Inactivation of PPX1 and PPN1 Genes Encoding Exopolyphosphatases of Saccharomyces cerevisiae Does not Prevent Utilization of Polyphosphates as Phosphate Reserve
L. P. Lichko*, T. V. Kulakovskaya, E. V. Kulakovskaya, and I. S. Kulaev
Skryabin Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, pr. Nauki 5, 142290 Pushchino, Moscow Region, Russia; fax: (495) 956-3370; E-mail: alla@ibpm.pushchino.ru* To whom correspondence should be addressed.
Received March 13, 2008; Revision received April 10, 2008
Cytosol polyphosphates (polyPs) are the main phosphate (Pi) reserve in the yeast Saccharomyces cerevisiae. In this work, the participation of cytosol polyPs and exopolyphosphatases in maintenance of Pi homeostasis under Pi deficit in the cultivation medium has been studied in different strains of S. cerevisiae. The growth of yeast strains with inactivated genes PPX1 and PPN1 encoding the yeast exopolyphosphatases and a strain with double mutations in these genes in a Pi-deficient medium is not disturbed. All the studied strains are able to maintain relatively constant Pi levels in the cytosol. In Pi-deficient medium, polyP hydrolysis in the cytosol of the parent and PPN1-deficient strains seems to be performed by exopolyphosphatase Ppx1 and proceeds without any change of the spectrum of polyP chain lengths. In the PPX1-deficient strain, long-chain polyPs are depleted first, and only then short-chain polyPs are hydrolyzed. In the double PPX1 and PPN1 mutant having low exopolyphosphatase activity, polyP hydrolysis in the cytosol starts with a notable delay, and about 20% of short-chain polyPs still remain after the polyP hydrolysis in other strains has almost been completed. This fact suggests that S. cerevisiae possesses a system, which makes it possible to compensate for inactivation of the PPX1 and PPN1 genes encoding exopolyphosphatases of the yeast cells.
KEY WORDS: Pi depletion, inorganic polyphosphates, exopolyphosphatase, cytosol, PPX1 and PPN1 mutants, Saccharomyces cerevisiaeDOI: 10.1134/S0006297908090046
Abbreviations: Pi) phosphate; polyPs) polyphosphates.
Inorganic polyphosphates (polyPs) are involved in many functions of
microbial cells [1-4] including
phosphate (Pi) storage in osmotically neutral form.
Limitation of Pi in the growth medium has been studied in
the yeast for a long time [5-7]. After prolonged Pi limitation, the
yeast is shown to utilize almost the complete polyP pool to support its
vitality [8]. There is increasing interest in the
study of mechanisms of Pi homeostasis in yeast cells [8-10]. However, it is still
unclear what enzymes are involved in the consumption of polyP reserve
in the yeast.
At present, only two genes encoding polyP-metabolizing enzymes are identified in Saccharomyces cerevisiae: PPX1 [11] and PPN1 [12, 13]. PPX1 encodes exopolyphosphatase splitting out Pi from the end of the polyP chain [11]. The enzyme encoded by PPN1 was first regarded as endopolyphosphatase splitting long polyP chains to shorter ones [12, 13]. However, this enzyme was shown to have an exopolyphosphatase activity as well [14-16]. The enzyme encoded by the PPX1 gene more effectively hydrolyzes short-chain polyPs, including polyP3, while the enzyme encoded by PPN1 preferentially hydrolyzes long-chain polyPs [16, 17].
The 45-kD exopolyphosphatase encoded by the PPX1 gene is localized in the cytosol, cell envelopes [18], and soluble mitochondrial fraction [17], while the enzymes encoded by the PPN1 gene and having high molecular masses (120-830 kD) are localized in the nuclei, vacuoles, and mitochondrial membranes of S. cerevisiae [19] and appear in the cytosol under PPX1 inactivation [20] and at early growth stages in Pi-rich medium [16]. The cytosol of yeast cell contains a significant amount of polyP and exopolyphosphatases encoded by the PPX1 and PPN1 genes depending on cultivation conditions [19].
In this connection, the objective of the present work was to study the effect of PPX1 and PPN1 inactivation on polyP utilization in the cytosol of S. cerevisiae under Pi limitation in the cultivation medium.
MATERIALS AND METHODS
Strains and culture conditions. Strains of the yeast S. cerevisiae--CRY (parent strain), CRX (a strain with inactivated PPX1 gene), CRN (a strain with inactivated PPN1 gene), and CNX (a strain with inactivated PPX1 and PPN1 genes)--were from the Kornberg's laboratory (Stanford University, USA) [13]. All strains were grown aerobically in a shaker at 30°C in YPD medium (1% yeast extract, 2% peptone, and 2% glucose) containing 5.7 mM orthophosphate (Pi-rich medium). The media (250 ml) were inoculated directly with the cultures picked from YPD agar slants. The cells were grown for 24 h to the stationary growth phase. Then the cells were separated by centrifugation at 5000g for 10 min, washed with sterile distilled water, and re-inoculated into 250 ml of Pi-rich or Pi-deficient YPD medium (2 g wet biomass for each strain). The Pi-deficient medium containing 0.08 mM Pi was prepared as described previously [21]. After cultivation for 1-5 h (exponential growth phase), the cells were centrifuged, washed with distilled water, and used for further analysis.
Preparation and characterization of cytosol. The cytosol fractions were prepared by disruption of spheroplasts in 0.1 M sorbitol followed by centrifugation at 100,000g for 3 h as described earlier [22]. The cytosol preparations had no activity of ATPases, which were sensitive to orthovanadate (inhibitor of plasma membrane ATPase), azide and oligomycin (inhibitors of mitochondrial ATPase), and nitrate (inhibitor of vacuolar ATPase), so they were considered free from contamination with these organelles.
The cytosol fractions were subjected to gel filtration on an FPLC Superose 6 column (Pharmacia, Sweden). The elution buffer was 20 mM Tris-HCl, pH 7.2, containing 0.1% Triton X-100, 2 mM MgSO4, and 100 mM NaCl. The molecular mass marker kit (Pharmacia) included thyroglobulin (669 kD), ferritin (440 kD), beta-amylase (200 kD), aldolase (160 kD), bovine serum albumin (67 kD), ovalbumin (43 kD), carbonic anhydrase (29 kD), and cytochrome c (13 kD).
Other methods. Preparation of spheroplasts, assay of phosphohydrolase activities, and extraction, assay, and electrophoresis of acid-soluble polyP have been described in detail in our previous paper [23]. The cytosol fractions for polyP assays were obtained in the presence of known exopolyphosphatase inhibitors: 4 mg/ml heparin and 20 mM EDTA [17]. Then equal volumes of 1 N HClO4 were added to the fractions and suspensions were centrifuged at 5000g for 20 min. The cytosol fractions for exopolyphosphatase assays were obtained without addition of heparin and EDTA. All experiments have been performed at least three times, and the mean values are presented.
RESULTS
No difference in the growth of all yeast strains was observed in Pi-deficient medium. Growth rates and biomass amounts were the same as in case of Pi-rich medium (not illustrated).
Exopolyphosphatase activities in the cytosol preparations of all strains were assayed after 5-h growth (exponential growth phase), both in Pi-rich and Pi-deficient medium (table). The highest activities were registered in cytosol of the parent CRY strain and in the PPN1-deficient mutant CRN in both cases. In Pi-deficient medium, this activity was half as high in the PPX1-deficient strain CRX and insignificant in the double mutant CNX (table). Except for the parent strain CRY, the levels of exopolyphosphatase activities detected in Pi-rich media were close to those found under Pi deficit. As for the CRY strain, it had lower exopolyphosphatase activity in Pi-deficient medium as compared with that in Pi-rich medium.
Exopolyphosphatase activities (mU/mg protein) in cytosol preparations of
S. cerevisiae in exponential growth phase (5 h)
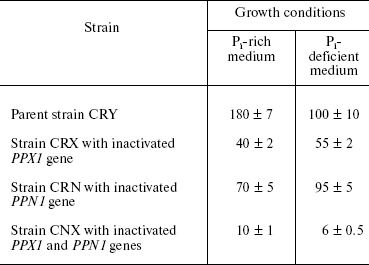
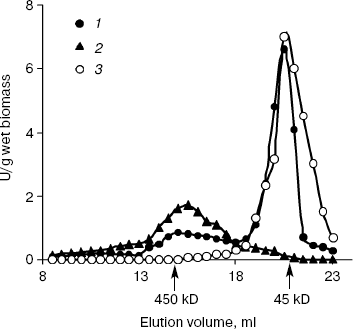
Figure 1 demonstrates the data on gel filtration of the cytosol preparations under Pi deficit. In the cytosol of PPN1-defficient strain CRN, exopolyphosphatase activity was represented by Ppx1 of ~45 kD. Just the same was observed for this strain in Pi-rich medium [19]. In the CRY cytosol, a minor amount of ~450 kD exopolyphosphatase activity was present in addition to Ppx1 (Fig. 1). The contribution of this enzyme to the total exopolyphosphatase activity was half that observed in Pi-rich medium [20]. Only exopolyphosphatase of ~450 kD was revealed in the cytosol of CRX, and Ppx1 was missing in the PPX1-deficient strain CRX both in Pi-deficient (Fig. 1) and Pi-rich media [20]. Minor exopolyphosphatase activity in the cytosol preparation of the double mutant CNX (table) was eluted in the region of >1000 kD (not illustrated).
Thus, at exponential growth of the yeast in Pi-deficient medium, exopolyphosphatase activities in the cytosols of strains CRY and CRN were represented mainly by the enzyme Ppx1. Ppn1 was responsible for this activity in the CRX strain.Fig. 1. Exopolyphosphatase activities in Superose 6 gel filtration fractions of the cytosol preparations from S. cerevisiae. The cytosols were isolated from cells grown to exponential phase (5 h) in Pi-deficient medium. 1) Parent strain CRY; 2) PPX1-deficient strain CRX; 3) PPN1-deficient strain CRN.
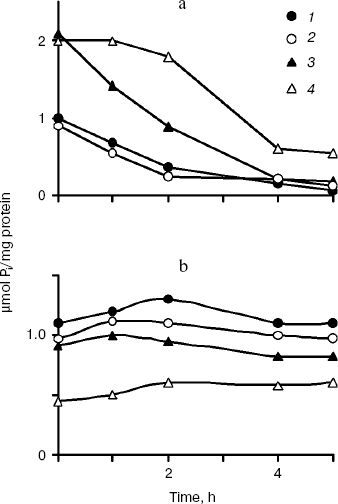
The polyP levels in the cytosol preparations of all cultures decreased on growth in Pi-deficient medium with rather constant Pi levels (Fig. 2). However, the polyP levels did not decrease similarly in strains with different enzymes in the cytosol. The dynamics of polyP levels in the CRY and CRN strains having the same enzyme Ppx1 was actually similar (Fig. 2). The spectrum of chain lengths was constant in time and similar for strains CRY and CRN (Fig. 3). This fact is consistent with data on substrate specificity of Ppx1 hydrolyzing polyPs with chain lengths of 9-200 phosphate residues at equal rates [22].
Fig. 2. PolyP (a) and Pi dynamics (b) in cytosol preparations of S. cerevisiae under exponential growth in Pi-deficient medium. 1) Parent strain CRY; 2) PPX1-deficient strain CRX; 3) PPN1-deficient strain CRN; 4) PPX1- and PPN1-deficient strain CNX. On isolation of cytosol preparations spheroplasts were lysed in the presence of heparin (4 mg/ml) and 20 mM EDTA, known inhibitors of exopolyphosphatases [17].
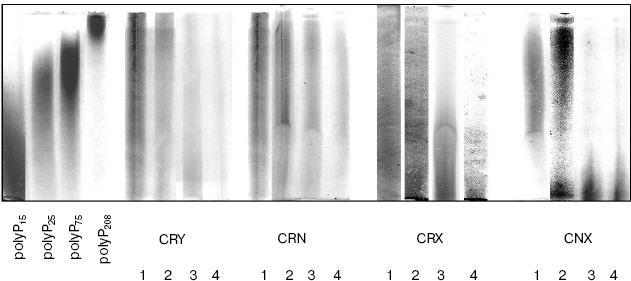
In the CRX strain, the rate of polyP hydrolysis was somewhat lower (Fig. 2) with accumulation of short-chain polyPs (Fig. 3) that were not so effectively hydrolyzed by Ppn1 as long-chain ones [16]. However, after long-chain polyPs had been depleted, short polymers were hydrolyzed as well. As a result, the 5-h growth in Pi-deficient medium was followed by polyP depletion in the cytosol preparations of the CRY, CRX, and CRN strains (Fig. 2). In contrast to these strains, about 20% of short-chain polyPs still remained in the cytosol of PPX1- and PPN1-deficient strain CNX after 5 h of growth (Fig. 2). In this strain, the polyP level moderately decreased within the first 2 h (Fig. 2), whereas the polyP chains were shortening (Fig. 3). Conceivably, fragmentation of long-chain polyPs could occur in this period due to the presence of a certain enzyme possessing endopolyphosphatase activity.Fig. 3. Electrophoresis of cytosol polyPs in 20% polyacrylamide gel. Chain lengths of polyP markers are indicated on the left. The numbers below indicate hours of growth of the strains in Pi-deficient medium.
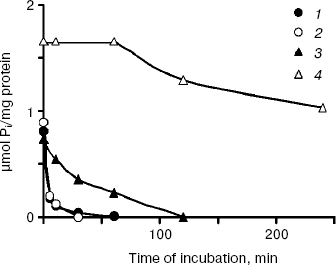
The polyP levels in the cytosol preparations of all cultures decreased not only under growth in Pi-deficient medium, but also under incubation of the preparations at 30°C (Fig. 4). In the CRY and CRN strains having the same enzyme Ppx1 in the cytosol, a sharp decrease of polyPs was observed in the very first minutes of incubation. PolyP hydrolysis in the CRX cytosol (Ppn1 enzyme) was slower and completed only in 120 min (Fig. 4). The polyP level in the cytosol of the double mutant CNX did not change within the first hour of incubation and decreased by ~40% in 4 h (Fig. 4).
Fig. 4. Polyphosphate dynamics in cytosol preparations of S. cerevisiae isolated from cells grown to stationary growth phase in Pi-rich medium under their incubation at 30°C: 1) parent strain CRY; 2) PPX1-deficient strain CRX; 3) PPN1-deficient strain CRN; 4) PPX1- and PPN1-deficient strain CNX. On isolation of cytosol preparations spheroplasts were lysed in the absence of inhibitors of exopolyphosphatases--heparin and EDTA.
DISCUSSION
The results suggest that the cytosol polyPs serve as Pi reserve and are effectively used under Pi limitation in the cultivation medium. Deficit of Pi in the cultivation medium has but a minor effect on exopolyphosphatase activities in the cytosol of all studied strains as compared with the same growth phase in Pi-rich medium (table), i.e. enzymes encoded by the PPX1 and PPN1 genes are not induced under Pi deficit. In this respect, the yeast differs from bacteria. Limitation of Pi in Escherichia coli and Acinetobacter johnsonii is known to induce expression of the ppx gene, followed by a drastic increase of exopolyphosphatase activity [24, 25].
As regards the cytosol of S. cerevisiae, the CRY and CRN strains possess mainly exopolyphosphatase Ppx1, which is the enzyme that hydrolyzes the cytosol polyPs as confirmed by the dynamics of chain-length changes. Exopolyphosphatase Ppn1 performs this function in the PPX1-deficient strain CRX. This enzyme hydrolyzes first of all the long-chain polyPs and then the short-chain polyPs, to which it has less affinity [16]. In the double mutant CNX lacking both enzymes, polyP hydrolysis proceeds with a notable delay but does not affect culture growth both in Pi-deficit and Pi-rich media. Exopolyphosphatase activity in the cytosol of this strain is insignificant, and polyP degradation is supposed to be performed by alternative enzymes. The problem of availability of polyphosphate kinase in S. cerevisiae, which is able to catalyze both polyP synthesis and degradation, still has to be solved, though this activity has been found recently in the yeast Candida humicola [26]. The question of the presence of endopolyphosphatase in S. cerevisiae is still unclear. In the double mutant, polyP degradation is followed by accumulation of short-chain polyPs, which might be hydrolyzed by a pyrophosphatase [27]. Such dynamics of polyP hydrolysis suggests the occurrence of endopolyphosphatase not encoded by the PPN1 gene.
The findings suggest that S. cerevisiae possesses a system that makes it possible to compensate for the inactivation of PPX1, PPN1, or both of these genes encoding exopolyphosphatases in the cytosol.
This work was supported by the Russian Foundation for Basic Research (grant 08-04-00472) and a grant of support for Leading Scientific Schools of the Russian Federation (NSh-1004.2008.4).
REFERENCES
1.Kulaev, I. S. (1979) The Biochemistry of
Inorganic Polyphosphates, Wiley, New York.
2.Reusch, R. N. (1992) FEMS Rev., 103,
119-130.
3.Kornberg, A., Rao, N., and Ault-Riche, D. (1999)
Ann. Rev. Biochem., 68, 89-125.
4.Kulaev, I. S., Vagabov, V. M., and Kulakovskaya, T.
V. (2004) The Biochemistry of Inorganic Polyphosphates,
Wiley, New York.
5.Liss, E., and Langen, P. (1962) Arch.
Microbiol., 41, 383-392.
6.Kulaev, I. S., and Vagabov, V. M. (1983) Adv.
Microbiol. Physiol., 24, 83-171.
7.Hofeler, H., Jensen, D., Pike, M. M., Delayre, J.
L., Cirillo, V. P., Springer, C. S., Jr., Fossel, E. T., and Balschi,
J. A. (1987) Biochemistry, 26, 4953-4962.
8.Vagabov, V. M., Trilisenko, L. V., and Kulaev, I.
S. (2000) Biochemistry (Moscow), 65, 349-354.
9.Persson, B. L., Lagerstedt, J. O., Pratt, J. R.,
Pattison-Granberg, J., Lundh, K., Shokrollahzadeh, S., and Lundh, F.
(2003) Curr. Genet., 43, 225-244.
10.Werner, T. P., Amhein, N., and Freimoser, F. M.
(2005) Arch. Microbiol., 184, 129-136.
11.Wurst, H., Shiba, T., and Kornberg, A. (1995)
J. Bacteriol., 177, 898-906.
12.Kumble, K. D., and Kornberg, A. (1996) J.
Biol. Chem., 271, 27146-27151.
13.Sethuraman, A., Rao, N. N., and Kornberg, A.
(2001) Proc. Natl. Acad. Sci. USA, 98, 8542-8547.
14.Shi, X., and Kornberg, A. (2005) FEBS
Lett., 579, 2014-2018.
15.Ogawa, N., DeRisi, J., and Brown, P. O. (2000)
Mol. Biol. Cell, 11, 4309-4321.
16.Andreeva, N. A., Kulakovskaya, T. V., and Kulaev,
I. S. (2006) Biochemistry (Moscow), 71, 975-977.
17.Lichko, L. P., Andreeva, N. A., Kulakovskaya, T.
V., and Kulaev, I. S. (2003) FEMS Yeast Res., 3,
233-238.
18.Lichko, L., Kulakovskaya, T., and Kulaev, I.
(2002) Biochim. Biophys. Acta, 1599, 102-105.
19.Lichko, L. P., Kulakovskaya, T. V., and Kulaev,
I. S. (2006) Biochemistry (Moscow), 71,
1171-1175.
20.Lichko, L., Kulakovskaya, T., and Kulaev, I.
(2004) Biochim. Biophys. Acta, 1674, 98-102.
21.Rubin, G. M. (1973) J. Biol. Chem.,
11, 3860-3875.
22.Andreeva, N. A., Kulakovskaya, T. V., Karpov, A.
V., Sidorov, I. A., and Kulaev, I. S. (1998) Yeast, 14,
383-390.
23.Lichko, L., Kulakovskaya, T., and Kulaev, I.
(2006) Yeast, 23, 735-740.
24.Nesmeyanova, M. A. (2000) Biochemistry
(Moscow), 65, 309-314.
25.Kortstee, J. J., Appeldoorn, K. J., Bonting, C.
F. C., van Niel, E. W. J., and van Veen, H. W. (2000) Biochemistry
(Moscow), 65, 332-340.
26.McGrath, J. W., Kulakova, A. N., Kulakov, L. A.,
and Quinn, J. P. (2005) Res. Microbiol., 156,
485-491.
27.Baykov, A. A., Cooperman, B. S., Goldman, A., and
Lahti, R. (1999) in Progress in Molecular and Subcellular Biology.
Inorganic Polyphosphates. Biochemistry. Biology. Biotechnology,
Vol. 23 (Schroder, H. C., and Muller, W. E. G., eds.) Springer-Verlag,
Berlin-Heidelberg-New York, pp. 127-150.