channel of the NPC (Fig. 1b). In spite of a rather large diameter of the tunnel, even some small proteins (less than 20-30 kD), such as histones, pass via the NPC only with mediators [12]. The major transport is realized by means of a Ran mediator (Ran-dependent transport). This kind of transport has been studied quite well. Its main distinction is GTP hydrolysis catalyzed by Ran. In addition to Ran, other transport factors are involved in this process.
Fig. 1. Simplified model of translocation through the nuclear envelope (modified from [58]). a) Schematic representation of the nuclear pore complex (NPC). Designations: NE, nuclear envelope; CF, cytoplasmic filament; NF, nuclear filament. The FG-surface is formed by FG nucleoporins. It is accepted that FG nucleoporins mediate active protein transport through the nuclear pore complex. The selective filter is unfolded hydrophobic polypeptide chains of nucleoporins lining the central channel of NPC. b) Ions and small neutral proteins can pass via the selective filter by diffusion. c) Large molecules or complexes penetrate via NPC only within transport complexes.
Ran-DEPENDENT NUCLEOCYTOPLASMIC TRANSPORT OF PROTEINS
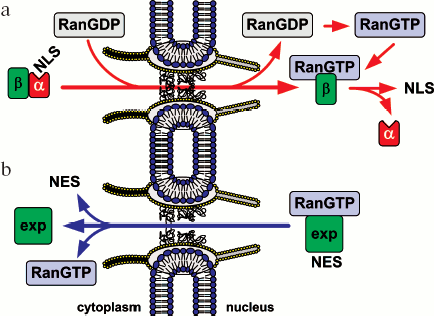
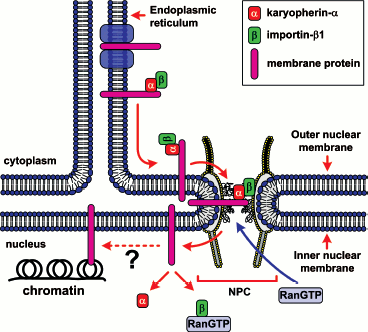
Import/export of most proteins, including membrane proteins, ribosomal subunits, and some types of RNAs is realized with the participation of a large evolutionarily conservative family of transport factors, karyopherins-beta. Most karyopherins-beta accomplish either nuclear import and are called importins or nuclear export and are called exportins. Only some of them take part in both export and import processes. Most of karyopherins-beta interact directly with their cargoes, and sometimes even via an adapter protein. Karyopherin-alpha, known also as importin-alpha, is the most-studied adapter protein. Importins bind to the nuclear localization signal (NLS) in the cargoes and move them to the nucleus. Exportins bind to the nuclear export signal (NES) in the cargoes and ensure its transport to the cytoplasm. In addition to the above, a whole set of transport proteins take part in the transporting process. Among them, a key protein is GTPase Ran. A simplified model of Ran-dependent nuclear transport is schematically shown in Fig. 2. The role of Ran in protein transport is described in detail below.
Signals of Nucleocytoplasmic Transport and Transport FactorsFig. 2. Simplified model of Ran-dependent nucleocytoplasmic transport of proteins. a) Nuclear import. Protein import with the nuclear localization signal (NLS) is mediated by the karyopherin-alpha/importin-beta1_heterodimer (designated as alpha and beta). Protein transport with importins-beta only is not shown. b) Nuclear export. A great part of export of NES-containing proteins is mediated by exportin Crm1 (designated as exp).
In every eukaryotic cell, a rapid directed shuttling of thousands of proteins and RNAs to the nucleus and from it takes place. The shuttling of most proteins is mediated by karyopherins-beta. Karyopherins-beta are a whole family of proteins. In human cells there are at least 20, and in yeast there are at least 14 (Table 1) [13, 14]. The significantly higher rate of cargoes compared to the number of karyopherins-beta raises the question: by what features do different karyopherins recognize their substrates?
Table 1. Members of the
karyopherin-beta family (modified from [14])
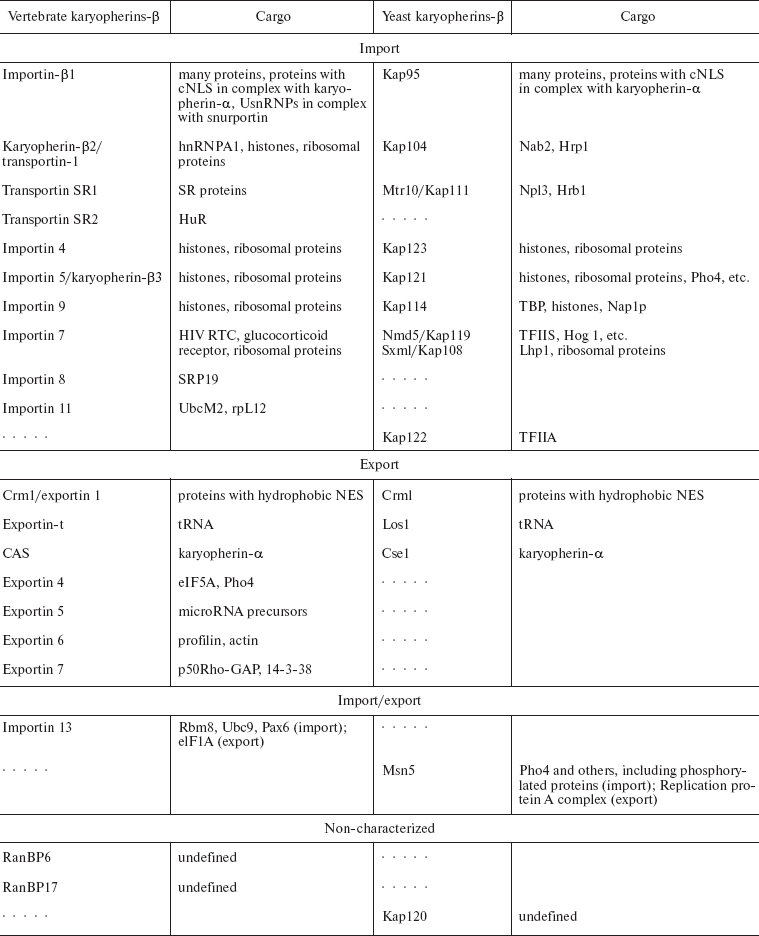
Note: Orthologs of karyopherins-beta and their vertebrate and
yeast substrates are compared. Dotted lines indicate that orthologs
have not been identified. Nmd5 and Sxm1 are orthologs of importin 7.
These proteins are highly homologous by their primary structure with
importin 7. Two names of one transportin are given with a slash. NES,
nuclear export signal; cNLS, classical nuclear localization signal.
Nuclear localization signals and karyopherins-alpha._Protein transport to the nucleus was first shown for nucleoplasmin and the virus SV40 large T antigen. The nucleoplasmin NLS consists of two clusters of positively charged residues separated by a spacer (KR-10aa-KKKL171), and the large T antigen NLS is a repeat of positively charged residues (PKKKRKV132) [15]. Signals of such type are very frequent and conservative. They are usually called classical or basic nuclear localization signals (cNLS).
Such signals are found in proteins CBP80 (RRR-11aa-KRRK20), BRCA1 (KRKRRP508 and PKKNNRLRRK615), DNA helicase Q1 (KK-15aa-KKRK645), LEF-1 (KKKKRKEK382), and many others [15]. Classical NLS are recognized by karyopherins-alpha. Human cells contain at least 6 homologous members of this family, which fall into three phylogenetic groups--alpha1, alpha2, and alpha3 [16].
Other NLS are also known that can be recognized by karyopherins-alpha. For example, in the protein Matalpha2 NLS polar residues are interspersed with non-polar ones (VRILESWFAKNIENPYLDT159) [17], or in the c-Myc NLS proline and aspartate at the boundary of the cluster consisting of positively charged residues are important for nuclear targeting (PAAKRVKLD328) [18].
Nuclear localization signals and importins. Most importins bind directly to cargoes and therefore do not rely on karyopherins-alpha [19, 20]. Usually it is very hard to reveal nuclear localization signals recognized by importins. In some cases, the NLS contains several positively charged amino acid residues, e.g. in core histones (H2A, H2B, H3, H4) [21-23] and ribosomal proteins (rpS7, rpL5, rpL23a) [24]. In some RNA-binding proteins (Npl3p, Nop1p, Sof1p) arginine-glycine-rich NLS have been found [25, 26]. Sometimes the NLS domain is relatively large. For example, the NLS M9 in hnRNP A1 consists of 38 amino acid residues, is glycine-rich, and contains a small number of positively charged residues [27]. In some cases, a very elongated segment of a protein molecule is identified as its NLS, which shows that the three-dimensional packing of the whole molecule is critical for being recognized by importins-beta [28].
As seen from the above, NLS are quite diverse so their identification is rather complicated and frequently requires an examination of the three-dimensional structure of the importin/NLS-protein complex. In structural studies of importin-beta1 with fragments of its different substrates (karyopherin-alpha, SREBP-2, PTHrP), it was shown that in every case various contacts are involved in the complex formation [29-31]. The studies also demonstrated that karyopherins-beta can have a number of different conformations, thus accommodating to a certain substrate. This explains how a limited number of importins can transport a vast number of various substrates that frequently have no similarity in amino acid sequences of their NLS.
Nuclear export signals and exportins. Exportins also recognize specific signals, i.e. nuclear export signals [19, 20]. The most common and best characterized is the hydrophobic leucine-rich NES. It is a nonconservative motif with 3 or 4 hydrophobic residues (e.g. LPPLERLTL83 in protein HIV Rev) [32]. The hydrophobic NES is found in all eukaryotes. At least 75 proteins containing NES of this type have been identified [33]. Hydrophobic NES were discovered in many transcription factors and cell cycle regulators, in protein Rev HIV, and in the protein kinase A inhibitor, in which the hydrophobic NES was first found [32, 34]. These NES are recognized by exportin Crm1. Like importin-beta1, Crm1 can move a number of substrates without or with adapter proteins [35-37]. Proteins lacking hydrophobic NES can also be exported from the nucleus. Their export is mediated by specific exportins. For example, proteins Pho4 and Mig1 without hydrophobic amino acids in the NES are exported by karyopherin Msn5p [38, 39]. In this case, the transport is connected with substrate phosphorylation, from which it can be concluded that the phosphorylation site can be included in the NES or phosphorylation contributes to the recognition of the NES by exportin [13, 40].
It is worthwhile to distinguish exportin CAS (Cse1p) that in complex with RanGTP (Ran in complex with GTP) transports karyopherin-alpha from the nucleus to the cytoplasm, thus providing for recycling of the factor.
In addition to the protein cargoes, several types of RNA are exported from the nucleus [41]. At least two exportins are responsible for their export. They are exportin-t, which ensures tRNA transport and recognizes a part of its structure as its NES, and exportin 5, which transports tRNA and microRNA precursors to the cytoplasm, recognizing the RNA hairpin structure with a 3´ overhang as the NES [42, 43]. The transport of most mRNAs proceeds without karyopherins, and proteins of the TAP/NFX family play their role [41].
Structure of Nuclear Pore Complex, Nucleoporins
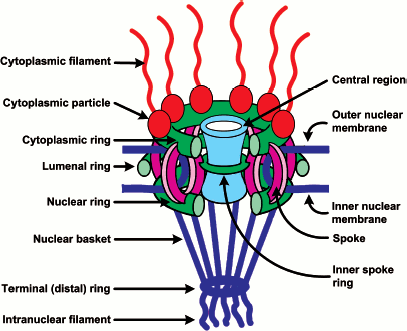
To answer the question what proteins form the nuclear pore complex (NPC), some systems from Saccharomyces cerevisiae, Xenopus laevis, and mammals were studied in detail. The reconstruction of the NPC structure based on high-resolution electron microscopy data showed that the architecture of all NPC is very conservative. The NPC has an 8-beam symmetry square to the membrane and is asymmetric to the membrane surface [44-47]. In a simplified way, the complex consists of three substructures: (1) the cytoplasmic filaments, (2) a central core, and (3) the nuclear basket (Fig. 3).
The central core contains eight spokes sandwiched between the nuclear and cytoplasmic rings. This spoke structure encircles the central region or the central channel (CGC, central gated channel) that mediates transport events. Such a structure is also inherent to yeast and vertebrate NPCs. However, vertebrate NPCs are larger in size and can have additional structures. The molecular mass of yeast NPC is 44,000 kD [1, 48], and that of vertebrate NPC is 60,000 kD [49]. The cryo-electron microscopy studies of yeast NPCs demonstrated that their central core has a simplified structure and does not contains an inner spoke ring [47]. The electron microscopy analysis showed that cytoplasmic fibrils about 50 nm long are branching from the cytoplasmic ring, and nuclear fibrils about 100 nm long are branching from the nuclear ring, and the latter merge at the distal ends, thus forming a basket-like structure [46]. It was proposed that the asymmetric filamentous formations are first implemented in the recognition and binding of transport complexes [50, 51].Fig. 3. Schematic representation of the NPC structure (modified from [4]). The main substructures of NPC are cytoplasmic filaments, the central core, and the nuclear basket. See their description in the text.
Proteomic studies of mammalian and yeast NPCs showed that an NPC consists of about 30 different nucleoporin proteins whose copies are present in a large quantity (as a rule, 8, 16, or 32 copies as follows from the 8-beam symmetry), their total number being 500-1000 per NPC [1, 48]. Among nucleoporins there are neither motor proteins nor ATPase/GTPase. Nucleoporins fall into a number of families. 1) Transmembrane nucleoporins (TM) fasten NPCs in the nuclear membrane. 2) FG-nucleoporins have FG repeats (GLFG, FXFG, or FG) and hydrophobic linkers (see also Fig. 1a). FN-nucleoporins with FN repeats (nucleoporins Nup35/Nup53) can be classified as a subgroup. 3) Recently a class of nucleoporins that contain WD repeats or a seven-bladed propeller motif has been discovered [4, 48, 52, 53]. It is believed that FG-nucleoporins make up half of the NPC mass (from 200 to 700 copies per NPC). The major part of them is distributed symmetrically to the nuclear membrane, and only some nucleoporins are only on the nuclear or cytoplasmic face of the NPC [1].
FG-nucleoporins have sites of binding to karyopherins. When passing through NPCs, all karyopherins bind to FG-nucleoporins. Just these interactions make the basis of the current model of nucleocytoplasmic transport [4]. It is assumed that asymmetrically located FG-nucleoporins play an important role in the transport directionality and the substrate release. Thus, importins often have a higher affinity to nucleoporins of the nuclear face of the NPC, and exportins to its cytoplasmic face [54-57]. The major component of cytoplasmic fibrils is nucleoporin RanBP2/Nup358 (21 molecules per fibril). The transport to the nucleus starts from the binding of the transport complex to this FG-nucleoporin [58]. The cytoplasmic ring is enriched with nucleoporin Nup214, and the nuclear ring and fibrils are enriched with nucleoporin Nup153. The central region of the nuclear pore complex is represented by so-called Nup62-complexes, whose major proteins are Nup62, Nup58, and Nup54. The data on all nucleoporins are summarized in Table 2.
Table 2. Yeast and vertebrate nucleoporins
(modified from [4])
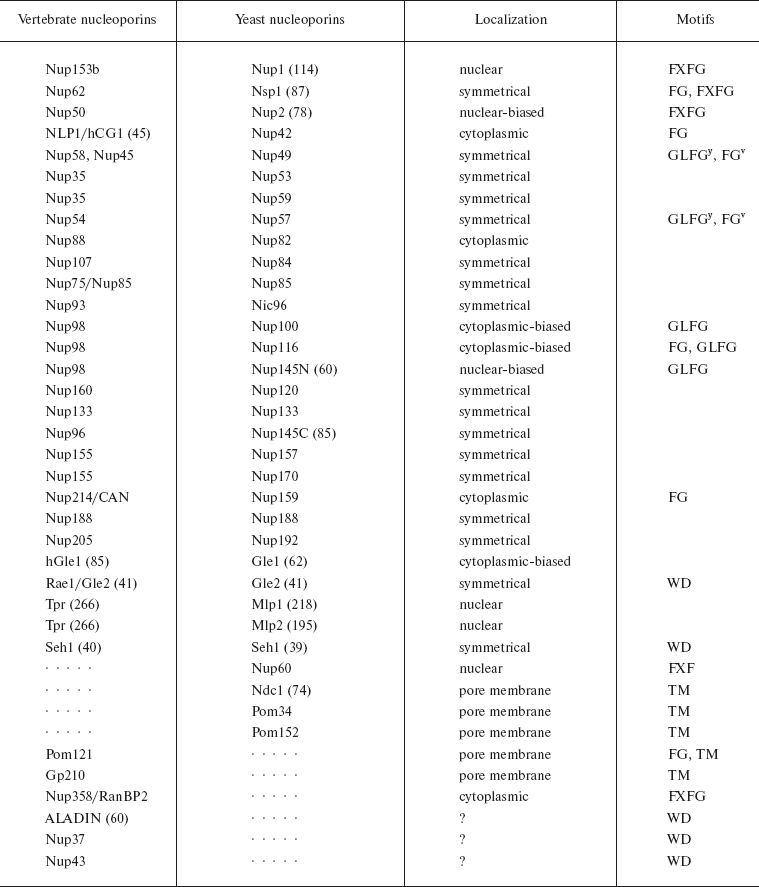
Note: Orthologs of basic yeast and vertebrate nucleoporins are
combined. The general nomenclature of nucleoporins has a numerical
designation showing the predicted molecular mass in kD. For some
nucleoporins with accepted names, the mass is given in parentheses. Two
names of one transportin are given with a slash. Dotted lines indicate
that orthologs have not been found. TM means transmembrane
nucleoporins. The localization of nucleoporins is indicated based
mainly on the data on yeast nucleoporins (y is used for
yeast nucleoporins, v for vertebrate nucleoporins).
Model of Ran-Dependent Transport
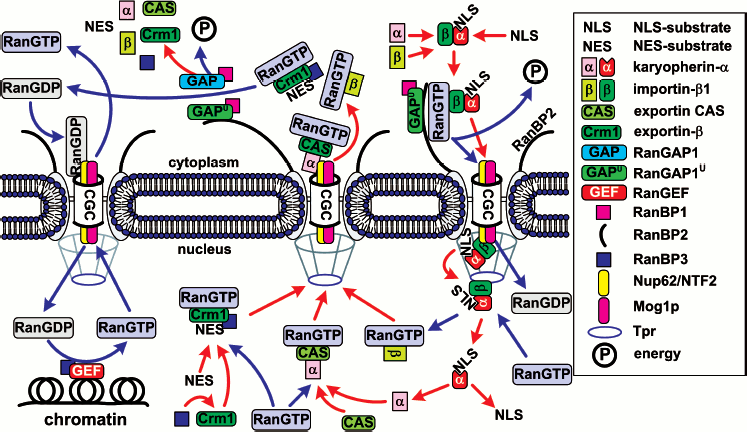
The hypothesis on the vital role of Ran in nuclear import of proteins was first based on two facts. (i) Ran mutations resulted in distortions of nuclear import of proteins in vivo [59, 60]. (ii) Ran was characterized as a cytoplasmic component required for trafficking of NLS-containing substrates across the nuclear membrane in vitro [61, 62]. A direct link between Ran and nuclear import was established after the discovery of Ran-binding proteins (RanBP) [63]. Characterization of these factors and their interaction with Ran has provided a general outline for nuclear import of proteins with a NLS (Fig. 4). This model is not final since step by step further participants are found and new interactions are detected. The discovery of novel Ran-binding proteins permits regular defining of the pathways of translocation of proteins that have no cNLS. More than that, a radically different trafficking mechanism (the Ran-independent one) has been discovered quite recently.
Ran and Ran regulators. The major nucleocytoplasmic transport is monitored by a small Ran GTPase, which plays the key role in the formation of diverse transporting complexes (Fig. 4). Ran is an abundant preferably nuclear 25 kD protein [64]. Like most GTPases [65], Ran hydrolyzes GTP very slowly, as a result of which its nucleotide binding is regulated by interaction with different regulatory proteins. The latter include (i) the protein activating the Ran GTPase - RanGAP1 (Ran GTPase activating protein), (ii) the protein activating RanGAP - RanBP1, (iii) the guanine nucleotide exchanging factor - RanGEF (Ran guanine exchange factor), and (iv) the factor activating RanGEF - RanBP3 (summarized in Table 3).Fig. 4. General scheme of nuclear import of ñNLS/NES-containing proteins. The conventional model of transition of cNLS/NES-bearing proteins through NPC includes the following stages. 1) Formation of the import karyopherin-alpha/importin-beta1/cNLS-protein complex in the presence of RanGAP1U and low concentration of RanGTP in the cytoplasm; docking of the import transport complex to cytoplasmic filaments (the top right corner of the figure). 2) Transition across the central channel (CGC). 3) RanGTP-stimulated release of karyopherin-alpha/cNLS-protein to the nucleoplasm. 4) Retention of importin-beta1 in NPC and transport of importin-beta1 into cytoplasm in complex with RanGTP (bottom right corner of the figure). 5) After the dissociation of the karyopherin-alpha/cNLS-protein complex, karyopherin-alpha is exported from the nucleus in complex with CAS/RanGTP. NES-containing proteins are exported in complex with Crm1/RanGTP (bottom central part of the figure). 6) On the way out of the NPC, all the exported transport complexes dissociate due to GTP hydrolysis in Ran stimulated by RanGAP1 or RanGAP1U (top central part of the figure). 7) Nuclear import of RanGDP is mediated by the NTF2 factor (top left corner of the figure). 8) When in the nucleus, RanGEF (Ran guanine exchange factor) transforms Ran from its GDP-bound form into the GTP-bound one. The minimal cytoplasmic pool of RanGTP is maintained due to the transport factor Mog1 (bottom left corner of the figure). The details of the scheme are given in the text.
Table 3. Ran-binding vertebrate proteins
(modified from [102])
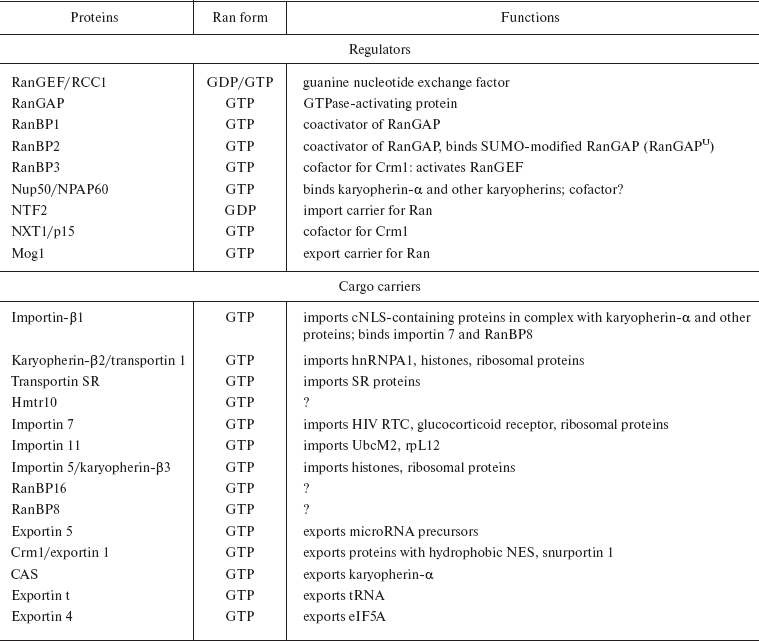
Note: Some of Ran-binding proteins are given. The Ran form is
indicated, to which the proteins bind, as well as known function. Two
names of one transportin are given with a slash.
RanGAP1 amplifies the GTPase activity of Ran about 10,000-fold, and in the presence of RanBP1 about 100,000-fold [66]. RanGAP1 is found only in the cytoplasm [67] or, after sumoylation (i.e. formation of conjugates with the SUMO-1 protein - RanGAP1U), binds to the Ran-binding domain (RanBD) of nucleoporin RanBP2 (Nup358) and is localized to fibrils of the NPC cytoplasmic surface (Figs. 4 and 5) [68]. RanBP1 shuttles between the nucleus and cytoplasm but is predominantly localized to the cytoplasm [69]. RanGEF (also named RCC1), that restores the pool of the RanGTP, is localized exclusively to the nucleus in the complex with histones H2A and H2B (approximately one RanGEF molecule per nucleosome) [70]. RanGEF accelerates the exchange of nucleotides for Ran by nearly 10,000-fold [71]. RanBP3 shuttles between the nucleus and cytoplasm but is localized mostly in the nuclei [72]. RanBP3 binds to RanGEF and stimulates its GEF activity in the exchange of RanGDP for RanGTP ten-fold [73].
Exact localization of RanGAP1 in the cytoplasm and RanGEF in the nucleus results in that Ran in complex with GTP (RanGTP) is for the most part in the nucleus, while that in complex with GDP (RanGDP) is in the cytoplasm. As a consequence, a RanGTP gradient appears which ensures the directionality of the transport.
Fitting of the karyopherin-alpha/importin-beta1/cNLS-protein complex into the NPC. In vitro nuclear import assays have identified two cytosolic components as sufficient for docking of cNLS-containing proteins at the nuclear envelope. These are karyopherin-alpha, which recognized cNLS, and importin-beta1, which bound to karyopherin-alpha and interacted with NPC through FG repeats of FG-nucleoporins [74-77]. When describing the mechanism of Ran-dependent import of proteins, we will focus on the transport of the karyopherin-alpha/importin-beta1/cNLS-protein complex, since the shuttling of proteins with non-classical NLS has only one main difference: karyopherin-alpha is not implicated in the trafficking, and karyopherin-beta is primarily responsible for the NLS-containing protein recognition.
The initial stage of docking to the NPC is the interaction of the karyopherin-alpha/importin-beta1/cNLS-protein complex with protein RanBP2 (Nup358), nucleoporin, which is a component of cytoplasmic fibrils [78, 79] (Fig. 3). RanBP2 contains FXFG repeats and four Ran-binding domains. The NPC also has other nucleoporins with FXFG repeats, with which the karyopherin-alpha/importin-beta1/cNLS-protein complex can interact. Nevertheless, at a low RanGTP content, the affinity of importin-beta1 to these nucleoporins is minor, whereas to RanBP2 it is rather high [80]. Thus, the specificity of binding of importin-beta1 to RanBP2 is RanGTP-dependent: apparently RanGTP is directly implicated in the interaction of importin-beta1 with RanBDs of nucleoporin RanBP2 [57].
It is known that the karyopherin-alpha/importin-beta1 dimer is unstable in the presence of RanGTP. Consequently, during docking of the karyopherin-alpha/importin-beta1/cNLS-protein complex to RanBP2, rapid hydrolysis of GTP should take place. As mentioned above, in addition to the soluble RanGAP1 there exists also RanGAP1U that is in complex with RanBP2. In a free form importin-beta1 significantly suppresses the activity of RanGAP1 [81, 82], but after interaction with the RanBDs of RanBP2 the inhibiting effect of importin-beta1 is relieved [82].
The data suggest that the docking of the transport complex to the NPC is a RanGTP-dependent process, which requires rapid hydrolysis of GTP for maintaining the stable state of the karyopherin-alpha/importin-beta1/cNLS complex. The minimal pool of RanGTP in the cytoplasm is provided due to the transport factor Mog1p, which has an affinity to GTP-bound Ran and transports it from the nucleus to the cytoplasm [83].
Transition of the karyopherin-alpha/importin-beta1/cNLS complex across the central channel of a nuclear pore. To reach the nuclear surface of a nuclear pore, the cNLS-containing protein should travel about 40 nm from the docking site at the tip of the cytoplasmic surface to the cytoplasmic face of the central gated channel (CGC) and additional 30 nm through the CGC to dock at its nucleoplasmic face. The initial 40 nm of the trek may be overcome by the bending of the cytoplasmic fibril. As a result of such bending, the fibril end with the transport complex approaches the CGC [80, 84]. After hydrolysis of GTP the transport complex is released in proximity to the CGC. Translocation of the complex across the CGC has not yet been studied in detail. Nonetheless, we can imagine a general sequence of events in a simplified way. Immediately after the release from the fibril, the transport complex binds to nucleoporin p62 and transport factor NTF2 (nuclear transport factor) on the cytoplasmic surface of the NPC [85-87]. NTF2 is a small homodimer protein that ensures a unidirectional import of the RanGDP-transport complex from the cytoplasm to the nucleus. A swift translocation of NTF2/RanGDP to the nucleus is explained by the fact that NTF2 interacts directly with FG-nucleoporins (of low affinity). NTF2 binds only to RanGDP: NTF2 binds to the Switch II region of RanGDP, whose conformation is drastically changed when Ran switches to the GTP-bound form [88]. This determines the unidirectionality of the transfer because Ran changes to the GTP-bound form practically just after its export from the NPC to the nucleus. Having passed across the CGC, on the nuclear surface of the nuclear gated complex the transport complex binds to Nup62, after which it shifts to the “dock”, nucleoporin Nup153, where it undergoes disassembling [57].
Disassembling of the karyopherin-alpha/importin-beta1/cNLS-protein complex. Disassembling of the transport complex proceeds at least in three stages. (i) Nup153, a nuclear basket protein, binds the transport complex by interactions of its FG-repeats with importin-beta1. In the presence of GTP and a soluble factor (probably Ran), the connection of Nup153 with importin-beta1 is damaged [57]. (ii) RanGTP, whose concentration in the nucleus is very high, binds to the transport complex and causes release of importin-beta1 from the complex with karyopherin-alpha/cNLS-protein [81, 89, 90]. (iii) The dissociation of importin-beta1 from the complex impairs the affinity of karyopherin-alpha to the cNLS-protein, as a result of which the latter is released into the nucleoplasm.
At present, it is not known whether the release of the complex from Nup153 and dissociation of importin-beta1 from karyopherin-alpha/cNLS-protein are coupled or separate events. Protein Tpr, which is a component of the nuclear basket of the NPC, is also implicated in the disassembling of the karyopherin-alpha/importin-beta1/cNLS-protein complex, but its role is less clear [57]. Like Nup153, Tpr also binds to the karyopherin-alpha/importin-beta1 complex; in the presence of GTP and a soluble factor (possibly Ran), this connection is disrupted. However, Tpr binds to the karyopherin-alpha/importin-beta1 complex in the absence of the cNLS-protein. It is postulated that Tpr serves to retain importin-beta1 in close proximity to the NPC. This is consistent with the finding that importin-beta1 does not distribute within the nucleoplasm upon crossing the nuclear envelope [76].
Translocation of proteins from the nucleus to the cytoplasm. The nucleocytoplasmic transport is a continuous process, therefore transport factors should return to their initial sites to provide for following rounds of transport.
The mechanism of such recycling was demonstrated for several transport factors. Importin-beta1 binds to RanGTP and in such complex returns to the cytoplasm [91]. This is a general principle of importin recycling, but in some cases importins return to the cytoplasm in complex with another protein substrate. For example, importin 13 imports proteins Rbm8, Ubc9, and Pax6 to the nucleus, and exports protein eIF1A from it [92].
After dissociation from importin-beta1 in the nucleus, karyopherin-alpha forms a trimeric complex with RanGTP and CAS (cellular apoptosis susceptibility gene), and as such it is again transported into the cytoplasm [93, 94].
The mechanism of translocation of NES-containing proteins from the nucleus to the cytoplasm is similar to the transport of proteins with non-classical NLS: for this only exporting karyopherin-beta and RanGTP are required. Crm1 is the most common exportin. RanBP3 as an additional factor is also involved in the translocation of NES-containing proteins with Crm1. After association with RanBP3, the affinity of Crm1 to RanGTP and proteins with hydrophobic NES increases greatly [95]. Such a quaternary complex is imported to the cytoplasm. Cargoes without hydrophobic NES are also found, and they have their particular exportins (see section “Ran-Dependent Nucleocytoplasmic Transport of Proteins”).
A specific feature of any type of Ran-dependent export is translocation through the NPC of the karyopherin-beta/RanGTP complex. The association of karyopherins-beta and RanGTP is very stable. It is disrupted only when Ran passes into the GDP-bound form; therefore, to release karyopherin-beta (and the translocated NES-containing protein), RanGAP1U (just upon the discharge from the nuclear pore complex) or RanGAP1 (in the cytoplasm) should be implicated. RanGAP1 activates the GTPase activity of Ran 100,000-fold [96]. However, RanGTP is not entirely accessible for RanGAP1 in the karyopherin-beta/RanGTP complex. The accessibility improves when RanBP1 takes part in the process, which amplifies the GTPase activity by another order of magnitude [96]. After GTP hydrolysis, the karyopherin-beta/RanGTP/(substrate)/(RanBP3) complex dissociates, and NTF2 mediates the return of RanGDP to the nucleus, RanBP3 getting back to the nucleus mainly in complex with importin-alpha3 [72].
The NTF2-mediated import of RanGDP to the nucleus is similar to the karyopherin-beta-mediated import, though it is not quite clear how NTF2 returns to the cytoplasm: no factors have been found that could be implicated in the export of NTF2. But NTF2 is a small 28-kD homodimer protein, and its translocation fits well the common model of translocation due to diffusion of proteins up to 60 kD.
Ran-binding proteins. A number of Ran-binding proteins that can be classified in three basic families have been identified using diverse approaches [97-100]. (i) Members of the first family contain a well-characterized RanBD--RanBP1, RanBP2, and RanBP3. (ii) Proteins of the second family in their N-terminal parts have about 150 amino acid residues that participate in the interaction with RanGTP, and this is the only relatedness between them. This family includes importin-beta1, RanBP5, RanBP7, RanBP8, Crm1, exportin-t, and CAS.
Proteins of the two families do not compete for the binding sites in RanGTP, which allows for the formation of ternary complexes with involvement of proteins from both families (e.g. RanBP1/RanGTP/RanBP5) [101]. (iii) The third family of proteins has no Ran-binding sequences characteristic of the first two families. Transportin-1 is a member of this family [27].
Ran-binding proteins of the first type participate in the translocation of proteins with NLS/NES in complex with karyopherin-beta. Their role in the process has been described above. Members of the other two families also play the role of transporters, but they directly interact with their cargo--they do not require adapter proteins such as karyopherin-alpha. Table 3 shows known functions of some Ran-binding proteins [102].
Translocation of Proteins of Inner Nuclear Membrane
Proteins of the inner nuclear membrane (INM) participate in the organization of lamins, chromatin, and nuclear membrane and function as transcription factors. To reach the internal nuclear membrane, INM proteins should pass through the NPC. After the synthesis on ribosomes, membrane proteins are in the membrane of the endoplasmic reticulum (ER). How do INM proteins get into the nucleus? Because the outer nuclear membrane (ONM) is contiguous and functionally equivalent to the ER, INM proteins can be transmitted by diffusion along the ER and ONM, reach the nuclear pore complex and due to diffusion pass through it, and when on the INM, they can be specifically retained in the nucleus through interactions with the nuclear architecture elements (Fig. 5) [103, 104].
This is the simplest explanation, but it should be remembered that the selective filter in the central channel significantly restricts diffusion through the nuclear pore complex. Recent studies have shown that the translocation of membrane proteins via the NPC requires energy [105], which confirms the existence of active transport in addition to or instead of passive transport due to diffusion. Energy might be necessary for remodeling of the nuclear pore complex when INM proteins are passing through it or for Ran-dependent transport. It has been demonstrated lately that proteins Heh1 and Heh2 are translocated via the NPC by the karyopherin-alpha/importin-beta1/Ran-dependent mechanism [3]. As in the case of soluble proteins, these proteins contain cNLS. Moreover, NLS sequences are found also in other INM proteins: in lamin B receptor, LAP1, LAP2beta, emerin, MAN1, and LEM2 [106]. It is believed that Nup170 is implicated in INM protein transport, because it plays the key role in regulation of permeability of the NPC channel [107].Fig. 5. Simplified scheme of transport of inner nuclear membrane proteins (modified from [104]).
Receptors of steroid hormones are imported into the nucleus after interaction with the hormone in the cytoplasm. These receptors work as transcription factors in the nucleus gathering transcription complexes on hormone-specific DNA elements. Androgen, estrogen, and glucocorticoid receptors have two NLS: the classical NLS-1 and NLS-2, which consists of 200 amino acid residues and overlaps with the ligand-binding domain (LBD) [108-114]. NLS-1 of the glucocorticoid receptor interacts with the karyopherin-alpha/importin-beta1 complex and importin 7, and NLS-2 interacts with importin 7 and importin 8. Interestingly, the binding of the karyopherin-alpha/importin-beta1 complex, importin 7, or importin 8 to the glucocorticoid receptor is ligand-independent [115]. Thus, the common model, according to which the binding to the ligand leads to dissociation from the receptor of chaperone Hsp90 masking NLS, proved to be erroneous. Instead, the binding of the ligand can regulate some chaperone-dependent processes in which the receptor and/or NPC are/is involved after recognition of NLS by transport factors [115-117]. It is also known that chaperones intensify interaction of the receptors and the cytoskeleton, i.e. facilitate their retention in the cytoplasm [118].
Signals and transport factors implicated in the receptor transfer have been insufficiently studied. Regions that can be NES have been found for many receptors [119, 120], but the analysis of sequences of fifty known receptors has revealed only a typical hydrophobic NES that can be recognized by Crm1 [121]. Nevertheless, the use of leptomycin B, that specifically inhibits Crm1, demonstrated that this karyopherin is implicated in the export of receptors. But the corresponding data are in discrepancy, and other researchers have obtained a conflicting result [122, 123]. Based on these data, it can be concluded that the receptor export can be both Crm1-dependent and Crm1-independent, which may be explained by cell conditions. It is assumed that receptors are recognized with the help of mediators. Thus, proteins of the 14-3-3 family and p160 coactivators, which bind to receptors, have a NES and can play the role of adapters providing for Ran-dependent (Crm1-dependent) export [124, 125]. In addition, export of some receptors can be Ca2+-dependent and take place with the help of calreticulin (see next section) [126].
Ran-INDEPENDENT NUCLEOCYTOPLASMIC TRANSPORT OF PROTEINS
The nucleocytoplasmic transport mediated by the karyopherin-alpha/importin-beta1_complex depends both on Ran and hydrolysis of GTP catalyzed by Ran. Alternative pathways of translocation different from the analyzed classical Ran-dependent transport have been described. For example, alternative translocation pathways have been described for beta-catenin, importin-beta1, transportin 1, Crm1, exportin-t, hnRNP K, calmodulin, ERK, NHP6A, importin-alpha, proteins of the STAT family, TIAR and TIA-1, cAMP-dependent kinase inhibitor (PKI, protein kinase A inhibitor), and glucocorticoid receptor [126-140].
It was shown in in vitro experiments that beta-catenin binding to the NPC does not require additional factors and is inhibited in the presence of importin-beta1. This provides evidence for overlapping of the binding sites of beta-catenin and importin-beta1 in the NPC [127]. Wheat germ agglutinin (WGA), i.e. lectin that binds to GlcNAc-modified nucleoporins such as Nup62, inhibited beta-catenin import. The in vitro experiments demonstrated that import of beta-catenin into the nucleus depends on GTP hydrolysis, but it can also go on in the absence of Ran. Another related GTPase might play the role of Ran in this process. Based on this data, it was proposed that such proteins as beta-catenin (with ARM, armadillo repeats) can bind to the NPC and be imported into the nucleus independent of the system of karyopherin-alpha/importin-beta1_and RanGTP [127].
In the absence of karyopherin-alpha and cNLS-containing substrate, importin-beta1 does not require Ran to be imported into the nucleus [130]. The transport is inhibited by WGA but is not affected by chilling, which is rather strange. Depletion of ATP reduces the translocation efficiency.
Translocation of transportin-1 to the cytoplasm and back does not depend on GTP and ATP, but is inhibited by WGA and chilling. Evidently, other energy sources are used in the translocation of transportin-1 [136]. The dependence of the process on temperature shows that certain temperature-dependent NPC conformations can be crucial for translocation of transportin-1. The inhibiting effect of WGA evidences that the transport occurs through the NPC.
The transport of Crm1 also has an alternative pathway, but its translocation has not been studied in detail [129]. Previously it was established that Crm1 binds to Nup214 [141]. Moreover it was found that namely this nucleoporin is the last binding site of the transport complex upon its export from the nucleus [142], and its removal from the NPC results in complete inhibition of Crm1-dependent export [143]. Inasmuch as no transport factors mediating nuclear import of Crm1 has been discovered, it may be assumed that Crm1 migrates due to its own affinity to nucleoporins.
Nuclear import of exportin-t and other exportins has been studied quite insufficiently [131]. It can be speculated that their import to the nucleus is not mediated specially because of their own affinity to nucleoporins of the cytoplasmic face of NPC. It was found that translocation of exportin-t does not require GTP/ATP hydrolysis [131].
The hnRNP K contains two NLS -- cNLS and “KNS” (hnRNP K nuclear shuttling domain) [134]. Owing to the cNLS the translocation can follow the classical karyopherin-alpha/importin-beta1 pathway. The KNS-dependent transport has the following specific features. (i) KNS is responsible for both import and export of hnRNP K. (ii) Nuclear import depends on RNA-polymerase II transcription. (iii) Nuclear import does not require additional factors [134].
The Ca2+-binding protein calmodulin (CaM) can penetrate into the nucleus by diffusion, and its allocation in the cytoplasm and nucleus might depend only on the concentration of Ca2+ in the cytoplasm and on the interaction with CaM-binding proteins. However, it has been shown that CaM import into the nucleus can be active and occur with additional factors. Thus, it is found that CaM import is inhibited by WGA and chilling, which confirms translocation via the NPC, but the process is energy-independent [137].
An alternative pathway of migration of proteins bearing cNLS from the cytoplasm into the nucleus, that is independent of Ran but is CaM-mediated, has been determined. Thus, intracellular concentrations of Ca2+, inhibiting classical nuclear transport, triggered alternative translocation of cNLS-containing proteins recognized by CaM. Such translocation is independent of GTP hydrolysis, but is ATP-mediated, inhibited by WGA and suppressed by chilling [144].
It was shown that nuclear transport of protein NHP6A does not require GTP hydrolysis and is not suppressed by chilling, which is in favor of Ran-independent transport [139].
In response to extracellular stimuli, kinase ERK translocates from the cytoplasm to the nucleus. At least two possible pathways of its translocation are known: passive (as a monomer due to diffusion) and active. It was demonstrated on a model system with GFP-fused ERK protein that cannot penetrate into the nucleus due to diffusion, that WGA, chilling, and an excess of importin-beta1 inhibited ERK import into the nucleus, but neither additional factors nor energy losses were required for the process. ERK was shown to interact directly with nucleoporin Nup214, which may explain its translocation without adapter proteins [133].
It was revealed that as a result of different stresses karyopherin-alpha migrates to the nucleus, which inhibits the classical transport of karyopherin-alpha/importin-beta1/cNLS [135]. Such translocation of karyopherin-alpha was inhibited by WGA, which is evidence for an active transport through NPC. Under the same conditions, the other model substrate GST-NLS-GFP was not imported to the nucleus. These data are evidence for importin-beta1/Ran-independent translocation.
As a rule, STAT proteins (signal transducers and activators of transcription) shuttle between the nucleus and the cytoplasm in a Ran-dependent way [145]. But lately it has been shown that the translocation may be Ran-independent; in this case it does not require additional carriers and is energy-independent [132]. It has been demonstrated that Stat1 can directly interact with FG-nucleoporins Nup153 and Nup214 but not with Nup62 [132].
Two closely related proteins, TIAR and TIA-1, have three RNA recognition motifs (RRM). However, it has been established that motif RRM2 is responsible for nuclear import of these proteins, and RRM3 is responsible for their nuclear export. The import to the nucleus is Ran-dependent, whereas the export is independent of GTP hydrolysis on Ran [140].
An interesting Crm1-independent export was found for PKI and the glucocorticoid receptor. The Ca2+-binding protein calreticulin is an alternative to Crm1 for migration of these proteins [126, 128]. It appeared that to reduce intracellular concentration of Ca2+, calreticulin can bind to the NES of the glucocorticoid receptor and in the complex with RanGTP be transported to the cytoplasm. In other words, this pathway is very similar to the classical nuclear Crm1-dependent export.
NUCLEOCYTOPLASMIC TRANSPORT REGULATION
The multistage character of translocation, the great number of mediators, the diversity of NES/NLS, and the multicomponent structure of NPC imply a possibility of regulation of the nucleocytoplasmic distribution of different proteins in response to stimuli (growth, proliferation, and differentiation) or the cell cycle stage. The number of works devoted to studying the nucleocytoplasmic transport regulation of a particular protein grows from year to year. The variety of examples can be represented by a limited number of similar mechanisms of regulation [15, 146, 147]. In this section we will analyze the currently known pathways of nucleocytoplasmic transport of some proteins.
Transport Regulation by Modulation of Importin/Exportin Interactions with the Substrate NLS/NES
The basic factor determining the nucleocytoplasmic distribution of proteins may be regulation of transport complexes upon their formation due to modulation of importin-NLS/exportin-NES interactions. These interactions are very sensitive to conformational changes in the NLS/NES regions and in the substrate-binding sites of karyopherins. Just this is the reason why the major part of known examples of nucleocytoplasmic distribution regulation affects the changes in the substrate modulating its NLS/NES [147]. Examples of similar regulation due to the changes in karyopherins are not numerous.
Masking of Substrate NLS/NES from Recognition by Importin/Exportin
The masking of NLS/NES is the most widespread mechanism of regulation of the nucleocytoplasmic transport.
Intramolecular masking of NLS/NES. Intramolecular masking consists in the following: upon introduction of charge or conformational changes to the NLS/NES-containing region of the protein the access of karyopherin to the NLS/NES vanishes (Fig. 6).
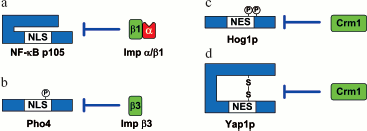
Subunit p50 of the dimer transcription factor NF-kappaB (nuclear factor kappa B) is synthesized as a p105 precursor form in which NLS, recognized by the karyopherin-alpha/importin-beta1 complex, is masked and inaccessible. At an immune response, p105 is specifically phosphorylated and its C-terminal part becomes degraded. As a result of this processing, NLS renders accessible up to p50 to be recognized by karyopherin-alpha/importin-beta1, and NF-kappaB can be imported to the nucleus (Fig. 6a) [148]. A similar mechanism was shown for integrase interactor 1 (INI1) from the human SNF5 chromatin-remodeling complex. The C-terminus of this protein masks NES recognized by Crm1 and thus prevents its nuclear export [149].Fig. 6. Examples of intramolecular masking (modified from [147]). a) When in the form of the p105 precursor, the nuclear localization signal of transcription factor NF-kappaB p50 is inaccessible for binding to the karyopherin-alpha/importin-beta1 complex. Proteasomal degradation of the C-terminal part of the p105 precursor after its phosphorylation unmasks NLS and leads to its nuclear import. b) Phosphorylation of Pho4 at Ser152 masks NLS from recognition by importin-beta3. Nuclear translocation is revealed after dephosphorylation of the residue. c) Phosphorylation of Hog1p at Thr174/Tyr176 masks NES from recognition by Crm1. Cytoplasmic translocation is found after dephosphorylation of the residues. d) Oxidative stress leads to disulfide linkage in protein Yap1p. The formation of this bond inhibits the binding of Crm1 to NES and leads to accumulation of the protein in the nucleus. The details of the scheme are given in the text.
Intramolecular masking can be caused by phosphorylation near NLS/NES or within them. Protein NF-AT2 (nuclear factor of activated T cell 2) contains two NLS, whose interaction with importins depends on the phosphorylation of the protein. At a low Ca2+ content in a cell, amino acid residues in both signals are phosphorylated, which leads to inhibition of the NF-AT2 nuclear import. With an increase in the Ca2+ concentration, calcineurin dephosphorylates the signals and NF-AT2 is imported into the nucleus [150]. In a similar way, at a high content of phosphates in a cell, the Pho80-Pho85 cyclin-dependent kinase complex phosphorylates the transcription factor Pho4 at Ser152 near NLS, which prevents importin-beta3 (Kap121/Pse1) binding and, as a consequence, inhibits nuclear import (Fig. 6b) [151].
Similar mechanisms have also been found for export regulation: under osmotic stress protein Hog1p (high osmolarity glycerol pathway-signaling protein) is phosphorylated by Pbs2p kinase at Thr174 and Tyr176, which renders NES inaccessible for binding to exportin-1 (Xpo1p) and leads to inhibiting the export of Hog1p from the nucleus (Fig. 6c) [152].
The masking of NLS/NES can be a result of conformational changes due to the formation of disulfide bonds between cysteine residues. Thus, under oxidative stress a disulfide bond between Cys598 and Cys620 is formed in the transcription factor Yap1p, which makes its NES inaccessible for interaction with exportin Crm1 (Fig. 6d) [153]. A similar mechanism of import-export regulation was also shown for the transcription factor Pap1 [154].
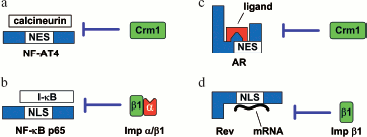
Intermolecular masking of NLS/NES. Such masking consists in that the distortion of importin-NLS/exportin-NES interactions is caused by the binding of the NLS/NES-containing protein to another protein or nucleic acid (Fig. 7).
At high Ca2+ concentration, calcineurin (Ca2+-responsive phosphatase) binds to the transcription factor NF-AT4 and masks its NES from interaction with Crm1, which suppresses nuclear export of the factor. At low Ca2+ concentration, calcineurin dissociates from NF-AT4 and unmasks its NES (Fig. 7a) [155].Fig. 7. Examples of intermolecular masking (modified from [147]). a) At a high concentration of Ca2+, calcineurin binds to NF-AT4 and masks its NES from interaction with Crm1. Cytoplasmic import of NF-AT4 can take place only after dissociation of calcineurin in response to decreasing concentration of Ca2+. b) The specific inhibitor I-kappaB masks the NLS of the NF-kappaB ğ65 from interaction with the karyopherin-alpha/importin-beta1 complex. Unmasking of NLS on NF-kappaB p65 and nuclear import can occur only after proteasomal degradation of I-kappaB. c) NES of the androgen receptor (AR) is localized to the ligand-binding domain. In the presence of the ligand (androgen), NES is masked and Crm1 cannot recognize it. Import of AR to the cytoplasm can take place only after androgen dissociation. d) The binding of the Rev arginine-rich motif to mRNA makes its NLS unable to interact with importin-beta1. Nuclear import of Rev can take place only after its dissociation from mRNA in the cytoplasm. The details of the scheme are given in the text.
A similar regulation mechanism of nuclear import was shown for NF-kappaB p65: its NLS is masked from interaction with the karyopherin-alpha/importin-beta1 complex by specific inhibitor protein I-kappaB [156]. Upon immune challenge, I-kappaB is phosphorylated, leading to its ubiquitination and the following degradation by the proteasome. This results in unmasking of NF-kappaB p65 NLS and NF-kappaB p65 nuclear import (Fig. 7b) [157].
The BRCA1-binding protein BRAP2 may operate analogously to I-kappaB: it interacts with NLS of not only BRCA but also with NLS of the SV40 virus large T antigen [158]; however the effect of such interactions on import has not been shown, and so their role in the nuclear transport regulation is not yet clear.
The nuclear localization of the tumor suppressor p53 is regulated by a number of mechanisms. One of them is associated with phosphorylation of Ser15/20 in p53 in response to DNA damage. This leads to masking of NES1. Another mechanism consists in tetramerization of protein p53 within the nucleus in response to the DNA damage: NES2 is masked in tetrameric p53. Dissociation of this tetramer is required for the export of the protein from the nucleus [159].
The binding of the ligand may also cause masking of NES/NLS as was demonstrated for the androgen receptor in which NES is in the ligand-binding domain [120]. In the presence of the ligand (androgen), NES is masked and Crm1 cannot recognize it. The receptor is translocated only after the dissociation of androgen (Fig. 7c).
Intermolecular masking of localization signals can take place when the protein binds to RNA or DNA. An example is virus HIV-1 protein Rev implicated in the HIV-1 mRNA translocation from the nucleus to the cytoplasm. When mRNA binds to the arginine-rich motif of Rev, its NLS looses the ability to interact with importin-beta1. The migration of Rev from the cytoplasm to the nucleus is possible only after the mRNA transported by it is released in the cytoplasm (Fig. 7d) [160].
The yeast transcription factor GAL4 [161, 162] and human chromatin-remodeling factor SRY both have NLS that overlap with their DNA-binding domains. It is clear that when in the nucleus, the binding to the DNA results in dissociation of proteins from the complex with importin-beta1. The physiological significance of this mechanism may be in the Ran-independent release of the transporting protein in the nucleus, when RanGTP is limited or the Ran activity is suppressed by cytosolic Ca2+ [163]. Thus, upon binding to DNA, STAT1 is released from the complex with karyopherin-alpha/importin-beta1 [164].
Transport Regulation by Amplified Binding of Importin/Exportin to the Substrate NLS/NES
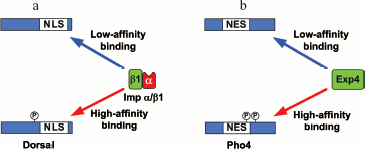
In contrast to the NLS/NES masking, as a result of which interactions of NLS with importin and NES are distorted, there is regulation when the importin/exportin binding to NLS/NES is amplified (Fig. 8).
An example of such regulation is the SV40 virus large T antigen: phosphorylation by kinase CK II at Ser111/112, flanking the NLS in the T antigen, strengthens the affinity of the T antigen to the karyopherin-alpha/importin-beta1 complex 100-fold and stimulates its nuclear import 50-fold [165]. The nuclear import of the Drosophila Dorsal is amplified in a similar way by phosphorylation at Ser312 located close to the N-terminal boundary of the NLS. Such phosphorylation catalyzed by kinase PKA causes higher affinity of Dorsal to the karyopherin-alpha/importin-beta1_complex (Fig. 8a) [166].Fig. 8. Examples of enhancement of NLS/NES (modified from [147]). a) Phosphorylation of Dorsal at Ser312 in response to stimulation of signal pathways enhances the affinity of the karyopherin-alpha/importin-beta1 complex to NLS and stimulates manifold Dorsal nuclear import. b) Phosphorylation of Pho at Ser114/128 enhances the RanGTP-dependent interaction of NES with exportin 4 and leads to its nuclear export. The details of the scheme are given in the text.
Phosphorylation may also enhance nuclear export in a similar manner. Phosphorylation of Pho4 at Ser114 and Ser128 raises its affinity to exportin-4 and stimulates nuclear export (Fig. 8b) [39].
Transport Regulation by Retention in the Cytoplasm or Nucleus
Another mechanism of nucleocytoplasmic transport regulation is realized through the binding of the NLS/NES-containing protein to the specific cytoplasmic or nuclear factors that retain proteins in the cytoplasm or nucleus.
For example, the glucocorticoid receptor is retained in the cytoplasm through complexation with Hsp90 in the absence of the ligand. When binding to the hormone (ligand), the glucocorticoid receptor is dissociated from Hsp90 and imported into the nucleus by the NLS-dependent mechanism [167]. Similarly, the tumor repressor p53 is retained in the cytoplasm by protein Parc (Parkin-like ubiquitin ligase). Suppression of the expression of protein Parc in the model system in the absence of stress actions led to nuclear import of p53 and activation of p53-dependent apoptosis [168].
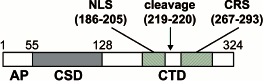
The NLS is more or less in the center of the molecule in the multifunctional nucleocytoplasmic DNA/RNA-binding protein YB-1. In this protein, closer to its C-terminus, there is an amino acid sequence that is responsible for its cytoplasmic localization, i.e. the cytoplasmic retention signal (CRS). The CRS dominates over the NLS and retains the protein in the cytoplasm [169]. It was supposed that the CRS provides for the cytoplasmic localization of YB-1 because of the interaction with the ligand (mRNA/proteins) that is retained in the cytoplasm. But it cannot be excluded that the CRS masks the NLS from the binding to transport factors. It was demonstrated that upon DNA-damage stress in the cell under the action of the proteasome the C-terminal portion of YB-1 is split off together with the retention signal in the cytoplasm, after which the truncated protein is imported to the nucleus (Fig. 9) [170]. A similar processing and nuclear transport of the truncated YB-1 was also observed in endothelial cells treated with thrombin [171]. YB-1 binds to karyopherin-beta2, but this complex is disrupted in the presence of RanGTP [172]. This is an evidence for the classical Ran-dependent nuclear transport of YB-1.
The HIV-1 transactivator Tat [173], angiogenin [174], and transcription factor IFI16 [175] are examples of proteins whose NLS-dependent nuclear transport is not mediated by additional factors and which are retained in the nucleus, at least in part, due to the interaction of NLS with components of the nucleus. In the case of angiogenin, that is rather small and can be imported to the nucleus by diffusion, NLS is not implicated in the interaction with importins, but takes part in interactions with nucleus components, thus inhibiting diffusion into the cytoplasm [174]. Nuclear or cytoplasmic retention can be regulated by phosphorylation. For example, the nuclear retention of IFI16 is enhanced after phosphorylation of NLS by kinase CK II [175]. The Tat protein NLS is also involved in the interaction with components of the nucleus, which retains Tat in the nucleus. Interestingly, the Tat NLS is also implicated in the Tat binding to the cytoplasmic retention factor, and the disruption of this complex (as well as the following nuclear import) requires ATP and GTP hydrolysis [173].Fig. 9. Structure and localization of YB-1 signal sequences (modified from [170]). YB-1 domains: AD, N-terminal alanine/proline-rich domain; CSD, cold-shock domain; CTD, C-terminal domain. Nuclear localization and cytoplasmic retention signals (NLS and CRS) are shown. The arrow indicates the splitting site of YB-1 by the 20S proteasome. The details of the scheme are given in the text.
It is clear that interaction with retention factors in the nucleus or the cytoplasm plays an important role in the regulation of nucleocytoplasmic distribution of proteins in response to stimulation of different signal pathways in the cell.
Transport Regulation by Cotransport and Changing the Cargo-Binding Properties of Karyopherin
As mentioned above, different karyopherins are implicated in protein import or export and only some of them are implicated in transport in both directions [13, 20, 176]. The small family of karyopherins provides for the shuttling of at least 1500 proteins. This shows that every karyopherin can bind a great number of various substrates. Currently the presence of specific substrate-karyopherin pairs is known only for about 40 substrates, and what is more, not all NLS/NES are recognized or identified accurately [13, 20, 176]. For some proteins, it has been demonstrated that they are transported in complex with other proteins. Such a complex is formed just after the synthesis of the protein prior to its binding to karyopherin [21, 22, 25, 177-179]. A simultaneous binding of karyopherins to a number of different substrates is also possible, which may confirm their cotranslocation by one karyopherin.
Sometimes an exact ratio of proteins or their simultaneous delivery to a certain site of the nucleus or cytoplasm is required for regulation of cell processes. Such a coordinated regulation of transport can be achieved due to a synchronous transition of two or more different proteins on the same karyopherin. This type of transport was described for histones H2A and H2B and their chaperone Nap1p by karyopherin Kap114p [21, 22]. It has been established that karyopherin Kap114p has overlapping substrate-binding sites for four different substrates: histones H2A and H2B, Sua7p and Nap1p, as well as for transport partners RanGTP and nucleoporins [180]. Inasmuch as complexes of karyopherin Kap114p containing both histones H2A/H2B and Sua7p could not be obtained, the authors suggested that these proteins have overlapping binding sites on karyopherin. On the other hand, Sua7p can bind to karyopherin simultaneously with Nap1p. This means that their sites do not overlap. At the same time, stable complexes of Kap114p with concurrent histones H2A/H2B and Sua7p were prepared in the presence of Kap114p/Nap1p/Sua7p as a primer for assembling. It is assumed that histones H2A/H2B can be involved in the complex due to their interaction with Nap1p [180]. It is not excluded that owing to some conformational rearrangements of karyopherin upon binding of one substrate, another substrate-binding site can be formed, probably with the participation of the first substrate.
Transport Regulation by Changes in the Variety of Importins and Exportins
The wealth of different factors recognizing signal sequences in transport proteins (Table 1) shows that transport of specific cell proteins can be regulated selectively. Since the factors have different substrate specificity, it is logical to suggest that transport of some proteins can be regulated due to changing the level of expression of the corresponding transport factor (importin or exportin).
The analysis of level of karyopherins-alpha mRNA expression in different tissues showed that the expression of karyopherin-alpha genes could be tissue-specific: the content of importin-alpha1 mRNA was low in various tissues, while content of importins-alpha4, -alpha5, and -alpha6 mRNAs was very high in testicles and lower in spleen [181, 182]. The analysis of the amount of proteins confirmed the data on tissue-specificity of karyopherins-alpha. Thus, human importin-alpha., which constitutes more than 1% of the proteins in skeletal muscles, is essentially absent in heart, spleen, and kidney [183]. A great amount of importin-alpha2 was found in heart, testicles, skeletal muscles, and ovaries, and the highest amount of importin-alpha3 was revealed in ovary [184]. Recently it has been demonstrated that the Drosophila heat-shock factor (dHSF), which is carried specifically by importin-alpha3, is imported into the nucleus only at late stages of development when importin-alpha3 expression begins [185].
There is a rather small amount of data on karyopherins-beta. It was shown that the content of Crm1 mRNA [186], transportin-1 mRNA [187], and importin-alpha3_mRNA [188] was practically constant in different tissues during maturation of Drosophila, whereas CAS mRNA was found in different tissues only at definite states of development [189].
Transport Regulation by Changing the Variety of Nucleoporins
Though NPC has no motor proteins and in effect is a passive partner in the transport, its content may vary. It is known that nucleoporins have different specificity (affinity) to various importins and exportins [190]. This suggests that a change in the expression of various nucleoporins can modulate the efficiency of translocation of some proteins through the NPC. For example, the transport of proteins with classical NLS, except for the ribosomal protein L23a, is selectively impaired in cells deficient in Nup98 [191].
There are also descriptions of tissue-specific differences in NPC. So the amount of the protein Npap60 in NPC, that facilitates nuclear import of some factors and complexes, was an order of magnitude higher in testis than in other tissues [192]. The nucleoporin Nup BS-63, that is a splice variant of Nup358, was also detected only in germ cells of testis. It was demonstrated that it can directly interact with Ran, importin-beta2, and the chromatin-remodeling factor aF10. Factors such as aF10 may access the nucleus through direct interaction with Nup BS-63 [193]. The functional significance of individual nucleoporins was also shown on the Drosophila Nup154 (which is a homolog of Nup155)--the normal gamete development was impaired in its absence [194].
An interesting result was obtained in vitro: a decrease in the concentration of Ca2+ impeded significantly the protein nuclear import even by diffusion. Apparently, a decreased concentration of Ca2+ results in conformational changes in NPC, which can be the reason for the hindered transport [195]. This effect is most likely caused by conformational changes in the integral NPC transmembrane nucleoporin gp210 that has a number of Ca2+-binding domains [196].
The first data demonstrating the possibility of protein shuttling between the cytoplasm and nucleus were obtained in the 1950s [197]. About thirty years later, the first shuttling protein, nucleoline, was identified [198]. During recent years, a real breakthrough has occurred in studying the nucleocytoplasmic transport. The discovery of Ran GTPase in 1993 was the beginning [61, 62], followed by an avalanche-like discovery of many other factors participating in nucleocytoplasmic transport. At present most of the key components of this process are known, which has permitted developing a common “rough” model of nucleocytoplasmic transport (Fig. 4). It should be realized that the nucleocytoplasmic transport is one of the most dynamically developing spheres of cell and molecular biology; therefore, the above model is far from being complete and the only one. Every year there appear data on new participants of the “classical” Ran-dependent transport as well as proofs that it can have an alternative pathway.
The current list of proteins shuttling between the cytoplasm and the nucleus includes transport receptors and adaptors, receptors of steroid hormones, transcription factors, cell cycle regulators, and a large number of RNA-binding proteins [5, 199-204]. An impression is created that just these nucleocytoplasmic shuttling proteins are key units in transmitting information between the two main compartments of the cell. The nucleocytoplasmic distribution of such proteins calls for steady regulation and coordination for normal cell functioning. Most frequently, it is found that the reasons of cancer degeneration of cells include distortions in the distribution of proteins between the nucleus and cytoplasm [147]. This may explain the attention paid lately to the studies of the nucleocytoplasmic transport including the search for its alternative mechanisms and detailed analyses of the already known regulation mechanisms of protein distribution between the cytoplasm and nucleus.
In this review we have tried to elucidate the basic aspects of nucleocytoplasmic transport of proteins (pore complex structure, nuclear localization signals of proteins and signals of export from the nucleus, model of the classical Ran-dependent mechanism of transport) as well as to attract readers' attention to alternative transport mechanisms and systematize the key regulation mechanisms of protein transport.
The authors are grateful to E. V. Serebrova for her help in writing this paper.
The work is supported by the Russian Foundation for Basic Research grant No. 07-04-00403-a and grants of the Presidium of the Russian Academy of Sciences within the programs “Molecular and cell biology” and “Fundamental science to medicine”.
REFERENCES
1.Rout, M. P., and Aitchison, J. D. (2000) Essays
Biochem., 36, 75-88.
2.Franke, W. W., and Scheer, U. (1974) Symp. Soc.
Exp. Biol., 249-282.
3.King, M. C., Lusk, C. P., and Blobel, G. (2006)
Nature, 442, 1003-1007.
4.Suntharalingam, M., and Wente, S. R. (2003) Dev.
Cell, 4, 775-789.
5.Gorlich, D., and Kutay, U. (1999) Annu. Rev.
Cell. Dev. Biol., 15, 607-660.
6.Keminer, O., and Peters, R. (1999) Biophys.
J., 77, 217-228.
7.Paine, P. L., Moore, L. C., and Horowitz, S. B.
(1975) Nature, 254, 109-114.
8.Pante, N., and Kann, M. (2002) Mol. Biol.
Cell, 13, 425-434.
9.Kiseleva, E. V., Goldberg, M. W., Allen, T. D., and
Akey, C. W. (1998) J. Cell. Sci., 111, 223-236.
10.Ribbeck, K., and Gorlich, D. (2001) Embo
J., 20, 1320-1330.
11.Siebrasse, J. P., and Peters, R. (2002) EMBO
Rep., 3, 887-892.
12.Breeuwer, M., and Goldfarb, D. S. (1990)
Cell, 60, 999-1008.
13.Mosammaparast, N., and Pemberton, L. F. (2004)
Trends Cell. Biol., 14, 547-556.
14.Pemberton, L. F., and Paschal, B. M. (2005)
Traffic, 6, 187-198.
15.Jans, D. A., Xiao, C. Y., and Lam, M. H. (2000)
Bioessays, 22, 532-544.
16.Goldfarb, D. S., Corbett, A. H., Mason, D. A.,
Harreman, M. T., and Adam, S. A. (2004) Trends Cell. Biol.,
14, 505-514.
17.Hall, M. N., Craik, C., and Hiraoka, Y. (1990)
Proc. Natl. Acad. Sci. USA, 87, 6954-6958.
18.Makkerh, J. P., Dingwall, C., and Laskey, R. A.
(1996) Curr. Biol., 6, 1025-1027.
19.Fried, H., and Kutay, U. (2003) Cell. Mol.
Life Sci., 60, 1659-1688.
20.Weis, K. (2003) Cell, 112,
441-451.
21.Mosammaparast, N., Guo, Y., Shabanowitz, J.,
Hunt, D. F., and Pemberton, L. F. (2002) J. Biol. Chem.,
277, 862-868.
22.Mosammaparast, N., Jackson, K. R., Guo, Y.,
Brame, C. J., Shabanowitz, J., Hunt, D. F., and Pemberton, L. F. (2001)
J. Cell Biol., 153, 251-262.
23.Muhlhausser, P., Muller, E. C., Otto, A., and
Kutay, U. (2001) EMBO Rep., 2, 690-696.
24.Jakel, S., and Gorlich, D. (1998) Embo J.,
17, 4491-4502.
25.Leslie, D. M., Zhang, W., Timney, B. L., Chait,
B. T., Rout, M. P., Wozniak, R. W., and Aitchison, J. D. (2004) Mol.
Cell. Biol., 24, 8487-8503.
26.Senger, B., Simos, G., Bischoff, F. R.,
Podtelejnikov, A., Mann, M., and Hurt, E. (1998) Embo J.,
17, 2196-2207.
27.Pollard, V. W., Michael, W. M., Nakielny, S.,
Siomi, M. C., Wang, F., and Dreyfuss, G. (1996) Cell, 86,
985-994.
28.Rosenblum, J. S., Pemberton, L. F., Bonifaci, N.,
and Blobel, G. (1998) J. Cell Biol., 143, 887-899.
29.Cingolani, G., Petosa, C., Weis, K., and Muller,
C. W. (1999) Nature, 399, 221-229.
30.Cingolani, G., Bednenko, J., Gillespie, M. T.,
and Gerace, L. (2002) Mol. Cell., 10, 1345-1353.
31.Lee, S. J., Sekimoto, T., Yamashita, E., Nagoshi,
E., Nakagawa, A., Imamoto, N., Yoshimura, M., Sakai, H., Chong, K. T.,
Tsukihara, T., and Yoneda, Y. (2003) Science, 302,
1571-1575.
32.Fischer, U., Huber, J., Boelens, W. C., Mattaj,
I. W., and Luhrmann, R. (1995) Cell, 82, 475-483.
33.La Cour, T., Gupta, R., Rapacki, K., Skriver, K.,
Poulsen, F. M., and Brunak, S. (2003) Nucleic Acids Res.,
31, 393-396.
34.Wen, W., Meinkoth, J. L., Tsien, R. Y., and
Taylor, S. S. (1995) Cell, 82, 463-473.
35.Johnson, A. W., Lund, E., and Dahlberg, J. (2002)
Trends Biochem. Sci., 27, 580-585.
36.Ohno, M., Segref, A., Bachi, A., Wilm, M., and
Mattaj, I. W. (2000) Cell, 101, 187-198.
37.Paraskeva, E., Izaurralde, E., Bischoff, F. R.,
Huber, J., Kutay, U., Hartmann, E., Luhrmann, R., and Gorlich, D.
(1999) J. Cell. Biol., 145, 255-264.
38.Hood, J. K., and Silver, P. A. (1999) Curr.
Opin. Cell Biol., 11, 241-247.
39.Komeili, A., and O'Shea, E. K. (1999)
Science, 284, 977-980.
40.Kaffman, A., and O'Shea, E. K. (1999) Annu.
Rev. Cell Dev. Biol., 15, 291-339.
41.Lei, E. P., and Silver, P. A. (2002) Dev.
Cell, 2, 261-272.
42.Kim, V. N. (2004) Trends Cell Biol.,
14, 156-159.
43.Zeng, Y., and Cullen, B. R. (2004) Nucleic
Acids Res., 32, 4776-4785.
44.Akey, C. W., and Radermacher, M. (1993) J.
Cell Biol., 122, 1-19.
45.Hinshaw, J. E., Carragher, B. O., and Milligan,
R. A. (1992) Cell, 69, 1133-1141.
46.Stoffler, D., Fahrenkrog, B., and Aebi, U. (1999)
Curr. Opin. Cell Biol., 11, 391-401.
47.Yang, Q., Rout, M. P., and Akey, C. W. (1998)
Mol. Cell., 1, 223-234.
48.Cronshaw, J. M., Krutchinsky, A. N., Zhang, W.,
Chait, B. T., and Matunis, M. J. (2002) J. Cell Biol.,
158, 915-927.
49.Reichelt, R., Holzenburg, A., Buhle, E. L., Jr.,
Jarnik, M., Engel, A., and Aebi, U. (1990) J. Cell Biol.,
110, 883-894.
50.Marelli, M., Dilworth, D. J., Wozniak, R. W., and
Aitchison, J. D. (2001) Biochem. Cell Biol., 79,
603-612.
51.Rout, M. P., and Aitchison, J. D. (2001) J.
Biol. Chem., 276, 16593-16596.
52.Devos, D., Dokudovskaya, S., Alber, F., Williams,
R., Chait, B. T., Sali, A., and Rout, M. P. (2004) PLoS Biol.,
2, e380.
53.Weirich, C. S., Erzberger, J. P., Berger, J. M.,
and Weis, K. (2004) Mol. Cell., 16, 749-760.
54.Ben-Efraim, I., and Gerace, L. (2001) J. Cell
Biol., 152, 411-417.
55.Pyhtila, B., and Rexach, M. (2003) J. Biol.
Chem., 278, 42699-42709.
56.Shah, S., and Forbes, D. J. (1998) Curr.
Biol., 8, 1376-1386.
57.Shah, S., Tugendreich, S., and Forbes, D. (1998)
J. Cell Biol., 141, 31-49.
58.Peters, R. (2005) Traffic, 6,
421-427.
59.Corbett, A. H., Koepp, D. M., Schlenstedt, G.,
Lee, M. S., Hopper, A. K., and Silver, P. A. (1995) J. Cell
Biol., 130, 1017-1026.
60.Schlenstedt, G., Wong, D. H., Koepp, D. M., and
Silver, P. A. (1995) Embo J., 14, 5367-5378.
61.Melchior, F., Paschal, B., Evans, J., and Gerace,
L. (1993) J. Cell Biol., 123, 1649-1659.
62.Moore, M. S., and Blobel, G. (1993)
Nature, 365, 661-663.
63.Stochaj, U., and Rother, K. L. (1999)
Bioessays, 21, 579-589.
64.Moore, M. S. (1998) J. Biol. Chem.,
273, 22857-22860.
65.Bourne, H. R., Sanders, D. A., and McCormick, F.
(1991) Nature, 349, 117-127.
66.Bischoff, F. R., Krebber, H., Smirnova, E., Dong,
W., and Ponstingl, H. (1995) Embo J., 14, 705-715.
67.Hopper, A. K., Traglia, H. M., and Dunst, R. W.
(1990) J. Cell Biol., 111, 309-321.
68.Mahajan, R., Delphin, C., Guan, T., Gerace, L.,
and Melchior, F. (1997) Cell, 88, 97-107.
69.Kunzler, M., Gerstberger, T., Stutz, F.,
Bischoff, F. R., and Hurt, E. (2000) Mol. Cell. Biol.,
20, 4295-4308.
70.Nemergut, M. E., Mizzen, C. A., Stukenberg, T.,
Allis, C. D., and Macara, I. G. (2001) Science, 292,
1540-1543.
71.Bischoff, F. R., and Ponstingl, H. (1991)
Nature, 354, 80-82.
72.Welch, K., Franke, J., Kohler, M., and Macara, I.
G. (1999) Mol. Cell. Biol., 19, 8400-8411.
73.Nemergut, M. E., Lindsay, M. E., Brownawell, A.
M., and Macara, I. G. (2002) J. Biol. Chem., 277,
17385-17388.
74.Adam, E. J., and Adam, S. A. (1994) J. Cell
Biol., 125, 547-555.
75.Gorlich, D., Prehn, S., Laskey, R. A., and
Hartmann, E. (1994) Cell, 79, 767-778.
76.Gorlich, D., Vogel, F., Mills, A. D., Hartmann,
E., and Laskey, R. A. (1995) Nature, 377, 246-248.
77.Gorlich, D., Kostka, S., Kraft, R., Dingwall, C.,
Laskey, R. A., Hartmann, E., and Prehn, S. (1995) Curr. Biol.,
5, 383-392.
78.Wu, J., Matunis, M. J., Kraemer, D., Blobel, G.,
and Coutavas, E. (1995) J. Biol. Chem., 270,
14209-14213.
79.Yokoyama, N., Hayashi, N., Seki, T., Pante, N.,
Ohba, T., Nishii, K., Kuma, K., Hayashida, T., Miyata, T., Aebi, U., et
al. (1995) Nature, 376, 184-188.
80.Delphin, C., Guan, T., Melchior, F., and Gerace,
L. (1997) Mol. Biol. Cell., 8, 2379-2390.
81.Floer, M., and Blobel, G. (1996) J. Biol.
Chem., 271, 5313-5316.
82.Lounsbury, K. M., and Macara, I. G. (1997) J.
Biol. Chem., 272, 551-555.
83.Oki, M., and Nishimoto, T. (2000) J. Biol.
Chem., 275, 32894-32900.
84.Pante, N., and Aebi, U. (1996) Science,
273, 1729-1732.
85.Moore, M. S., and Blobel, G. (1994) Proc.
Natl. Acad. Sci. USA, 91, 10212-10216.
86.Nehrbass, U., and Blobel, G. (1996)
Science, 272, 120-122.
87.Paschal, B. M., and Gerace, L. (1995) J. Cell
Biol., 129, 925-937.
88.Stewart, M., Kent, H. M., and McCoy, A. J. (1998)
J. Mol. Biol., 277, 635-646.
89.Gorlich, D., Pante, N., Kutay, U., Aebi, U., and
Bischoff, F. R. (1996) Embo J., 15, 5584-5594.
90.Rexach, M., and Blobel, G. (1995) Cell,
83, 683-692.
91.Chi, N. C., Adam, E. J., and Adam, S. A. (1997)
J. Biol. Chem., 272, 6818-6822.
92.Mingot, J. M., Kostka, S., Kraft, R., Hartmann,
E., and Gorlich, D. (2001) Embo J., 20, 3685-3694.
93.Hood, J. K., and Silver, P. A. (1998) J. Biol.
Chem., 273, 35142-35146.
94.Kunzler, M., and Hurt, E. C. (1998) FEBS
Lett., 433, 185-190.
95.Lindsay, M. E., Holaska, J. M., Welch, K.,
Paschal, B. M., and Macara, I. G. (2001) J. Cell Biol.,
153, 1391-1402.
96.Bischoff, F. R., Klebe, C., Kretschmer, J.,
Wittinghofer, A., and Ponstingl, H. (1994) Proc. Natl. Acad. Sci.
USA, 91, 2587-2591.
97.Dingwall, C., Kandels-Lewis, S., and Seraphin, B.
(1995) Proc. Natl. Acad. Sci. USA, 92, 7525-7529.
98.Gorlich, D., Dabrowski, M., Bischoff, F. R.,
Kutay, U., Bork, P., Hartmann, E., Prehn, S., and Izaurralde, E. (1997)
J. Cell Biol., 138, 65-80.
99.Hartmann, E., and Gorlich, D. (1995) Trends
Cell Biol., 5, 192-193.
100.Mueller, L., Cordes, V. C., Bischoff, F. R.,
and Ponstingl, H. (1998) FEBS Lett., 427, 330-336.
101.Deane, R., Schafer, W., Zimmermann, H. P.,
Mueller, L., Gorlich, D., Prehn, S., Ponstingl, H., and Bischoff, F. R.
(1997) Mol. Cell. Biol., 17, 5087-5096.
102.Macara, I. G. (2001) Microbiol. Mol. Biol.
Rev., 65, 570-594.
103.Soullam, B., and Worman, H. J. (1995) J.
Cell Biol., 130, 15-27.
104.Kutay, U., and Muhlhausser, P. (2006)
Nature, 442, 991-992.
105.Ohba, T., Schirmer, E. C., Nishimoto, T., and
Gerace, L. (2004) J. Cell Biol., 167, 1051-1062.
106.Horton, P., and Nakai, K. (1997) Proc. Int.
Conf. Intel. Syst. Mol. Biol., 5, 147-152.
107.Shulga, N., and Goldfarb, D. S. (2003) Mol.
Cell. Biol., 23, 534-542.
108.Picard, D., Kumar, V., Chambon, P., and
Yamamoto, K. R. (1990) Cell Regul., 1, 291-299.
109.Picard, D., and Yamamoto, K. R. (1987) Embo
J., 6, 3333-3340.
110.Poukka, H., Karvonen, U., Yoshikawa, N.,
Tanaka, H., Palvimo, J. J., and Janne, O. A. (2000) J. Cell
Sci., 113 (Pt. 17), 2991-3001.
111.Savory, J. G., Hsu, B., Laquian, I. R., Giffin,
W., Reich, T., Hache, R. J., and Lefebvre, Y. A. (1999) Mol. Cell.
Biol., 19, 1025-1037.
112.Simental, J. A., Sar, M., Lane, M. V., French,
F. S., and Wilson, E. M. (1991) J. Biol. Chem., 266,
510-518.
113.Ylikomi, T., Bocquel, M. T., Berry, M.,
Gronemeyer, H., and Chambon, P. (1992) Embo J., 11,
3681-3694.
114.Zhou, Z. X., Sar, M., Simental, J. A., Lane, M.
V., and Wilson, E. M. (1994) J. Biol. Chem., 269,
13115-13123.
115.Freedman, N. D., and Yamamoto, K. R. (2004)
Mol. Biol. Cell., 15, 2276-2286.
116.Davies, T. H., Ning, Y. M., and Sanchez, E. R.
(2002) J. Biol. Chem., 277, 4597-4600.
117.Pratt, W. B., and Toft, D. O. (1997) Endocr.
Rev., 18, 306-360.
118.Shank, L. C., and Paschal, B. M. (2005)
Crit. Rev. Eukaryot. Gene Exp., 15, 49-73.
119.Black, B. E., Holaska, J. M., Rastinejad, F.,
and Paschal, B. M. (2001) Curr. Biol., 11, 1749-1758.
120.Saporita, A. J., Zhang, Q., Navai, N., Dincer,
Z., Hahn, J., Cai, X., and Wang, Z. (2003) J. Biol. Chem.,
278, 41998-42005.
121.Katagiri, Y., Takeda, K., Yu, Z. X., Ferrans,
V. J., Ozato, K., and Guroff, G. (2000) Nat. Cell Biol.,
2, 435-440.
122.Itoh, M., Adachi, M., Yasui, H., Takekawa, M.,
Tanaka, H., and Imai, K. (2002) Mol. Endocrinol., 16,
2382-2392.
123.Liu, J., and DeFranco, D. B. (2000) Mol.
Endocrinol., 14, 40-51.
124.Amazit, L., Alj, Y., Tyagi, R. K., Chauchereau,
A., Loosfelt, H., Pichon, C., Pantel, J., Foulon-Guinchard, E.,
Leclerc, P., Milgrom, E., and Guiochon-Mantel, A. (2003) J. Biol.
Chem., 278, 32195-32203.
125.Kino, T., Souvatzoglou, E., de Martino, M. U.,
Tsopanomihalu, M., Wan, Y., and Chrousos, G. P. (2003) J. Biol.
Chem., 278, 25651-25656.
126.Holaska, J. M., Black, B. E., Rastinejad, F.,
and Paschal, B. M. (2002) Mol. Cell. Biol., 22,
6286-6297.
127.Fagotto, F., Gluck, U., and Gumbiner, B. M.
(1998) Curr. Biol., 8, 181-190.
128.Holaska, J. M., Black, B. E., Love, D. C.,
Hanover, J. A., Leszyk, J., and Paschal, B. M. (2001) J. Cell
Biol., 152, 127-140.
129.Kehlenbach, R. H., Dickmanns, A., and Gerace,
L. (1998) J. Cell Biol., 141, 863-874.
130.Kose, S., Imamoto, N., Tachibana, T.,
Shimamoto, T., and Yoneda, Y. (1997) J. Cell Biol., 139,
841-849.
131.Kutay, U., Lipowsky, G., Izaurralde, E.,
Bischoff, F. R., Schwarzmaier, P., Hartmann, E., and Gorlich, D. (1998)
Mol. Cell., 1, 359-369.
132.Marg, A., Shan, Y., Meyer, T., Meissner, T.,
Brandenburg, M., and Vinkemeier, U. (2004) J. Cell Biol.,
165, 823-833.
133.Matsubayashi, Y., Fukuda, M., and Nishida, E.
(2001) J. Biol. Chem., 276, 41755-41760.
134.Michael, W. M., Eder, P. S., and Dreyfuss, G.
(1997) Embo J., 16, 3587-3598.
135.Miyamoto, Y., Saiwaki, T., Yamashita, J.,
Yasuda, Y., Kotera, I., Shibata, S., Shigeta, M., Hiraoka, Y.,
Haraguchi, T., and Yoneda, Y. (2004) J. Cell Biol., 165,
617-623.
136.Nakielny, S., and Dreyfuss, G. (1998) Curr.
Biol., 8, 89-95.
137.Pruschy, M., Ju, Y., Spitz, L., Carafoli, E.,
and Goldfarb, D. S. (1994) J. Cell Biol., 127,
1527-1536.
138.Schmalz, D., Hucho, F., and Buchner, K. (1998)
J. Cell Sci., 111 (Pt. 13), 1823-1830.
139.Yen, Y. M., Roberts, P. M., and Johnson, R. C.
(2001) Traffic, 2, 449-464.
140.Zhang, T., Delestienne, N., Huez, G., Kruys,
V., and Gueydan, C. (2005) J. Cell Sci., 118,
5453-5463.
141.Fornerod, M., van Deursen, J., van Baal, S.,
Reynolds, A., Davis, D., Murti, K. G., Fransen, J., and Grosveld, G.
(1997) Embo J., 16, 807-816.
142.Kehlenbach, R. H., Dickmanns, A., Kehlenbach,
A., Guan, T., and Gerace, L. (1999) J. Cell Biol., 145,
645-657.
143.Hutten, S., and Kehlenbach, R. H. (2006)
Mol. Cell. Biol., 26, 6772-6785.
144.Sweitzer, T. D., and Hanover, J. A. (1996)
Proc. Natl. Acad. Sci. USA, 93, 14574-14579.
145.Meyer, T., and Vinkemeier, U. (2004) Eur. J.
Biochem., 271, 4606-4612.
146.Hogarth, C., Itman, C., Jans, D. A., and
Loveland, K. L. (2005) Bioessays, 27, 1011-1025.
147.Poon, I. K., and Jans, D. A. (2005)
Traffic, 6, 173-186.
148.Riviere, Y., Blank, V., Kourilsky, P., and
Israel, A. (1991) Nature, 350, 625-626.
149.Craig, E., Zhang, Z. K., Davies, K. P., and
Kalpana, G. V. (2002) Embo J., 21, 31-42.
150.Beals, C. R., Sheridan, C. M., Turck, C. W.,
Gardner, P., and Crabtree, G. R. (1997) Science, 275,
1930-1934.
151.Kaffman, A., Rank, N. M., and O'Shea, E. K.
(1998) Genes Dev., 12, 2673-2683.
152.Ferrigno, P., Posas, F., Koepp, D., Saito, H.,
and Silver, P. A. (1998) Embo J., 17, 5606-5614.
153.Kuge, S., Arita, M., Murayama, A., Maeta, K.,
Izawa, S., Inoue, Y., and Nomoto, A. (2001) Mol. Cell. Biol.,
21, 6139-6150.
154.Kudo, N., Taoka, H., Toda, T., Yoshida, M., and
Horinouchi, S. (1999) J. Biol. Chem., 274,
15151-15158.
155.Zhu, J., and McKeon, F. (1999) Nature,
398, 256-260.
156.Beg, A. A., Ruben, S. M., Scheinman, R. I.,
Haskill, S., Rosen, C. A., and Baldwin, A. S., Jr. (1992) Genes
Dev., 6, 1899-1913.
157.Traenckner, E. B., Wilk, S., and Baeuerle, P.
A. (1994) Embo J., 13, 5433-5441.
158.Li, S., Ku, C. Y., Farmer, A. A., Cong, Y. S.,
Chen, C. F., and Lee, W. H. (1998) J. Biol. Chem., 273,
6183-6189.
159.Stommel, J. M., Marchenko, N. D., Jimenez, G.
S., Moll, U. M., Hope, T. J., and Wahl, G. M. (1999) Embo J.,
18, 1660-1672.
160.Fineberg, K., Fineberg, T., Graessmann, A.,
Luedtke, N. W., Tor, Y., Lixin, R., Jans, D. A., and Loyter, A. (2003)
Biochemistry, 42, 2625-2633.
161.Chan, C. K., and Jans, D. A. (1999) FEBS
Lett., 462, 221-224.
162.Chan, C. K., and Jans, D. A. (2001) Gene
Ther., 8, 166-171.
163.Argentaro, A., Sim, H., Kelly, S., Preiss, S.,
Clayton, A., Jans, D. A., and Harley, V. R. (2003) J. Biol.
Chem., 278, 33839-33847.
164.McBride, K. M., Banninger, G., McDonald, C.,
and Reich, N. C. (2002) Embo J., 21, 1754-1763.
165.Hubner, S., Xiao, C. Y., and Jans, D. A. (1997)
J. Biol. Chem., 272, 17191-17195.
166.Briggs, L. J., Stein, D., Goltz, J., Corrigan,
V. C., Efthymiadis, A., Hubner, S., and Jans, D. A. (1998) J. Biol.
Chem., 273, 22745-22752.
167.Tago, K., Tsukahara, F., Naruse, M., Yoshioka,
T., and Takano, K. (2004) Mol. Cell Endocrinol., 213,
131-138.
168.Nikolaev, A. Y., Li, M., Puskas, N., Qin, J.,
and Gu, W. (2003) Cell, 112, 29-40.
169.Bader, A. G., and Vogt, P. K. (2005) Mol.
Cell. Biol., 25, 2095-2106.
170.Sorokin, A. V., Selyutina, A. A., Skabkin, M.
A., Guryanov, S. G., Nazimov, I. V., Richard, C., Th'ng, J., Yau, J.,
Sorensen, P. H., Ovchinnikov, L. P., and Evdokimova, V. (2005) Embo
J., 24, 3602-3612.
171.Stenina, O. I., Poptic, E. J., and DiCorleto,
P. E. (2000) J. Clin. Invest., 106, 579-587.
172.Lee, B. J., Cansizoglu, A. E., Suel, K. E.,
Louis, T. H., Zhang, Z., and Chook, Y. M. (2006) Cell,
126, 543-558.
173.Efthymiadis, A., Briggs, L. J., and Jans, D. A.
(1998) J. Biol. Chem., 273, 1623-1628.
174.Lixin, R., Efthymiadis, A., Henderson, B., and
Jans, D. A. (2001) Biochem. Biophys. Res. Commun., 284,
185-193.
175.Briggs, L. J., Johnstone, R. W., Elliot, R. M.,
Xiao, C. Y., Dawson, M., Trapani, J. A., and Jans, D. A. (2001)
Biochem. J., 353, 69-77.
176.Harel, A., and Forbes, D. J. (2004) Mol.
Cell., 16, 319-330.
177.Mosammaparast, N., Ewart, C. S., and Pemberton,
L. F. (2002) Embo J., 21, 6527-6538.
178.Titov, A. A., and Blobel, G. (1999) J. Cell
Biol., 147, 235-246.
179.Yoshida, K., and Blobel, G. (2001) J. Cell
Biol., 152, 729-740.
180.Hodges, J. L., Leslie, J. H., Mosammaparast,
N., Guo, Y., Shabanowitz, J., Hunt, D. F., and Pemberton, L. F. (2005)
Mol. Biol. Cell, 16, 3200-3210.
181.Nadler, S. G., Tritschler, D., Haffar, O. K.,
Blake, J., Bruce, A. G., and Cleaveland, J. S. (1997) J. Biol.
Chem., 272, 4310-4315.
182.Tsuji, L., Takumi, T., Imamoto, N., and Yoneda,
Y. (1997) FEBS Lett., 416, 30-34.
183.Nachury, M. V., Ryder, U. W., Lamond, A. I.,
and Weis, K. (1998) Proc. Natl. Acad. Sci. USA, 95,
582-587.
184.Kohler, M., Ansieau, S., Prehn, S., Leutz, A.,
Haller, H., and Hartmann, E. (1997) FEBS Lett., 417,
104-108.
185.Fang, X., Chen, T., Tran, K., and Parker, C. S.
(2001) Development, 128, 3349-3358.
186.Collier, S., Chan, H. Y., Toda, T., McKimmie,
C., Johnson, G., Adler, P. N., O'Kane, C., and Ashburner, M. (2000)
Genetics, 155, 1799-1807.
187.Norvell, A., Kelley, R. L., Wehr, K., and
Schupbach, T. (1999) Genes Dev., 13, 864-876.
188.Mathe, E., Bates, H., Huikeshoven, H., Deak,
P., Glover, D. M., and Cotterill, S. (2000) Dev. Biol.,
223, 307-322.
189.Tekotte, H., Berdnik, D., Torok, T., Buszczak,
M., Jones, L. M., Cooley, L., Knoblich, J. A., and Davis, I. (2002)
Dev. Biol., 244, 396-406.
190.Allen, N. P., Huang, L., Burlingame, A., and
Rexach, M. (2001) J. Biol. Chem., 276, 29268-29274.
191.Wu, X., Kasper, L. H., Mantcheva, R. T.,
Mantchev, G. T., Springett, M. J., and van Deursen, J. M. (2001)
Proc. Natl. Acad. Sci. USA, 98, 3191-3196.
192.Fan, F., Liu, C. P., Korobova, O., Heyting, C.,
Offenberg, H. H., Trump, G., and Arnheim, N. (1997) Genomics,
40, 444-453.
193.Cai, Y., Gao, Y., Sheng, Q., Miao, S., Cui, X.,
Wang, L., Zong, S., and Koide, S. S. (2002) Mol. Reprod. Dev.,
61, 126-134.
194.Gigliotti, S., Callaini, G., Andone, S.,
Riparbelli, M. G., Pernas-Alonso, R., Hoffmann, G., Graziani, F., and
Malva, C. (1998) J. Cell Biol., 142, 1195-1207.
195.Perez-Terzic, C., Jaconi, M., and Clapham, D.
E. (1997) Bioessays, 19, 787-792.
196.Perez-Terzic, C., Pyle, J., Jaconi, M.,
Stehno-Bittel, L., and Clapham, D. E. (1996) Science,
273, 1875-1877.
197.Goldstein, L. (1958) Exp. Cell Res.,
15, 635-637.
198.Borer, R. A., Lehner, C. F., Eppenberger, H.
M., and Nigg, E. A. (1989) Cell, 56, 379-390.
199.Hache, R. J., Tse, R., Reich, T., Savory, J.
G., and Lefebvre, Y. A. (1999) J. Biol. Chem., 274,
1432-1439.
200.Cartwright, P., and Helin, K. (2000) Cell.
Mol. Life Sci., 57, 1193-1206.
201.Pines, J. (1999) Nat. Cell. Biol.,
1, 73-79.
202.Yang, J., and Kornbluth, S. (1999) Trends
Cell Biol., 9, 207-210.
203.Nakielny, S., and Dreyfuss, G. (1999)
Cell, 99, 677-690.
204.Shyu, A. B., and Wilkinson, M. F. (2000)
Cell, 102, 135-138.