REVIEW: Bacterial 5S rRNA-Binding Proteins of the CTC Family
G. M. Gongadze*, A. P. Korepanov, A. V. Korobeinikova, and M. B. Garber
Institute of Protein Research, Russian Academy of Sciences, Institutskaya ul. 4. 142290 Pushchino, Moscow Region, Russia; fax: (495) 632-7871; E-mail: gongadze@vega.protres.ru* To whom correspondence should be addressed.
Received February 2, 2008; Revision received February 26, 2008
The presence of CTC family proteins is a unique feature of bacterial cells. In the CTC family, there are true ribosomal proteins (found in ribosomes of exponentially growing cells), and at the same time there are also proteins temporarily associated with the ribosome (they are produced by the cells under stress only and incorporate into the ribosome). One feature is common for these proteins - they specifically bind to 5S rRNA. In this review, the history of investigations of the best known representatives of this family is described briefly. Structural organization of the CTC family proteins and their occurrence among known taxonomic bacterial groups are discussed. Structural features of 5S rRNA and CTC protein are described that predetermine their specific interaction. Taking into account the position of a CTC protein and its intermolecular contacts in the ribosome, a possible role of its complex with 5S rRNA in ribosome functioning is discussed.
KEY WORDS: 5S rRNA-binding proteins, CTC proteins, ribosome, translation, BacteriaDOI: 10.1134/S0006297908130038
Abbreviations: CTC, catabolite controlled, a protein whose gene (ctc) is controlled by catabolism products. Names of the individual ribosomal proteins are given according to their belonging to the ribosome subparticle (“L”, Large) and serial number on the two-dimensional electrophoregram (e.g. L25). The first letter of the organism name (“T”, Thermus) has been added to the name of ribosomal protein TL5.
In any cell, protein synthesis is realized by the ribosome. Universality
of the key steps of protein synthesis is determined by conservatism of
the ribosome structural elements. However, environment and the
evolutional history of an organism leave their imprint on the structure
and functioning of the organism's components. The ribosome, as well as
any cell component, possesses evolutional features of the particular
organism, a group of related organisms, or the domain of life in
general. Modern ribosomes include about 60-80 different ribosomal
proteins, but only 34 of them are conserved in all the domains of life
[1]. The rest of the ribosomal proteins are
evolutionary acquisitions of the Archaea, Bacteria, or Eukarya. The
role of these proteins in the functioning of the translation apparatus
remains poorly studied. Investigation of properties of the proteins of
this group is an important and promising task in both basic and applied
aspects.
A CTC protein is an example of such an evolutional feature of the translation apparatus of bacteria. This protein is found in the majority of taxons of Bacteria and is not found in Archaea and Eukarya. There are ribosomal and stress proteins among the representatives of the CTC family. Ribosomal 5S RNA is the only known target for a CTC protein. Despite almost forty years of researching into some of the proteins belonging to this family, their function in the cell and particular role in the work of translation apparatus are still open questions.
In present review, collected data regarding proteins of the CTC family are put together and arranged for the first time. A short historical reference to the known proteins of the family is given. Questions of structural organization of the CTC proteins and their occurrence in representatives of different known bacterial taxons are discussed. Structural features of bacterial 5S rRNA and CTC protein, which determine their specific interaction, are described. Taking into account the position in the ribosome occupied by CTC protein and its intermolecular contacts, the possible role of its complex with a 5S rRNA in bacterial ribosome functioning is discussed.
CTC FAMILY AND ITS REPRESENTATIVES
In 1996, we showed that the ribosomal 5S rRNA-binding protein TL5 of Thermus thermophilus is homologous to the general stress protein CTC of Bacillus subtilis through the entire amino acid sequence [2]. Besides, it turned out that N-terminal parts of the above-mentioned proteins are homologous to the ribosomal protein L25 of Escherichia coli. That work is a keystone to joining these proteins and their homologs into the CTC family. Then, it has been shown that genomes of the majority of the known bacteria contain one gene encoding a protein of this family [3, 4]. Furthermore, it turned out that the CTC family proteins were unique to bacteria, since a ctc gene was not found in the genomes of known Archaea and Eukarya [5]. Although the CTC family now includes more than 300 representatives, the only thing is known about most of them is an open reading frame in the genome. Properties of a few proteins belonging to the family may be considered to be dissected. One of the first found and examined representatives, which has given its own name to the entire family, is the general stress protein CTC of B. subtilis. In 1979, Haldenwang and Losick discovered a minor σ-subunit (σ37) of the RNA-polymerase of B. subtilis, which has been activated under stress conditions and during early stages of sporulation [6, 7]. A specific promoter recognized by this σ-subunit of RNA-polymerase has been identified [8, 9]. The promoter as well as the gene under its control was named ctc (catabolite controlled) (see [10] for a review). Afterwards, Volker and colleagues showed that one of the general stress proteins of B. subtilis, GSP10, is a product of the ctc gene [11]. The CTC protein is one of the proteins that are synthesized in B. subtilis cells in response to different stresses [12]. Not long ago, it was shown that this protein binds specifically to a 5S rRNA [13] and can be found in the ribosomal fraction under stress [14]. Thus, one may consider the CTC protein of B. subtilis to be temporarily associating with the ribosome. Recently, CTC protein synthesized in the cell in response to different stresses was found in one more representative of the class Bacilli, Listeria monocytogenes [15, 16]. It is likely that CTC protein of this organism is also able to associate with the ribosome under stress conditions. In addition, it has been shown that knockout of the ctc gene causes depressed cell growth under stress conditions and sporulation process defects in B. subtilis [17]. Thus, one may suggest that one of the functions of the CTC family proteins is keeping ribosomes working under stress conditions or at sporulation in some bacteria. At the same time, several proteins of the family are found in the ribosomes of exponentially growing cells, i.e. they are true ribosomal proteins. The ribosomal protein L25 of E. coli was the first discovered representative of the CTC family. In 1972, it has been shown in the work of Horne and Erdmann that one of the ribosomal proteins of E. coli specifically binding to the 5S rRNA is L25 [18]. Another protein of the CTC family, ribosomal 5S rRNA-binding protein TL5 of T. thermophilus, was identified by us in 1993 [19]. This multi-domain protein (206 residues) is as much as twice larger than the ribosomal protein L25 (94 residues) and is comparable to the general stress protein CTC (204 residues) of B. subtilis [2, 20, 21]. It turned out that the N-terminal part of the TL5 protein, homologous to the L25 protein, is responsible for binding to 5S rRNA [22]. Furthermore, a multi-domain CTC protein (238 residues) comparable in size with TL5 of T. thermophilus and CTC of B. subtilis has been found in the ribosome of Deinococcus radiodurans [23]. This protein also interacts with 5S rRNA by means of its N-terminal domain. Furthermore, CTC proteins of Enterococcus faecalis and Nostoc sp. were recently demonstrated to be able to bind specifically to the 5S rRNA [24].
Thus, one common property for all the above-mentioned CTC proteins is obvious--they are capable of forming a specific complex with the ribosomal 5S RNA.
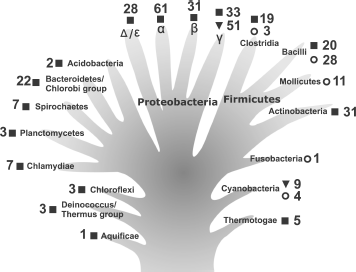
During the past decade, the number of deciphered bacterial genomes has increased significantly [3, 4]. There are now data regarding genomes of several hundred representatives of different bacterial taxons. This allows us to compare the main characteristics of the CTC family proteins and to obtain information about their distribution in different phylogenetic groups. As seen in Fig. 1, multi-domain CTC proteins are found in the majority of indicated bacterial taxons, including the most ancient ones (Aquificae, Thermotogae, and Deinococcus-Thermus group). Apparently, the first CTC proteins were multi-domain when they appeared in bacteria. All the known CTC proteins possess a so-called “5S rRNA-binding domain” [25]. The only exception could be Aquifex aeolicus, in which the ctc gene encodes a protein lacking the first 50 amino acid residues of the 5S rRNA-binding domain according to the NCBI GenBank [3, 26]. This protein has been purified and it appeared to be incapable of binding 5S rRNA [13]. However, during careful analysis of the corresponding genome region of this organism we have found a mistake (missed nucleotide). Due to this mistake, the position of the start codon was defined incorrectly. In fact, this gene encodes a protein possessing a full-sized 5S rRNA-binding domain. In addition, it has been shown before that L25 protein lacking the first 20 amino acid residues and N-terminal fragment of TL5 protein (91 amino acid residues) lacking 10 C-terminal residues lose their 5S rRNA-binding capacity [22, 27]. It follows from the above-said that only CTC protein having a domain proportionate to L25 (5S rRNA-binding domain) is able to interact with 5S rRNA. Therefore, taking into account that 5S rRNA is a main target for CTC proteins, it is reasonable to assume that function of the first CTC proteins was already related to the work of the translational apparatus.
It is worth paying attention to one more interesting feature of the CTC family proteins that is shown in Fig. 1. Only representatives of phylum Cyanobacteria and γ-subdivision of Proteobacteria possess a single-domain CTC protein (e.g. Nostoc sp. and E. coli). Such a protein form prevails in these taxonomic groups. Apparently, preserving only the so-called 5S rRNA-binding domain was sufficient for the CTC protein to perform its function in these bacteria. Furthermore, in genomes of the majority of phylum Firmicutes representatives, a gene encoding CTC protein is not found at all (Fig. 1). We have already mentioned at the beginning of the review that in such representatives of the class Bacilli as B. subtilis and L. monocytogenes CTC protein is only produced in response to exposure to different stresses. At the same time, knock out of the ctc gene in B. subtilis does not affect its growth under normal conditions [17]. All these observations suggest the “regressive evolution” of the CTC proteins in representatives of certain bacterial taxons. Most probably, changing environmental conditions have allowed certain bacteria to live (completely or partially) without a CTC protein.Fig. 1. Occurrence of CTC family proteins among representatives of a number of bacterial taxons. The information for analysis was taken from the NCBI GenBank [3] for completely deciphered genomes only. The data are presented against the background of the hypothetical evolutionary tree for greater clarity. Symbols indicate the presence of the genes encoding multi-domain (|) and single-domain (?) proteins or absence of such genes in genomes (?). Numbers show the number of deciphered genomes. In most cases, the taxonomic groups presented are restricted to superphylum or phylum (excluding phyla Proteobacteria and Firmicutes which are presented by classes). Symbols α, β, γ, and Δ/ε indicate subdivisions (classes) of the phylum Proteobacteria.
In the first works [28, 29], where sequence alignment for a few known CTC family proteins was performed, their main features were established. Despite lack of any prominent homology, several amino acid residues are identical in proteins of this family. In addition, most of them are situated in an N-terminal (so-called 5S rRNA-binding) domain (R10, R19, P25, Y29, G30, H85, and D87 by TL5 nomenclature). We have aligned sequences of 300 proteins of the family from all the studied taxons using ClustalW [30, 31] software for more impartial evaluation of the conservation of certain residues. In this case, the pattern of strictly conserved residues did not change. The sequence alignment for CTC proteins of typical representatives of large bacterial taxonomic groups, where these proteins were found, is presented in Fig. 2. This figure represents the picture that is valid for all 300 proteins. It can be seen that only seven amino acid residues of the N-terminal domain are strictly conserved in CTC proteins of the vast majority of the taxons. Five of them are the residues participating in interaction of proteins L25 and TL5 with 5S rRNA [25] (see section “5S rRNA-Binding Properties of CTC Proteins” for more information). All this suggests the significant conservatism in interaction between CTC proteins and the 5S rRNA in the majority of bacteria. The complete information about frequency for these five residues in known CTC proteins is presented in the table. We have already published similar result for 150 CTC proteins [25]. Frequency of these residues in the family proteins is 95-99%. It is seen that rare exceptions are not randomly distributed among different bacteria. Most of them are found in the proteins from organisms belonging to certain taxonomic groups. Such exceptions revealed to be common for all known CTC proteins of phylum Cyanobacteria, certain representatives of class Bacilli, and α-subdivision of Proteobacteria (see section “5S rRNA-Binding Properties of CTC Proteins” for more information). The indicated exceptions confirm the previously suggested idea of evolutionary isolation of certain groups of bacteria.
Frequency of occurrence for amino acid residues of the 5S rRNA-recognition module in 300 CTC family proteinsFig. 2. Sequence alignment for the typical representatives of the CTC family from a number of taxonomic groups of bacteria where they have been found so far. ClustalW software [30, 31] was used for the alignment. Sequences are: Mycob. tub., Mycobacterium tuberculosis, Actinobacteria (UniProtKB/SwissProt, P66121); Rhod. balt., Rhodopirellula baltica, Planctomycetes (UniProtKB/SwissProt, Q7UKU9); Therm. mar., Thermotoga maritima, Thermotogae (UniProtKB/SwissProt, Q9X1W2); Ther. ther., Thermus thermophilus, Deinococcus/Thermus group (UniProtKB/SwissProt, P56930); Ros. sp. RS1, Roseiflexus sp. RS-1, Chloroflexi (UniProtKB/TrEMBL, A5UQF4); Chlor. tep., Chlorobium tepidum, Bacteroidetes/Chlorobi group (UniProtKB/SwissProt, Q8KCQ1); Neiss. men., Neisseria meningitides, β-subdivision of Proteobacteria (UniProtKB/SwissProt, Q9JUX7); Esche. coli, Escherichia coli, γ-subdivision of Proteobacteria (UniProtKB/SwissProt, P68919); Ricket. pr., Rickettsia prowazekii, α-subdivision of Proteobacteria (UniProtKB/SwissProt, Q9ZCV3); Solib. usi., Solibacter usitatus, Acidobacteria (UniProtKB/TrEMBL, Q01QR2); Parach. sp., Parachlamydia sp. UWE25, Chlamydiae (UniProtKB/SwissProt, Q6MAT1); Aquif. aeo., Aquifex aeolicus, Aquificae (UniProtKB/SwissProt, O66678); Desul. psy., Desulfotalea psychrophila, Δ/ε-subdivision of Proteobacteria (UniProtKB/SwissProt, Q6AJL8); Flav. psyc., Flavobacterium psychrophilum, Bacteroidetes/Chlorobi group (UniProtKB/SwissProt, A6GVZ1); Bacil. sub., Bacillus subtilis, Bacilli (UniProtKB/SwissProt, P14194); Clost. per., Clostridium perfringens, Clostridia (UniProtKB/SwissProt, Q8XJ87); Helic. hep., Helicobacter hepaticus, Δ/ε-subdivision of Proteobacteria (UniProtKB/SwissProt, Q7VG30); Trep. pall., Treponema pallidum, Spirochaetes (UniProtKB/SwissProt, O83387); Thermo. el., Thermosynechococcus elongates, Cyanobacteria (UniProtKB/SwissProt, Q8DLG3). Identical or strictly conserved (>90%) amino acid residues (*) as well as conserved (<90%) ones (:) are indicated at the bottom line. Conserved residues interacting with the 5S rRNA are shown by white symbols.
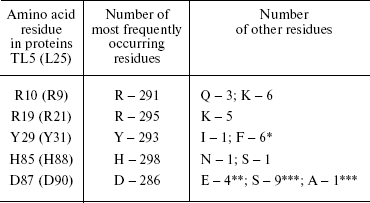
Note: These residues are present in CTC proteins of representatives of: * α-subdivision of Proteobacteria; ** class Bacilli; *** phylum Cyanobacteria.
In conclusion of this chapter we wish to comment about C-terminal parts (additional domains) of the multi-domain CTC family proteins. The family is represented mostly by proteins of this type. Sequence alignment for these proteins (Fig. 2) shows that in contrast to the “5S rRNA-binding domain” of a CTC protein, its additional domains possess more variability and have virtually no conserved distinct amino acid residues or their groups. However, it has been stated that residues forming hydrophobic core of the second domain of TL5 are preserved in other proteins [29]. This suggests that unique spatial structure of the second domain may remain in other representatives of the family.
5S rRNA-BINDING PROPERTIES OF CTC PROTEINS
5S rRNA is an intrinsic part of ribosomes of all studied organisms. Several ribosomal proteins form stable and specific complex with isolated 5S rRNA (see [32] for a review). In particular, in E. coli there are three proteins--L5, L18, and L25 [18, 33]. Homologs of two of them, L5 and L18, have been found in representatives of all the domains of life [5]. Homologs of the third 5S rRNA-binding protein L25 of E. coli, proteins of the CTC family, turned out to be unique to bacteria [3-5]. Which structural features of a CTC protein and 5S rRNA define specificity of their interaction?
E-Loop Is a CTC Protein-Binding Site in 5S rRNA
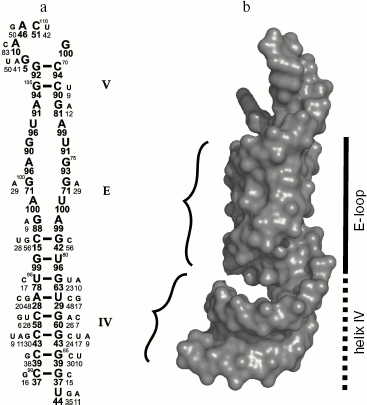
There have been repeated attempts to locate L25 protein-binding site in 5S rRNA using different methodological approaches (see reviews [32, 34-36]). The most precise result was obtained by Douthwaite and colleagues in 1979. They showed that ribosomal protein L25 binds specifically to 40-nucleotide (69-87/90-110) fragment of 5S rRNA (Fig. 3a) and protects it against hydrolysis by ribonucleases [37]. Later it was shown that the other proteins of the family such as ribosomal protein TL5 from T. thermophilus, general stress protein CTC of B. subtilis, and CTC proteins from E. faecalis and Nostoc sp. specifically bind to the same 5S rRNA fragment [13, 22, 24]. This RNA fragment includes the unique internal E-loop. As seen in Fig. 3a, the nucleotide sequence of the E-loop is strictly conserved in bacterial 5S rRNAs [38]. At the same time, the nucleotide sequence of the corresponding region of 5S rRNA from Archaea and Eukarya differs significantly from the E-loop of Bacteria [38, 39]. Besides, CTC family proteins can form stable hybrid complexes with different bacterial 5S rRNAs [13, 18, 19], while L25 protein does not bind to yeast 5S rRNA [40]. Therefore, strict conservation of the nucleotide sequence of 5S rRNA E-loop is required for specific interaction with CTC proteins. Later it was determined, however, that uniqueness of bacterial 5S rRNA E-loop is defined not only by conservatism of its nucleotide sequence, but more by the nature of its spatial structure [41, 42]. This region of 5S rRNA was called E-loop in the first works studying its secondary structure [39, 43] assuming that double helix can not be formed in this region since both strands are saturated with purines. However, it turned out that stability of the spatial structure is maintained by multiple intramolecular bonds, including non-canonical base pairing [42]. These intramolecular bonds lead to formation of distorted RNA double helix, which differs significantly from classical A-form (Fig. 3b). Major groove becomes narrower while minor groove becomes wider. Bivalent cations and bound water molecules play an important role in stabilization of the unique spatial structure of the RNA. It has been suggested that the unique surface of the RNA is one of the most important conditions for specific interaction with the protein [41]. It was shown later that unique spatial structure of the 5S rRNA E-loop is not changed during interaction with the protein [29, 44]. In addition, observed complementarity of interacting surfaces of RNA and protein [29] shows beyond doubt the significance of the unique E-loop structure for specific contact with CTC family proteins.
Intermolecular Interactions in CTC-5S rRNA ComplexFig. 3. Structure of the second domain of E. coli 5S rRNA. a) Secondary structure of the E. coli 5S rRNA fragment that is protected by L25 against hydrolysis with RNase A [37]. The loop and the helices are designated by the letter and the Roman numerals, accordingly. Frequency of occurrence for nucleotides in corresponding positions of 5S rRNA in 460 analyzed bacteria [38] is shown in percent under the symbol. b) The spatial structure of the same 5S rRNA fragment indicating its surface relief. Minor grooves of the double helix are shown with curly brackets. Spatial structure of the 5S rRNA fragment (PDB code: 364D) was used to build this model.
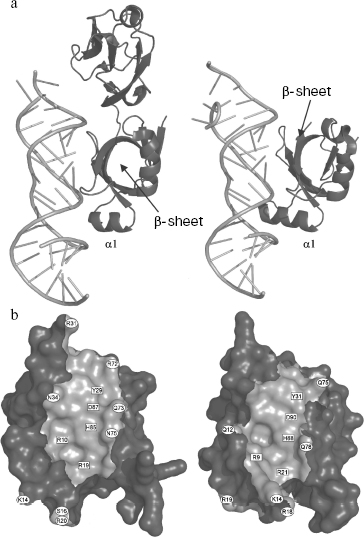
Spatial structures of two proteins of the family, L25 of E. coli and TL5 of T. thermophilus, complexed with specific 5S rRNA fragment, were independently determined by several groups at high resolution in 1999-2001 [29, 44, 45]. At the same time, spatial structure of one more representative of the family, CTC of D. radiodurans, was determined as a part of 50S ribosomal subunit at 3.1 Å resolution [23]. It turned out that despite low sequence homology (see section “CTC Family and Its Representatives”) between these proteins, their spatial structures and mode of interaction with RNA are very similar [23, 29] (Fig. 4a). Structural data have confirmed completely the results of biochemical experiments obtained earlier [22, 37]. TL5 protein indeed interacts with RNA by means of its N-terminal domain (Fig. 4a), while all the indicated proteins bind to the same region of 5S rRNA, E-loop. Amino acid residues belonging to one of the β-sheets of the CTC proteins form tight contact with sugar-phosphate backbone and bases of minor groove of 5S rRNA in the E-loop region, while residues of helix α1 and adjacent β1-α1 loop interact with sugar-phosphate backbone of the major groove of RNA. Comparison of TL5-5S rRNA and L25-5S rRNA complexes revealed identical structural elements of RNA and proteins participating in intermolecular contacts, which, apparently, should play a key role in specific interaction of these macromolecules [25, 29]. All the amino acid residues of L25 and TL5 proteins forming hydrogen bonds with RNA have been divided onto two groups for convenience--non-conserved and conserved [25]. Residues of the first group are situated on the periphery of the protein surface contacting with RNA (Fig. 4b) and form hydrogen bonds that are accessible to solvent. The second group of residues (R9, R21, Y31, H88, and D90 for L25; R10, R19, Y29, H85, and D87 for TL5) are situated in the central part of the protein surface contacting with RNA (Fig. 4b) and form intermolecular hydrogen bonds that are inaccessible to solvent. It turned out that simultaneous change to alanine of several non-conserved amino acid residues in the RNA-binding region of the protein does not significantly affect stability of the TL5-5S rRNA complex [24]. Thus, removing a significant number of solvent accessible intermolecular hydrogen bonds does not affect formation and stability of the complex. At the same time, single replacement of each of the five conserved residues belonging to the RNA-binding region of either TL5 or L25 leads to significant destabilization or even impossibility of formation of the complex [24, 25]. It has been suggested that these five above-mentioned residues form a 5S rRNA recognition module in TL5 and L25 proteins. Recently, in the work of Nevskaya and colleagues on the base of analysis of structures of several ribosomal RNA-protein complexes, it has been suggested that recognition modules on the surface of the interacting molecules should be formed by their conserved residues forming conserved hydrogen bonds [46]. This proposed principle is perfectly confirmed by the results obtained for the CTC family proteins.
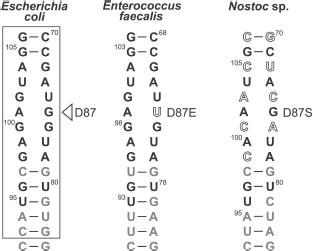
As already mentioned in section “CTC Family and Its Representatives”, the five amino acid residues (two arginines, tyrosine, histidine, and aspartate) indicated above are strictly conserved in CTC family proteins [25]. Therefore, it is reasonable to conclude that the RNA-recognition module found in TL5 and L25 proteins should be representative for all the CTC family proteins. It is likely that such RNA-recognition module is a feature of the vast majority of the proteins of the family, but there is no rule without an exception. During comparative sequence analysis for three hundred representatives of the CTC family (table), a few cases of natural changes of residues of the RNA-recognition module were found [25]. So, conserved aspartate in the CTC proteins of class Bacilli is changed to glutamate, while in the proteins of phylum Cyanobacteria it is changed to serine or alanine. Such substitutions introduced artificially into TL5 and L25 [24, 25] resulted in the impossibility of formation of stable RNA-protein complex. One might suggest that CTC proteins of certain representatives of class Bacilli or phylum Cyanobacteria have lost their ability to interact with ribosomal 5S RNA, but these organisms have preserved the gene encoding such a protein. However, this would seem to be an unacceptable dissipation for a bacterial organism. It turned out that indicated changes in the protein RNA-recognition module are accompanied by certain changes in the interacting with CTC region of 5S rRNA of the corresponding organism (Fig. 5). It is the guanine, the base of which interacts with the conserved aspartate of TL5 and L25 proteins that is changed to uracil in the E. faecalis (Bacilli) 5S rRNA. Increase in size of one of the interacting residues is accompanied by decrease in size of the other one. Such simultaneous changes in protein and RNA can be compensatory. A more complicated situation is observed in the cyanobacterial 5S rRNA E-loop. Almost half of the nucleotides in the corresponding region are changed here (Fig. 5). However, the fact that in the indicated proteins and RNAs of all the known cyanobacteria similar changes have occurred suggests that these changes are compensatory as well. This question has been answered recently. CTC proteins and 5S rRNAs from E. faecalis and Nostoc sp. have been isolated. Their ability to form specific complexes has been tested [24]. It appears that 5S rRNA and the CTC protein from the same organism can form stable and specific complex.Fig. 4. a) Spatial structure models of TL5 (left) and L25 (right) complexed with the specific 5S rRNA fragment. The protein structural elements forming contacts with RNA are indicated. b) Contact areas of TL5 (left) and L25 (right) with 5S rRNA. The contact areas on the protein surfaces are light gray. Positions of the conserved and non-conserved amino acid residues forming H-bonds with RNA are marked with rectangles and ovals, correspondingly. TL5-5S rRNA (PDB code: 1FEU) and L25-5S rRNA (PDB code: 1DFU) structures were used for building the models.
Summarizing the data presented above, several main conclusions can be drawn. First, the unique surface of the E-loop composed of distorted double helix of 5S rRNA is a requirement for specific interaction with the CTC family protein. Second, the 5S rRNA-recognition module composed of the five strictly conserved residues of proteins TL5 and L25, forming solvent inaccessible hydrogen bonds with RNA, is representative for the majority of known CTC proteins. Third, simultaneous changes in the contact regions of CTC proteins and 5S rRNAs that occurred in these molecules during evolution of certain bacterial groups aim at preservation of their ability to form a stable complex.Fig. 5. Natural changes in contacting regions of several 5S rRNAs and CTC proteins [25]. Fragments of 5S rRNAs (helices IV and V and E-loop) of representatives of Bacilli and Cyanobacteria are presented. The corresponding region of E. coli 5S rRNA is given for comparison. The RNA region contacting with the protein is framed. Nucleotides that are strictly conserved in bacterial 5S rRNAs (>80%), are black, non-conserved ones (<80%) are gray. Changes in conserved nucleotides in these RNAs are shown in open typeface. Changes of the conserved aspartate in the corresponding CTC protein are indicated to the right of the RNA.
A CTC PROTEIN IN THE RIBOSOME
It follows from the previous chapters of this review that, first, the CTC family protein which is unique to Bacteria is present in the majority of known bacteria. Second, this protein can specifically bind to the 5S rRNA and can be permanently or temporary incorporated into the ribosome. In connection with this, the questions arise: for what reason has the CTC protein appeared in Bacteria and why is it preserved by most of them, and what role does it play in ribosome functioning? Apparently, by now these questions can only be answered partially, and this is what we will try to do in the following parts of this review.
Location of the 5S rRNA-Protein Complex in the Ribosome
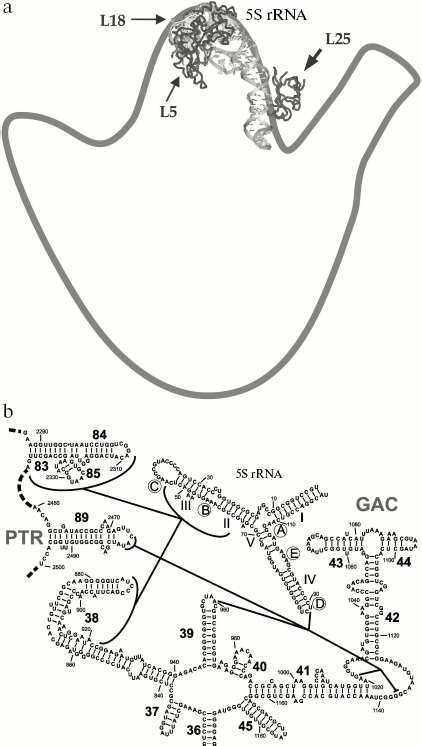
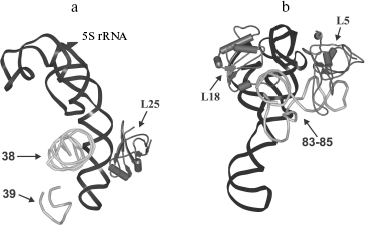
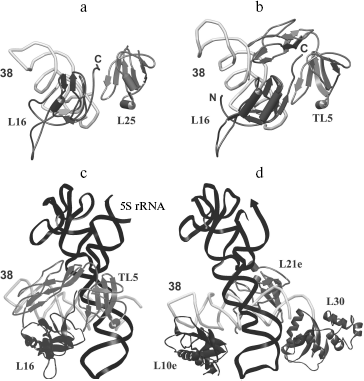
The location of L25 protein as well as the other components of 5S rRNA-protein complex in the ribosome were defined long ago. In the early eighties, by means of electron microscopy, it was shown that 5S rRNA is situated in central protuberance of the 50S ribosomal subunit of E. coli [47-49]. It has been shown that apical C-loop is situated at the top of the protuberance, while 3′-5′-terminal helix is located in the middle of the protuberance on the solvent facing surface. Later, all the three 5S rRNA-binding proteins have been located at the same region of 50S subunit [50, 51]. The L25 protein is situated at base of the central protuberance of 50S ribosomal subparticle on the side facing L7/L12 protuberance (Fig. 6a). The proteins L5 and L18 were located almost on top of the central protuberance on the side of L1 protuberance. The 5S rRNA consists of two domains (hairpins) situated along the main axis of the molecule [23, 52-54]. The first domain of the 5S rRNA molecule consists of helices 2 and 3 and loops B and C, while the second domain includes helices 4 and 5 and loops D and E [39, 55]. Afterwards, the location of 5S rRNA (its structural elements) and the proteins specifically binding to it were defined more exactly using intermolecular cross-links (Fig. 6b). Thus, it has been shown that nucleotide U89 of 5S rRNA D-loop (second domain) can be cross-linked to nucleotides of domain II (U958, A960, G1022, G1138) and domain V (C2475, U2477) of 23S rRNA [56-58]. Furthermore, several nucleotides of helices II and III of 5S rRNA (first domain) were cross-linked to regions of domain II (helix 38) and domain V (helices 83-85) of 23S rRNA [59]. Excluding the proteins of 5S rRNA-protein complex, there were no ribosomal proteins found to be cross-linkable to 5S rRNA in the ribosome [60]. At the same time, proteins L5 and L18 interacting with the first domain of 5S rRNA [23, 61] were cross-linked to a region of domain V (helices 84 and 85) of 23S rRNA [60]. The 5S rRNA is the only molecule found to be cross-linkable to L25 in the ribosome [62]. Thus, the main neighbors of the 5S rRNA found the ribosomal 50S subunits using the indicated technique are domains II and V of 23S rRNA and proteins L5, L18, and L25. These data indicate that in contrast to 5S rRNA, which has a number of adjacent 23S rRNA regions, L25 in the ribosome is rather isolated. This conclusion is supported by the results of in vitro assembly of 5S rRNA-protein complex and 50S ribosomal subunit of E. coli. First, L25 protein was shown to bind to 5S rRNA independently from the other proteins [63]. Second, 5S rRNA-protein complex assembles before incorporation into the ribosomal subunit [64, 65]. Spatial structures of 50S ribosomal subunits from three bacteria, D. radiodurans, E. coli, and T. thermophilus, have been defined during the last six years [23, 53, 54]. This allowed us to analyze the intermolecular contacts of CTC protein and 5S rRNA in the ribosome and to compare such contacts in different bacterial ribosomes. The results of crystallographic studies confirm most of the above presented intermolecular cross-linking data. It is worth drawing special attention to several intermolecular contacts of CTC protein and 5S rRNA in the ribosome, which could be visualized due to the crystallographic studies (Figs. 7 and 8). As mentioned above, one side of the 5S rRNA E-loop interacts with L25. At the same time, the opposite side of the E-loop, but not helices II and III of 5S rRNA [59], forms tight contact with the helix 38 of 23S rRNA (Fig. 7a). It follows from this that the second domain of 5S rRNA in the E-loop region is “jammed” between CTC protein and helix of 23S rRNA. The other extensive intermolecular contact is formed by the first domain of 5S rRNA and helices 83-85 of 23S rRNA. This interaction is realized via L5 and L18 proteins (Fig. 7b). Thus, the two above-mentioned groups of intermolecular contacts of 5S rRNA, previously observed in the ribosome of an archaea Haloarcula marismortui [52, 66, 67], are conserved in bacterial ribosomes as well (D. radiodurans, E. coli, and T. thermophilus). One more conserved though a single contact between 5S rRNA and 23S rRNA has been found to be typical for all the studied ribosomes (Fig. 7a). The conserved bulged uracil of helix 39 of 23S rRNA forms a hydrogen bond with D-loop of 5S rRNA. Beside these, there are no other known conserved intermolecular contacts of 5S rRNA found in the ribosome. Thus, one may assume that the indicated conserved intermolecular contacts define the unique position occupied by the 5S rRNA in the ribosome. These results support the assumption that 5S rRNA might be a linkage between domains II and V of 23S rRNA [35]. In addition, intermolecular contacts of one more ribosomal protein in the discussed area of 50S ribosomal subunit should be noted, which are conserved in bacterial and archaeal ribosomes. In the early works, L16 protein (L10e by archaeal nomenclature) has been reckoned among the key ribosomal proteins. The large ribosomal subunit lacking this protein exhibited defects in peptidyl transferase activity [68, 69] and in aminoacyl-tRNA binding [70]. Results of recent structural studies reveal multiple intermolecular contacts between L16 and ribosomal RNA [53, 54, 67, 71, 72]. In particular, L16 (L10e) protein forms tight contact with helix 38 of the 23S rRNA (Fig. 8). In addition, single contacts of this protein with helix IV of 5S rRNA have been identified in both bacterial and archaeal ribosomes [52, 67, 73]. We can add here that the C-terminal region of L16 protein interacts with CTC protein in bacterial ribosomes. Furthermore, in the case of TL5 protein this contact is formed with two domains, which significantly enlarges contact area as compared to the single-domain protein L25 (Fig. 8, a and b). At the same time, the second domain of TL5 forms several hydrogen bonds with helix 38 (Fig. 8b). Thus, one of the possible functions of bacterial CTC protein can be fixation (stabilization) of the contacts between 5S rRNA, helix 38 of 23S rRNA, and L16 protein (Fig. 8c). In addition, in the archaeal ribosome, which lacks CTC protein, an additional stabilization of contact between helix 38 of 23S rRNA and 5S rRNA [52, 67] is realized by an alternative mechanism (Fig. 8d). In this ribosome, proteins L30 (archaeal L30 proteins is as much as twice larger than its bacterial homolog, which does not interact with 5S rRNA) and L21e (this protein is an evolutionary acquisition of Archaea and Eukarya [5]) are responsible for such stabilization.
Fig. 6. a) Position of the 5S rRNA-protein complex in the 50S ribosomal subunit of E. coli. An outline drawing of the 50S subunit (view from the side facing the 30S subunit) is built on the basis of electron microscopy data [34, 47, 48]. The spatial structure of the 5S rRNA-protein complex is presented as a ribbon model. The structure of E. coli 50S subunit (PDB code: 2AW4) was used to build this model. Location of the proteins belonging to the 5S rRNA-protein complex is indicated with arrows. b) A scheme showing chemical cross-links between regions of 5S rRNA (D-loop and helices II and III) and 23S rRNA (domains II and V) is drawn on the basis of data published in [35, 56-59]. The helices and loops of the 5S rRNA are indicated with the Roman numerals and letters, correspondingly. Helices of the 23S rRNA are indicated with the Arabic numerals. The 23S rRNA regions participating in formation of ribosomal functional centers are indicated: PTR (peptidyl transferase ring) - a part of peptidyltransferase center, and GAC (GTPase-associated center).
Fig. 7. Models illustrating positional relationship between the 5S rRNA-protein complex and several 23S rRNA regions in the E. coli ribosome. The E. coli 50S subunit structure (PDB code: 2AW4) was used to build this model. a) 5S rRNA, L25 protein, and helices 38 and 39 of the 23S rRNA. b) 5S rRNA, proteins L5 and L18, and helices 83-85 of the 23S rRNA. Positions of the proteins and RNA are indicated with arrows.
CTC Protein and Ribosome FunctioningFig. 8. Models showing relative positions of L16 protein (or its archaeal homolog L10e) and other ribosomal components of E. coli (a), T. thermophilus (b and c), and H. marismortui (d). Structures of 50S subunits from E. coli (PDB code: 2AW4), T. thermophilus (PDB code: 2J01), and H. marismortui (PDB code: 1JJ2) were used to build these models. Positions of ribosomal proteins, 5S rRNA, helix 38 of the 23S rRNA, and N- and C-termini of L16 protein are indicated.
The question of the necessity of certain ribosomal components has been raised simultaneously with the appearance of the protein biosynthesis theme as such. Researchers have tried to answer this question using different approaches--identification of ribosomal proteins and rRNA regions interacting with protein-synthesizing system components (tRNA, mRNA, translation factors, etc.) and dissection of changes in functional properties of the ribosome lacking one of its components. The 5S rRNA and L25 have not been found among the ribosomal RNAs and proteins contacting translational apparatus components [54, 74-85]. On the basis of these data, one may conclude that 5S rRNA and L25 protein (5S rRNA-binding domain of CTC protein) do not directly interact with translational apparatus components. At the same time, lack of 5S rRNA-protein complex during in vitro assembly of 50S subunit of E. coli led to dramatic consequences [65, 86]. Such a ribosomal subunit possessed basic morphological features of the intact 50S subunit and was able to associate with the ribosome [87] and cleave GTP [65]. However, 50S subunit lacking 5S rRNA-protein complex was unable to synthesize in vitro either artificial or natural polypeptide [65, 86]. Significant decrease in growth rate has been observed after inactivation of some of the 5S rRNA genes from the bacterial chromosome [88, 89]. The conclusion has been made that 5S rRNA is required for ribosome functioning. As to necessity of the individual 5S rRNA-binding proteins for assembly of functional bacterial ribosome, there was no unambiguous answer until recently. Among tens of studied spontaneous mutants lacking 1-2 ribosomal proteins, no mutants lacking one of the 5S rRNA-binding proteins have been found [90]. These data suggest that 5S rRNA-binding proteins are also required for assembly and functioning of the ribosome. However, direct evidence to support this assumption was absent until recently.
In 2007, we showed that the ribosomal proteins L5 and L18 are required for survival of E. coli cells [91]. Knockout of genes of these proteins in E. coli chromosome was lethal for cells. These results are in agreement with data showing that ribosomal proteins L5 and L18 are required for incorporation of 5S rRNA into the ribosomal subunit during in vitro assembly [92]. At the same time, it turned out that E. coli cells survive after knockout of gene of the ribosomal protein L25, though they grow slower than the parental strain [91]. Ribosomes lacking L25 were able to synthesize natural polypeptide both in vitro and in vivo, but the total amount of protein synthesized by these ribosomes was less than that synthesized by the control ribosomes. This difference is apparently responsible for the slow growth of ΔL25 strain. Thus, L25 protein is not essential for survival of E. coli cells but is nevertheless required for normal functioning of the translation apparatus. At the same time, normal growth of the mutant strain was restored by the expression of the gene of L25 protein or its homolog (CTC protein from B. subtilis) from a plasmid [91]. Ribosomes of the ΔL25 strain lacking only CTC family protein restore their activity after incorporation of such a protein in vivo.
Though there is no direct evidence of involvement of the CTC protein in certain steps of translation, the data presented in this chapter allows several suppositions concerning this. On one hand, CTC protein forms conservative contacts with 5S rRNA and L16 protein, while additional domains of TL5 of T. thermophilus and CTC of D. radiodurans form contacts with helix 38 of 23S rRNA. On the other hand, it follows from the results of crystallographic studies that the above-mentioned molecules (proteins L5 and L16 and helix 38 of 23S rRNA) interacting directly with 5S rRNA, are in touch with tRNA in the A- and P-sites [53, 54, 71, 72, 93]. Therefore, taking into account intermolecular contacts of CTC protein in the ribosome, one can propose that it participates in stabilization and normal work of these functional centers of the bacterial 50S ribosomal subunit. A similar assumption was made by Harms and colleagues [23], who have shown that the middle domain of the CTC protein from D. radiodurans interacts with helix 38 of 23S rRNA, while its C-terminus reaches the A-site of the 50S ribosomal subunit. In connection with the above-said, we would address the reader to the works of Bogdanov's group [35, 56-58]. On the base of results of cross-linking between 5S rRNA and domain II (GTPase-associated center, GAC) and domain V (peptidyl transferase center, PTC), they suggested that 5S rRNA might be an intermediary between these ribosomal functional centers. In consideration of the position of 5S rRNA in the ribosome and its intermolecular contacts revealed in the recent crystallographic studies [53, 54, 71, 72, 93], we suggest that 5S rRNA may also coordinate work of two tRNA-binding sites (the A-site and the P-site) of the 50S ribosomal subunit.
To complete this review, we would like to formulate the main conclusions and to say several words regarding perspectives of further investigations in this field. First, we believe that 5S rRNA is the main target for CTC proteins, which appeared in bacteria long ago. This indicates that the protein is required for efficient work of the translational apparatus in bacteria. At the same time, known examples of so-called “regressive evolution” of proteins of this family in Bacteria as well as absence of these proteins in Archaea and Eukarya suggest that there is an alternative way to reach the goal which is entrusted to CTC protein in most bacteria. Second, the position of CTC protein in the ribosome and its intermolecular contacts suggest that one of the possible functions this protein is stabilization of an important “structural junction” directly related to functioning of the A- and the P-sites. Of course, there are a number of unsolved questions regarding the role of the CTC protein in the life of bacterial cells. Some of them were touched on in the review. For example, it is unknown what role additional domains of CTC protein play in functioning of the bacterial translational apparatus and why some organisms have rejected them. What is the special function of the CTC protein of B. subtilis under stress and at early sporulation stages? Taking into account the experimental material already accumulated and novel methodological opportunities for researchers that have appeared during recent years, one may be sure that these and other questions regarding the role of the CTC protein in the functioning of the bacterial translational apparatus will be answered soon. Finally, we would like to dedicate several lines to a possible use of accumulated knowledge about CTC family proteins for applied purposes. As mentioned, CTC family proteins are characteristic feature of domain Bacteria and their main target is strictly conserved E-loop of the ribosomal 5S RNA. At the same time, knockout of gene of the L25 protein leads to significant depression of cell growth. Therefore, it seems reasonable to use either CTC proteins as such or their specific site on 5S rRNA as a cellular target for a new type of antimicrobial agents.
This work was supported by the Russian Foundation for Basic Research (grant No. 08-04-00459) and the Program of President of the Russian Federation for support of Leading Scientific Schools (NSh-751.2008.4).
REFERENCES
1.Lecompte, O., Ripp, R., Thierry, J. C., Moras, D.,
and Poch, O. (2002) Nucleic Acids Res., 30,
5382-5390.
2.Gryaznova, O. I., Davydova, N. L., Gongadze, G. M.,
Jonsson, B. H., Garber, M. B., and Liljas, A. (1996) Biochimie,
78, 915-919.
3.Benson, D. A., Karsch-Mizrashi, I., Lipman, D. J.,
Ostell, J., and Wheeler, D. L. (2006) Nucleic Acids Res.,
34, 16-20.
4.Frishman, D., Mokrejs, M., Kosykh, D.,
Kostenmuller, G., Kolesov, G., Zubrzycki, I., Gruber, C., Geier, B.,
Kaps, A., Albermann, K., Volz, A., Wagner, C., Fellenberg, M., Heumann,
K., and Mewes, H. W. (2003) Nucleic Acids Res., 31,
207-211.
5.Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi,
I., Appel, R. D., and Bairoch, A. (2003) Nucleic Acids Res.,
31, 3784-3788.
6.Haldenwang, W. G., and Losick, R. (1979)
Nature, 282, 256-260.
7.Haldenwang, W. G., and Losick, R. (1980) Proc.
Natl. Acad. Sci. USA, 77, 7000-7004.
8.Moran, C. P. Jh., Lang, N., Banner, C. D.,
Haldenwang, W. G., and Losick, R. (1981) Cell, 25,
783-791.
9.Moran, C. P. Jh., Lang, N., and Losick, R. (1981)
Nucleic Acids Res., 9, 5979-5990.
10.Haldenwang, W. G. (1995) Microbiol. Rev.,
59, 1-30.
11.Volker, U., Engelmann, S., Maul, B., Riethdorf,
S., Volker, A., Schmid, R., Mach, H., and Hecker, M. (1994)
Microbiology, 140, 741-752.
12.Hecker, M., and Volker, U. (1990) FEMS
Microbiol. Ecol., 74, 197-214.
13.Korepanov, A. P., Gongadze, G. M., and Garber, M.
B. (2004) Biochemistry (Moscow), 69, 607-611.
14.Schmalisch, M., Langbein, I., and Stulke, J.
(2002) J. Mol. Microbiol. Biotechnol., 4, 495-501.
15.Duche, O., Tremoulet, F., Glaser, P., and
Labadie, J. (2002) Appl. Environ. Microbiol., 68,
1491-1498.
16.Kazmierczak, M. G., Mithoe, S. C., Boor, K. J.,
and Wiedmann, M. (2003) J. Bacteriol., 185,
5722-5734.
17.Truitt, C. L., Weaver, E. A., and Haldenwang, W.
G. (1988) Mol. Gen. Genet., 212, 166-171.
18.Horne, J. R., and Erdmann, V. A. (1972) Mol.
Gen. Genet., 119, 337-344.
19.Gongadze, G. M., Tishchenko, S. V., Sedelnikova,
S. E., and Garber, M. B. (1993) FEBS Lett., 330,
46-48.
20.Dovgas, N. V., Markova, L. F., Mednikova, T. A.,
Vinokurov, L. M., Alakhov, Y. B., and Ovchinnikov, Y. A. (1975) FEBS
Lett., 53, 351-354.
21.Igo, M., Lampe, M., and Losick, R. (1988)
Genetics and Biotechnology of Bacilli (Ganesan, A. T., and Hoch,
J. A., eds.) Vol. 2, Academic Press, San Diego, CA, pp. 151-156.
22.Gongadze, G. M., Meshcheryakov, V. A., Serganov,
A. A., Fomenkova, N. P., Mudrik, E. S., Jonsson, B.-H., Liljas, A.,
Nikonov, S. V., and Garber, M. B. (1999) FEBS Lett., 451,
51-55.
23.Harms, J., Schluenzen, F., Zarivach, R., Bashan,
A., Gat, S., Agmon, I., Bartels, H., Franceschi, F., and Yonath, A.
(2001) Cell, 107, 679-688.
24.Korobeinikova, A. V., Gongadze, G. M., Korepanov,
A. P., Eliseev, B. D., Bazhenova, M. V., and Garber, M. B. (2008)
Biochemistry (Moscow), 73, 156-163.
25.Gongadze, G. M., Korepanov, A. P., Stolboushkina,
E. A., Zelinskaya, N. V., Korobeinikova, A. V., Ruzanov, M. V.,
Eliseev, B. D., Nikonov, O. S., Nikonov, S. V., Garber, M. B., and Lim,
V. I. (2005) J. Biol. Chem., 280, 16151-16156.
26.Deckert, G., Warren, P. V., Gaasterland, T.,
Young, W. G., Lenox, A. L., Graham, D. E., Overbeek, R., Snead, M. A.,
Keller, M., Aujay, M., Huber, R., Feldman, R. A., Short, J. M., Olsen,
G. J., and Swanson, R. V. (1998) Nature, 392,
353-358.
27.Newberry, V., and Garrett, R. A. (1980)
Nucleic Acids Res., 8, 4131-4142.
28.Stoldt, M., Wohnert, J., Gorlach, M., and Brown,
L. R. (1998) EMBO J., 17, 6377-6384.
29.Fedorov, R., Meshcheryakov, V., Gongadze, G.,
Fomenkova, N., Nevskaya, N., Selmer, M., Laurberg, M., Kristensen, O.,
Al-Karadaghi, S., Liljas, A., Garber, M., and Nikonov, S. (2001)
Acta Crystallogr. D, 57, 968-976.
30.Thompson, J. D., Higgins, D. G., and Gibson, T.
J. (1994) Nucleic Acids Res., 22, 4673-4680.
31.Chenna, R., Sugawara, H., Koike, T., Lopez, R.,
Gibson, T. J., Higgins, D. G., and Thompson, J. D. (2003) Nucleic
Acids Res., 31, 3497-3500.
32.Barciszewska, M. Z., Erdmann, V. A., and
Barciszewski, J. (1996) Biol. Rev. Camb. Phylos. Soc.,
71, 1-25.
33.Chen-Schmeisser, U., and Garrett, R. A. (1977)
FEBS Lett., 74, 287-291.
34.Stoffler, G., and Stoffler-Meilicke, M. (1984)
Ann. Rev. Biophys. Bioeng., 13, 303-330.
35.Bogdanov, A. A., Dontsova, O. A., Dokudovskaya,
S. S., and Lavrik, I. N. (1995) Biochem. Cell Biol., 73,
869-876.
36.Moore, P. B. (1996) Ribosomal RNA:
Structure, Evolution, Processing, and Function in
Protein Biosynthesis (Zimmermann, R., and Dahlberg, A., eds.) CRC
Press, Boca Raton-New York-London-Tokyo, pp. 199-236.
37.Douthwaite, S., Garrett, R. A., Wagner, R., and
Feunteun, J. (1979) Nucleic Acids Res., 6, 2453-2470.
38.Szymanski, M., Barciszewska, M. Z., Erdmann, V.
A., and Barciszewska, J. (2002) Nucleic Acids Res., 30,
176-178.
39.Kjems, J., Olesen, S. O., and Garrett, R. A.
(1985) Biochemistry, 24, 241-250.
40.Wrede, P., and Erdmann, V. A. (1977) Proc.
Natl. Acad. Sci. USA, 74, 2706-2709.
41.Dallas, A., and Moore, P. B. (1997)
Structure, 5, 1639-1653.
42.Correll, C. C., Freeborn, B., Moore, P. B., and
Steitz, T. A. (1997) Cell, 91, 705-712.
43.MacKey, R. M., Spencer, D. F., Schnare, M. N.,
Doolittle, W. F., and Gray, M. W. (1982) Can. J. Biochem.,
60, 480-489.
44.Lu, M., and Steitz, T. A. (2000) Proc. Natl.
Acad. Sci. USA, 97, 2023-2028.
45.Stoldt, M., Wohnert, J., Ohlenschlager, O.,
Gorlach, M., and Brown, L. R. (1999) EMBO J., 18,
6508-6521.
46.Nevskaya, N. A., Nikonov, O. S., Revtovich, S.
V., Garber, M. B., and Nikonov, S. V. (2004) Mol. Biol.
(Moscow), 38, 789-798.
47.Shatsky, I. N., Evstafieva, A. G., Bystrova, T.
F., Bogdanov, A. A., and Vasiliev, V. D. (1980) FEBS Lett.,
121, 97-100.
48.Stoffler-Meilicke, M., Stoffler, G., Odom, O. W.,
Zinn, A., Kramer, G., and Hardesty, B. (1981) Proc. Natl. Acad. Sci.
USA, 78, 5538-5542.
49.Evstafieva, A. G., Shatsky, I. N., Bogdanov, A.
A., and Vasiliev, V. D. (1982) FEBS Lett., 185,
57-62.
50.Stoffler-Meilicke, M., Noah, M., and Stoffler, G.
(1983) Proc. Natl. Acad. Sci. USA, 80, 6780-6784.
51.Lotti, M., Noah, M., Stoffler-Meilicke, M., and
Stoffler, G. (1989) Mol. Gen. Genet., 216, 245-253.
52.Ban, N., Nissen, P., Hansen, J., Moore, P. B.,
and Steitz, T. A. (2000) Science, 289, 905-920.
53.Schuwirth, B. S., Borovinskaya, M. A., Hau, C.
W., Zhang, W., Vila-Sunjurjo, A., Holton, J. M., and Cate, J. H. (2005)
Science, 310, 827-834.
54.Selmer, M., Dunham, C. M., Murphy, F. V. IV,
Weixlbaumer, A., Petry, S., Kelley, A. C., Weir, J. R., and
Ramakrishnan, V. (2006) Science, 313, 1935-1942.
55.Christiansen, J., Douthwaite, S. R.,
Christiansen, A., and Garrett, R. A. (1985) EMBO J., 4,
1019-1024.
56.Dontsova, O., Tishkov, V., Dokudovskaya, S.,
Bogdanov, A., Doring, T., Rinke-Appel, J., Thamm, S., Greuer, B., and
Brimacombe, R. (1994) Proc. Natl. Acad. Sci. USA, 91,
4125-4129.
57.Dokudovskaya, S., Dontsova, O., Shpanchenko, O.,
Bogdanov, A., and Brimacombe, R. (1996) RNA, 2,
146-152.
58.Sergiev, P., Dokudovskaya, S., Romanova, E.,
Topin, A., Bogdanov, A., Brimacombe, R., and Dontsova, O. (1998)
Nucleic Acids Res., 26, 2519-2525.
59.Osswald, M., and Brimacombe, R. (1999) Nucleic
Acids Res., 27, 2283-2290.
60.Osswald, M., Greuer, B., and Brimacombe, R.
(1990) Nucleic Acids Res., 18, 6755-6760.
61.Gongadze, G. M., Perederina, A. A.,
Meshcheryakov, V. A., Fedorov, R. V., Moskalenko, S. E., Rak, A. V.,
Serganov, A. A., Shcherbakov, D. V., Nikonov, S. V., and Garber, M. B.
(2001) Mol. Biol. (Moscow), 35, 521-526.
62.Szymkowiak, C., and Wagner, R. (1985) Nucleic
Acids Res., 13, 3953-3968.
63.Spierer, P., and Zimmermann, R. A. (1978)
Biochemistry, 17, 2474-2479.
64.Nierhaus, K. H., and Dohme, F. (1974) Proc.
Natl. Acad. Sci. USA, 71, 4713-4717.
65.Dohme, F., and Nierhaus, K. H. (1976) Proc.
Natl. Acad. Sci. USA, 73, 2221-2225.
66.Nissen, P., Ippolito, J. A., Ban, N., Moore, P.
B., and Steitz, T. A. (2001) Proc. Natl. Acad. Sci. USA,
98, 4899-4903.
67.Klein, D. J., Moore, P. B., and Steitz, T. A.
(2004) J. Mol. Biol., 340, 141-177.
68.Moore, V. G., Atchison, R. E., Thomas, G., Moran,
M., and Noller, H. F. (1975) Proc. Natl. Acad. Sci. USA,
72, 844-848.
69.Hampl, H., Schulze, H., and Nierhaus, K. H.
(1981) J. Biol. Chem., 256, 2284-2288.
70.Kazemie, M. (1976) Eur. J. Biochem.,
67, 373-378.
71.Yusupov, M. M., Yusupova, G. Z., Baucom, A.,
Lieberman, K., Earnest, T. N., Cate, J. H., and Noller, H. F. (2001)
Science, 292, 883-896.
72.Nissen, P., Hansen, J., Ban, N., Moore, P. B.,
and Steitz, T. A. (2000) Science, 289, 920-930.
73.Nishimura, M., Yoshida, T., Shirouzu, M., Terada,
T., Kuramitsu, S., Yokoyama, S., Ohkudo, T., and Kobayashi, Y. (2004)
J. Mol. Biol., 344, 1369-1383.
74.Stark, H., Rodnina, M. V., Rinke-Appel, J.,
Brimacombe, R., Wintermeyer, W., and van Heel, M. (1997) Nature,
389, 403-406.
75.Wilson, K. S., and Noller, H. F. (1998)
Cell, 92, 131-139.
76.Wilson, K. S., Ito, K., Noller, H. F., and
Nakamura, Y. (2000) Nat. Struct. Biol., 7, 866-870.
77.Stark, H., Rodnina, M. V., Wieden, H. J., Zemlin,
F., Wintermeyer, W., and van Heel, M. (2002) Nat. Struct. Mol.
Biol., 9, 849-854.
78.Lancaster, L., Kiel, M. C., Kaji, A., and Noller,
H. F. (2002) Cell, 111, 129-140.
79.Marzi, S., Knight, W., Brandi, L., Caserta, E.,
Soboleva, N., Hill, W. E., Gualerzi, C. O., and Lodmell, J. S. (2003)
RNA, 9, 958-969.
80.Klaholz, B. P., Myasnikov, A. G., and van Heel,
M. (2004) Nature, 427, 862-865.
81.Wilson, K. S., and Nechifor, R. (2004) J. Mol.
Biol., 337, 15-30.
82.Scarlett, D.-J. G., McCaughan, K. K., Wilson, D.
N., and Tate, W. P. (2005) J. Biol. Chem., 278,
15095-15104.
83.Wilson, D. N., Schluenzen, F., Harms, J. M.,
Yoshida, T., Ohkubo, T., Albrecht, R., Buerger, J., Kobayashi, Y., and
Fucini, P. (2005) EMBO J., 24, 251-260.
84.Petry, S., Brodersen, D. E., Murphy, F. V. IV,
Dunham, C. M., Selmer, M., Tarry, M. J., Kelley, A. C., and
Ramakrishnan, V. (2005) Cell, 123, 1255-1266.
85.Weixlbaumer, A., Petry, S., Dunham, C. M.,
Selmer, M., Kelley, A. C., and Ramakrishnan, V. (2007) Nat. Struct.
Mol. Biol., 14, 733-737.
86.Erdmann, V. A., Fahnestock, S., Higo, K., and
Nomura, M. (1971) Proc. Natl. Acad. Sci. USA, 68,
2932-2936.
87.Selivanova, O. M., Gongadze, G. M., Gudkov, A.
T., and Vasiliev, V. D. (1986) FEBS Lett., 197,
79-83.
88.Ammons, D., Rampersad, J., and Fox, G. E. (1999)
Nucleic Acids Res., 27, 637-642.
89.Ammons, D., and Rampersad, J. (2001) Curr.
Microbiol., 43, 89-92.
90.Dabbs, E. R. (1991) Biochimie, 73,
639-645.
91.Korepanov, A. P., Gongadze, G. M., Garber, M. B.,
Court, D. L., and Bubunenko, M. G. (2007) J. Mol. Biol.,
366, 1199-1208.
92.Rohl, R., and Nierhaus, K. H. (1982) Proc.
Natl. Acad. Sci. USA, 79, 729-733.
93.Bashan, A., Agmon, I., Zarivach, R., Schluenzen,
F., Harms, J., Berisio, R., Bartels, H., Franceschi, F., Auerbach, T.,
Hansen, H. A. S., Kossoy, E., Kessler, M., and Yonath, A. (2003)
Mol. Cell, 11, 91-102.