REVIEW: D-Amino Acid Oxidase: Physiological Role and Applications
S. V. Khoronenkova1,2 and V. I. Tishkov1,3*
1Chemistry Faculty, Lomonosov Moscow State University, 119992 Moscow, Russia; E-mail: vitishkov@gmail.com; vit@enz.chem.msu.ru2Innovations and High Technologies MSU Ltd., 109559 Moscow, Russia
3Bach Institute of Biochemistry, Russian Academy of Sciences, Leninsky pr. 32, 119071 Moscow, Russia
* To whom correspondence should be addressed.
Received June 27, 2008; Revision received August 13, 2008
D-Amino acids play a key role in regulation of many processes in living cells. FAD-dependent D-amino acid oxidase (DAAO) is one of the most important enzymes responsible for maintenance proper level of D-amino acids. The most interesting and important data for regulation of the nervous system, hormone secretion, and other processes by D-amino acids as well as development of different diseases under changed DAAO activity are presented. The mechanism of regulation is complex and multi-parametric because the same enzyme simultaneously influences the level of different D-amino acids, which can result in opposing effects. Use of DAAO for diagnostic and therapeutic purposes is also considered.
KEY WORDS: D-amino acid oxidase, physiological role, practical applicationDOI: 10.1134/S0006297908130105
Abbreviations: 7-ACA, 7-aminocephalosporanic acid; DAAO, D-amino acid oxidase; NMDA, N-methyl-D-aspartate; MTOB, 4-methylthio-2-oxobutyrate; pkDAAO, TvDAAO, and RgDAAO, D-amino acid oxidases from porcine kidney, Trigonopsis variabilis, and Rhodotorula gracilis, respectively.
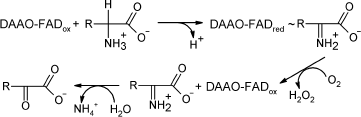
D-Amino acid oxidase (EC 1.4.3.3, DAAO) is a FAD-containing enzyme that
catalyzes oxidative deamination of D-amino acids yielding hydrogen
peroxide and an imino acid. The latter is further non-enzymatically
hydrolyzed to an α-keto acid and ammonium (see Scheme).
Scheme
A major characteristic of all DAAOs is their high specificity towards D-isomers of amino acids: they are almost inactive towards the corresponding L-isomer.
DAAO was first described by Krebs in 1935 [1]. The enzyme is widespread in nature, from microorganisms to mammals. The latter have DAAO localized in various tissues, e.g. brain, kidney, and liver. The presence of DAAO in liver depends on the species: mouse has DAAO in kidney only, while pig has both, in liver and kidney. Until 1980s, no systematic studies on DAAO of vertebrates were performed because of their low content in tissues and low stability. Microbial DAAO, except that of the yeast Trigonopsis variabilis, had no any practical importance. All studies were focused on the enzymes from porcine kidney (pkDAAO) and T. variabilis (TvDAAO). The pkDAAO was studied as a model FAD-containing enzyme, while TvDAAO was used to develop biocatalysts for the production of 7-aminocephalosporanic acid (7-ACA) from the natural antibiotic cephalosporin C. However, low operational stability restricted the use of the biocatalyst.
In the mid 1990s, interest in DAAO was revived due to a number of factors. First, an import physiological role of DAAO in metabolic control became appreciated; second, development of genetic engineering resulted in the construction of recombinant strains producing DAAO and thus, the problem of enzyme production in commercial quantities has been solved; and third, the genomic era opened the opportunity for identification and sequencing of novel DAAO genes. A number of reviews on DAAO catalytic mechanism and structure have been published in the past five years [2, 3]. The last review on the properties of microbial DAAOs was published in 2008 [4]. However, a review summarizing information about the physiological role of D-amino acids and DAAOs together with enzyme application trends for analysis and medical diagnostics was missing. In this review, we attempt to fill in this information gap and to analyze the most recent data in the field.
PHYSIOLOGICAL ROLE OF D-AMINO ACIDS IN THE EUKARYOTIC CELL AND
REGULATION OF THEIR LEVEL WITH D-AMINO ACID OXIDASE
DAAOs provide microorganisms with exogenous D-amino acids as a source of carbon, nitrogen, and energy [5]. In eukaryotic cells, DAAO supports the physiological level of D-amino acids that play an important role in the regulation of many processes such as aging, neural signaling, hormone secretion, etc. Changes in the levels of D-amino acids have a major impact on the organism as a whole. For instance, excess of some D-amino acids in mouse brain tissues provides long-term potentiation1 in the hippocampus and supports spatial learning. Experiments were performed on wild-type and daao knock-out mice in the Morris labyrinth [6], which demonstrated progressive learning of the mutant mouse compared to the control. Other important examples on the participation of DAAO in vertebrate physiology are discussed below.
1 Long-term potentiation (LTP) is an increase of synaptic transmission between two neurons maintained for a long time after the effect on the synaptic pathway. LTP participates in mechanisms of synaptic plasticity determining the adaptability of the nervous system of living organisms to the changing environment. Regulation of D-serine level. Nearly half of the research articles on DAAO published in the last 5-7 years have been devoted to the role of D-serine as a neuromodulator of NMDA (N-methyl-D-aspartate)-receptors, which are at play in many pathological processes. The allele combination of G72 and daao genes increases the probability of developing schizophrenia [7-9]. The rise in expression levels of G72 [10] increases DAAO activity in human brain, resulting in the decreasing levels of D-serine, which is known to bind to the glycine-binding site of NMDA-receptors [11]. Therefore, the decreased levels of D-serine lower the functional activity of NMDA-receptors, which is supposed to be one of the reasons for the development of schizophrenia [12-15]. Currently, this is a working hypothesis supported by many researchers [16]. It should be noted that NMDA-receptors are equally important for other physiological processes such as learning and memory formation [17], development of epilepsy [18], etc. At the same time, other researchers could not find a correlation between daao and pGL72 protein gene polymorphism and predisposition to schizophrenia [19, 20].
Regulation of hormone secretion. D-Aspartate is one of the most important regulators of hormone secretion. It is present in significant quantities in brain tissues, and its concentration increases with aging by up to 0.14% per year [21, 22]. The highest concentration of D-Asp is observed in the secreting glands [23, 24]. D-Asp regulates the secretion of melatonin [25], prolactin [26], testosterone [27], luteinizing hormone, and growth hormone [28]. The content of D-Asp in crystalline, dentin, knee cartilage, and white matter increases with aging [29-32].
Regulation of arterial hypertension. NG-Nitroarginine exists in the cell as a mixture of L- and D-isomers: initially, NG-nitro-D-Arg is synthesized, and then it is converted into the L-form. Both compounds are relevant to arterial hypertension, but the organism reacts much less and slower to the D-isomer than to the L-form [33, 34]. Thorough studies showed that NG-nitro-D-Arg effectiveness is based on its potent inhibition of NO synthase [35], a key regulator of intercellular processes including arterial hypertension [36]. Prolonged action of NG-nitro-D-Arg is provided by its slow racemization in kidney. One of the key enzymes catalyzing its racemization is the kidney DAAO [37].
Regulation of D-alanine level. Significant levels of D-Ala have been found in the tissues of Crustacea and Bivalvia. Under conditions of salt stress, this amino acid accumulates in all tissues. Abe et al. [38] proposed that D-Ala plays an important role in the regulation of intracellular osmotic pressure. The content of D-Ala in the gray matter of Alzheimer's patients is 2.2 times higher than in control groups [39]. In addition to D-Ala, the patients show increased content of free D-Asp and D-Ser and overall D-amino acids in the spinal fluid [40]. These observations are explained in terms of a combined effect of decreased DAAO activity and increased activity of the corresponding racemases compared to the control groups.
Regulation of D-proline level. D-Pro and D-Leu content is in the fourth place, after D-Ser, D-Asp, and D-Ala [41, 42]. Currently, the physiological role of D-Pro is actively discussed in the literature. In long-living tissues such as dentine, tooth enamel, crystalline, etc., there is a clear parallel between the human age and content of D-oxyproline and D-aspartate [22, 31]. Data on neuro-, hepato-, and nephrotoxicity of D-proline in rats is a focus of discussion [43, 44]. Studies on D-Pro content in different tissues of daao knock-out mice show that the latter accumulate significant amounts of D-Pro in the kidneys, which is then cleared naturally [45].
PRACTICAL APPLICATION OF D-AMINO ACID OXIDASE
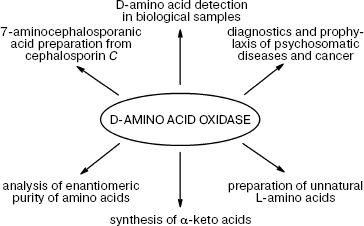
DAAO is finding more and more practical applications (Fig. 1).
Quantification of D-amino acids and DAAO assay. The important role played by D-amino acids in metabolism is now well established, i.e. their content as well the activity of DAAO changes in response to various pathological conditions in brain tissues, serum, and spinal fluid. Therefore, DAAO assay and quantification of the levels of D-amino acids is an important problem whose solution may help tremendously in early diagnostics and disease monitoring. However, only recently some efforts in this direction have been reported: two papers were published in 2008. The first by Pernot et al. describes the development of a micro-biosensor for determination of D-serine in vivo [42]: the biosensor (25 × 150 µm) is a cylindrical platinum microelectrode covered by a layer of poly-m-phenylenediamine, a selective mediator for H2O2. The enzyme from Rhodotorula gracilis (RgDAAO) and pkDAAO for comparison were physically adsorbed on the electrode surface. For in vivo experiments, the enzyme layer was protected with an additional Nafion membrane.Fig. 1. Practical application of D-amino acid oxidase.
The biosensor based on RgDAAO exhibited higher sensitivity for D-Ser (9.2 pA/mM in the range of 0.1-500 µM) compared to that for pkDAAO (6.5 pA/mM). The theoretical detection limit for D-Ser for the former was 16 nM. The major advantage of the former was the selective detection of D-Ser originated from the substrate specificity of RgDAAO: the ratio of the activities towards D-Ser and the other D-amino acids was 100 : 5.5 and 100 : 104 for D-Ser/D-Asp and D-Ser/D-Ala pairs, respectively. In the case of pkDAAO the corresponding activity ratios were 100 : 36.8 and 100 : 165 for D-Ser/D-Asp and D-Ser/D-Ala, respectively. Of note, the equal RgDAAO activities towards D-Ser and D-Ala restrict the selectivity of D-Ser detection in the presence of equal concentrations of D-Ala. High selectivity of D-Ser in vivo detection is the result of the low content of D-Ala in brain tissues, i.e. 7% of that of D-Ser content. Obviously, in the case of pathologies resulting in increased levels of D-Ala and decreased levels of D-Ser, the correct determination of the concentration of D-Ser will not be possible. For these conditions, one has to develop an enzyme that has no activity towards D-Ala and D-Asp.
We have performed site-directed mutagenesis of the T. variabilis enzyme (TvDAAO) [46]. The wild-type enzyme was known to be inactive towards D-Asp, being however very active with D-Ala [47]. Our studies resulted in the construction of three mutant forms inactive towards D-Ala. In addition, one of the mutants lost activity towards D-Pro, which also has a tendency to accumulate under pathological conditions (see above). Compared to the wild-type enzyme, two mutants show no decrease in the specific activity towards D-Ser, while one demonstrated a 30% increase. Thus, the created mutant forms of TvDAAO are perfectly suitable for selective determination of D-Ser in excess of D-Ala, D-Asp, and D-Pro [48].
Broad substrate specificity of DAAO is a plus when considering the determination of overall concentration of D-isomers of amino acids in food and drinks. D-Amino acids, and in particular D-Ala, are components of bacterial cell walls. Their appearance in foods is a marker of bacterial contamination [49]. A detailed discussion on previously published research (to 2006) can be found in [4]. In 2007, an amperometric biosensor with DAAO immobilized on a graphite electrode whose surface had been modified with Prussian blue was described [50]. To enhance the electrode response, the surface of the electrode was additionally modified by carbon nanotubes. The magnitude of the registered signal was linearly dependent on D-Ala concentration in the range of 5 to 200 µM.
A number of methods have been developed for detection of DAAO activity [51-61]. Most are based on the coupling of the DAAO-catalyzed reaction with that of horseradish peroxidase [52]. The great variety of peroxidase substrates allows various types of assay to be performed: UV-Vis [56] and fluorimetric [60], in gel [52, 53], and directly in tissue slices [57, 58]. Very recently, a highly sensitive DAAO assay in complex mixtures, including screening of recombinant libraries of E. coli, has been developed [61]. The sensitivity of detection was as low as 0.13 ng per sample (1.6⋅10-15 mole).
Diagnostics and prophylaxis of psychosomatic diseases and cancer. As discussed above, some psychosomatic diseases such as schizophrenia and Alzheimer's and Parkinson's diseases result in significant changes in the levels of some D-amino acids (D-Ser, D-Ala, D-Asp) in serum, white and gray matters, and spinal fluid. The determination of D-amino acids under normal and pathological conditions created a basis for the development of diagnostic criteria and monitoring of these diseases [39, 40, 62-64].
Studies on daao knock-out mice demonstrated the absence of compensatory effects for the null DAAO activity. This observation opens new horizons for the treatment of schizophrenia with enzyme inhibitors whose application will normalize levels of D-Ser to stimulate NMDA-receptors [65] (for comparison, a common approach is direct injections of D-Ser in addition to anti-psychic prophylaxis [66-68]). To realize the new approach, the authors developed a high throughput screening protocol for cell-based DAAO assay [69]: the primary screen resulted in the selection of 1966 hits for further studies. The others, based on a similar approach, came up with 5-methylpyrazol-3-carbonic acid [70] as a drug candidate. Its injection into rats results in significant increase of D-Ser in cerebral cortex and midbrain; moreover, its continuous injection for 4 weeks gave no side effects.
Dopamine is a well-known prophylaxis and treatment medication for Parkinson's disease [71]. Very recently, it has been shown that D-3,4-dihydroxyphenylalanine (D-DOPA) is a better substrate for human DAAO than D-Ser [72]. The catalytic efficiency (kcat/Km) for D-DOPA is 14-fold higher than that for D-Ser. Thus, human DAAO provides an alternate metabolic route for D-DOPA conversion into dopamine.
Another promising application for DAAO is its relevance to cancer diagnostics and possibly treatment. It has been demonstrated that parenteral injection of D-amino acid-containing solutions into cancer patients results in improved nutritional status and inhibits cancer cell growth [73, 74]. Later, research performed on rats showed no DAAO activity in cancer cells [75].
Japanese authors [76, 77] proposed polyethylene conjugated DAAO for treatment of cancer. Accumulation of injected pkDAAO in the tumor has been shown in mice, thus showing the targeted delivery of the anticancer agent. The mechanism of exogenously administered DAAO into cancer cells only is unknown. One can speculate that higher metabolic level of cancer cells results in increased delivery rates of any compound. “Targeted delivery” of this type has been described for many compounds. Administration of the enzyme followed by the injection of D-proline (which is the best substrate for pkDAAO) resulted in significant inhibition of the cancer cell growth because of increased production of hydrogen peroxide and other products of oxidative metabolism in cancer cells, while not affecting the metabolism in healthy cells. To prolong DAAO lifetime in the body, the enzyme was attached to polyethylene glycol.
The next step was the use of recombinant pkDAAO expressed in E. coli [78]. Its high efficiency as an anticancer treatment in combination with D-Pro was shown for various cancer cell lines and models. It has also been demonstrated that the activity of catalase, which degrades peroxide, is 1-2 orders of magnitude lower in cancer cells than in healthy cells. Thus, targeted delivery of DAAO into cancer cells combined with the much lower activity of catalase increases the burden of cytotoxic compounds in cancer cells specifically. Therefore, stable DAAO conjugates can be considered as promising anticancer treatments in the future.
To conclude this chapter, we must say that the development of medical treatment for neurodegeneration and cancer based on DAAO regulation and monitoring of D-amino acid content is not a trivial task, mainly because of the multiple roles played by DAAO in the cell. One and the same enzyme changes the levels of various D-amino acids, which in turn exhibit different effects depending on the amino acid nature and place in metabolism. For example, increase in D-Ser stimulates NDMA-receptors, while increase in D-Ala is linked to Alzheimer's disease. Tissue-specific gene therapy could be the solution for this uncertainty in predicting the drug effects. Tissue-specific mutagenesis of DAAO resulting in the desired substrate specificity profiles in a particular tissue will allow one to selectively affect only one D-amino acid content, without changes in the enzyme biosynthesis levels. This approach may be realized in the near future: (i) the possibility of tissue-specific effects on the DAAO gene was demonstrated in mice [45], and (ii) our results show that site-directed mutagenesis yield desired mutant forms with the necessary profile of substrate specificity.
Synthesis of physiologically active compounds. As noted above, DAAO is highly specific to D-isomers of amino acids. Depending on the task to be solved, this particular property of the enzyme can be modulated to produce a racemic mixture or the major reaction product, α-keto acids, or natural or non-natural L-amino acids of high optical purity (ee > 99). Both α-keto acids and non-natural L-amino acids are widely used as precursors for various drugs.
Pig kidney DAAO was first used in 1971 to make L-pipecolic acid from a racemic mixture via D-isomer oxidation [79]. L-Pipecolic acid is an intermediate of L-Lys catabolism and accumulates in the organism as a result of peroxisome dysfunction [80]. This amino acid plays a very important role in the organism, and particularly in the development of various diseases of the nervous system [81]. L-Pipecolic acid derivatives are components of medications used as immunodepressants and anticancer and neuroprotective drugs [82]. Later, pkDAAO was replaced with a more stable and active enzymes from the yeasts T. variabilis [83, 84] and Rhodotorula gracilis [85]. TvDAAO was also used to produce optically active L-methionine from D-amino acid with the yield of 100% in a cascade system of four enzymes [86]. Production of phenyl pyruvate from D-phenylalanine using recombinant TvDAAO and RgDAAO with the yield of 99% is described in [87, 88]. Ionic liquids were used to increase the substrate solubility and oxygen concentration [89].
4-Methylthio-2-oxobutyric acid (MTOBA) is an important compound in pharmaceutics. Being a metabolic precursor of methional, a strong inducer of apoptosis, MTOBA is used as an anticancer drug. In methionine-dependent cancer cells, the MTOBA content is lower than in normal cells. MTOBA can be easily produced via oxidative deamination of D-methionine. Very recently, a more cost-effective method for MTOBA production was proposed based on the treatment of a racemic mixture of D,L-methionine [90]. D-Methionine conversion in the reaction was ~98%.
Another important substance produced with the help of DAAO is L-6-hydroxynorleucine. This chiral compound is used for the synthesis of Omapatrilat and a whole series of metalloproteinase inhibitors [91]. Omapatrilat is an inhibitor of angiotensin-converting enzyme and neutral endopeptidase and is effective as an antihypertensive drug. The method for L-6-hydroxynorleucine stereo-isomer production from a racemic mixture of 6-hydroxynorleucine with the yield of 97% and purity of 98% is described in [92].
L-Amino acids containing the naphthalene group are of great interest for new drug development. For instance, L-2-naphthylalanine is a component of the peptide drug Nafarelin [93]. To produce this amino acid from a racemate of 2-naphthylalanine, a method based on a three-enzyme system, e.g. RgDAAO, catalase, and L-aspartate aminotransferase, was developed [94]. To increase the RgDAAO specific activity toward D-2-naphthylalanine, both site-directed mutagenesis [95] and random mutagenesis (“directed evolution”) methods were used [95], but in the latter case the best multi-point mutants were less active than single-point mutant RgDAAO Met213Gly generated using the former approach [96].
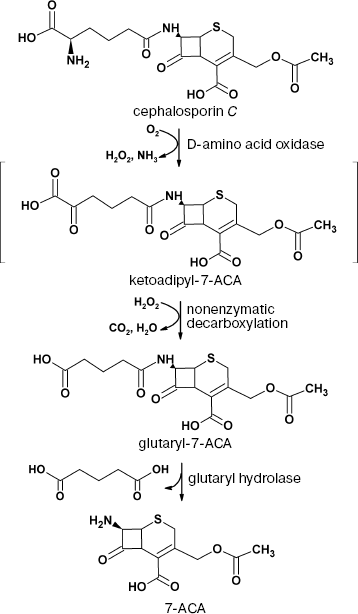
Cephalosporin C conversion. Development of antibiotic resistance is a well-known property of microorganisms [97]. Resistance toward penicillin-based antibiotics made cephalosporin-based antibiotics of different generations the most popular ones used in practice: more than half of antibiotics used worldwide are cephalosporins. 7-Aminocephalosporanic acid (7-ACA) is a starting compound for the production of various semi-synthetic cephalosporins of different generations, and until recently it has been produced by chemical hydrolysis of the natural antibiotic cephalosporin C. The disadvantage of this method is multiple steps, low yield, and use of various organic solvents.
Alternatively, 7-ACA can be produced using a biocatalytic method based on DAAO and glutaryl hydrolase. The scheme of the process is shown in Fig. 2.
Currently, there are many various methods for biocatalytic oxidation of cephalosporin C. Detailed analysis of these methods can be found in [4, 98]. Analysis proves that being relatively expensive, the enzyme does not exhibit the operational stability required for commercial production.Fig. 2. Production of 7-aminocephalosporanic acid (7-ACA) from cephalosporin C using the two-enzyme system D-amino acid oxidase/glutaryl hydrolase.
The solution of this problem could be based on the development of enzyme mutant forms with improved catalytic activity and thermal and operational stability. Such mutant forms of TvDAAO have recently been produced in our laboratory [46, 48]. Two forms among all that were produced exhibited a 4- and 2-fold higher catalytic effectiveness with respect to cephalosporin C compared to the wild-type enzyme. Half-inactivation period for one of the mutants at 54°C was 2.2 times longer than for the original DAAO [99]. Of note, the effect was achieved using single-point mutations based on the structural model build in our laboratory [3]. In addition, methods for recombinant TvDAAO immobilization with activity yield of 80% have been developed; for comparison, the best analogs described in the literature provide no more than 30% activity yield upon immobilization.
STRUCTURAL STUDIES OF DAAO FROM Trigonopsis
variabilis
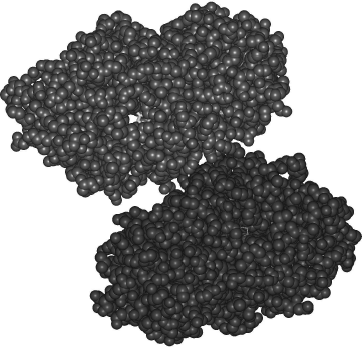
One of the single-point mutants of TvDAAO was crystallized and the structure was solved at 1.8 Å resolution. Of note, currently only three DAAO crystal structures are available: pig kidney [100], R. gracilis [101], and human [102], the latter appearing in 2006 at resolution of 2.5 Å. In the case of wild-type TvDAAO, many attempts during the last 30 years have given no enzyme crystals suitable for structural analysis.
The structure of mutant TvDAAO is shown in Fig. 3. Like R. gracilis DAAO, TvDAAO is a dimer; however, the interface between the subunits is 20% smaller than that for RgDAAO. The active site has two entries and has a significant volume to bind bulky molecules such as cephalosporin C. The upper subunit in Fig. 3 has a hollow channel with a clearly seen portion of an FAD molecule (shown in gray color). Detailed analysis of the enzyme structure-activity relationship can be found in [99]. The solution of crystal structure of TvDAAO gives an instrument to analyze the structure-function relationships of the enzyme by means of site-directed mutagenesis and construction of new enzyme forms with desired properties.
Analysis of the literature on D-amino acid oxidase shows that the interest in this enzyme is continuously growing: the number of publications on its physiological roles and practical applications rises by 15-20% a year. Significant success in fundamental studies on DAAO has been achieved in the last five years. In addition to the crystal structures of pkDAAO and RgDAAO, two other very important enzymes, human and T. variabilis DAAO, have had their structures resolved in 2006 and 2008. Genomic and proteomic approaches allow a completely new level of research on physiological roles of DAAO. From the practical point of view, more biosensors based on mutant DAAO with specifically designed profile of substrate specificity will appear. The major requirement for the realization of these ambitious plans will be the development of approaches to govern the changes in DAAO properties; it is likely that covalently attached FAD will become the first priority, as has happened in the case of other oxidases.Fig. 3. Dimeric structure of mutant D-amino acid oxidase from T. variabilis.
This work was supported by the Russian Foundation for Basic Research (grant 08-04-01703) and Federal Agency for Science and Innovations (State contract No. 02.512.12.2002).
REFERENCES
1.Krebs, H. A. (1935) Biochem. J., 29,
1620-1644.
2.Pilone, M. S. (2000) Cell. Mol. Life Sci.,
57, 1732-1747.
3.Tishkov, V. I., and Khoronenkova, S. V. (2005)
Biochemistry (Moscow), 70, 40-54.
4.Pollegioni, L., Molla, G., Sacchi, S., Rosini, E.,
Verga, R., and Pilone, M. S. (2008) Appl. Microbiol.
Biotechnol., 78, 1-16.
5.La Rue, T. A., and Spencer, J. F. T. (1967) Can.
J. Microbiol., 13, 777-788.
6.Maekawa, M., Watanabe, M., Yamaguchi, S., Konno,
R., and Hori, Y. (2005) Neurosci. Res., 53, 34-38.
7.Schumacher, J., Abon Jamra, R., Freudenberg, J.,
Becker, T., Ohlraun, S., Otte, A. C. J., Tullius, M., Kovalenko, S.,
van den Bogaert, A., Maier, W., Rietschel, M., Propping, P., Nothen, M.
M., and Cichon, S. (2004) Mol. Psychiatry, 9,
203-207.
8.Chumakov, I., Blumenfeld, M., Guerassimenko, O.,
Cavarec, L., Palicio, M., Abderrahim, H., Bougueleret, L., Barry, C.,
Tanaka, H., la Rosa, P., Puech, A., Tahri, N., Cohen-Akenine, A.,
Delabrosse, S., Lissarrague, S., Picard, F.-P., Maurice, K., Essioux,
L., Millasseau, P., Grel, P., Debailleul, V., Simon, A.-M., Caterina,
D., Dufaure, I., Malekzadeh, K., Belova, M., Luan, J.-J., Bouillot, M.,
Sambucy, J.-L., Primas, G., Saumier, M., Boubkiri, N., Martin-Saumier,
S., Nasroune, M., Peixoto, H., Delaye, A., Pinchot, V., Bastucci, M.,
Guillou, S., Chevillon, M., Sainz-Fuertes, R., Meguenni, S.,
Aurich-Costa, J., Cherif, D., Gimalac, A., van Duijn, C., Gauvreau, D.,
Ouellette, G., Fortier, I., Raelson, J., Sherbatich, T., Riazanskaia,
N., Rogaev, E., Raeymaekers, P., Aerssens, J., Konings, F., Luyten, W.,
Macciardi, F., Sham, P. C., Straub, R. E., Weinberger, D. R., Cohen,
N., and Cohen, D. (2002) Proc. Natl. Acad. Sci. USA, 99,
13675-13680.
9.Owen, M. J., Craddock, N., and O'Donovan, M. C.
(2005) Trends Genet., 21, 518-525.
10.Korostishevsky, M., Kaganovich, M., Cholostoy,
A., Ashkenazi, M., Ratner, Y., Dahary, D., Bening-Abu-Shach, U., Ben
Asher, E., Lancet, D., Ritsner, M., and Navon, R. (2004) Biol.
Psychiatry, 56, 169-176.
11.Nishikawa, T. (2005) Biol. Pharm. Bull.,
28, 1561-1565.
12.Cloninger, C. R. (2002) Proc. Natl. Acad. Sci.
USA, 99, 13365-13367.
13.Harrison, P. J., and Owen, M. J. (2003)
Lancet, 361, 417-419.
14.Hashimoto, A., Yoshikawa, M., Andoh, H., Yano,
H., Matsumoto, H., Kawaguchi, M., Oka, T., and Kobayashi, H. (2007)
Eur. J. Pharmacol., 555, 17-22.
15.Corvin, A., Donohoe, G., McGhee, K., Murphy, K.,
Kenny, N., Schwaiger, S., Nangle, J. M., Morris, D., and Gill, M.
(2007) Neurosci. Lett., 426, 97-100.
16.MacDonald, A. W., and Chafee, M. V. (2006)
Dev. Psychopathol., 18, 853-876.
17.Collingridge, G. (1987) Nature,
330, 604-605.
18.Meldrum, B. S., Akbar, M. T., and Chapman, A. G.
(1999) Epilepsy Res., 36, 189-204.
19.Chung, S., Jung, J., Chung, H. Y., Yoo, H. K.,
Kim, C. Y., Joo, Y. H., Choi, S. E., and Hong, J. P. (2007)
Psychiatr. Genet., 17, 313-319.
20.Vilella, E., Costas, J., Sanjuan, J., Guitart,
M., de Diego, Y., Carracedo, A., Martorell, L., Valero, J., Labad, A.,
de Frutos, R., Najera, C., Molto, M. D., Toirac, I., Guillamat, R.,
Brunet, A., Valles, V., Perez, L., Leon, M., de Fonseca, F. R.,
Phillips, C., and Torres, M. (2008) J. Psychiatr. Res.,
42, 278-288.
21.Man, E. H., Fisher, G. H., Payan, I. L.,
Cadilla-Perezrios, R., Garcia, N. M., Chemburkar, R., Arends, G., and
Frey, W. H. (1987) J. Neurochem., 48, 510-515.
22.Poinar, H. N., Hoss, M., Bada, J. L., and Paabo,
S. (1999) Science, 272, 864-866.
23.Dunlop, D. S., Neidle, A., McHale, D., Dunlop, D.
M., and Lajtha, A. (1986) Biochem. Biophys. Res. Commun.,
141, 27-32.
24.Furuchi, T., and Homma, H. (2005) Biol. Pharm.
Bull., 28, 1566-1570.
25.Takigawa, Y., Homma, H., Lee, J.-A., Fukushima,
T., Santa, T., Iwatsubo, T., and Imai, K. (1998) Biochem. Biophys.
Res. Commun., 248, 641-647.
26.D'Aniello, G., Tolino, A., D'Aniello, A., Errico,
F., Fisher, G. H., and di Fiore, M. M. (2000) Endocrinology,
141, 3862-3870.
27.D'Aniello, A., di Cosmo, A., di Cristo, C.,
Annunziato, L., Petrucelli, L., and Fisher, G. (1996) Life Sci.,
59, 97-104.
28.D'Aniello, A., di Fiore, M. M., Fisher, G. H.,
Milone, A., Seleni, A., D'Aniello, S., Perna, A. F., and Ingrosso, D.
(2000) FASEB J., 14, 699-714.
29.Helfman, P. M., and Bada, J. L. (1975) Proc.
Natl. Acad. Sci. USA, 72, 2891-2894.
30.Man, E. H., Sandhouse, M., Burg, J., and Fisher,
G. H. (1983) Science, 220, 1407-1408.
31.Ohtani, S., Matsushima, Y., Ohira, H., and
Watanabe, A. (1995) Growth Dev. Aging, 59, 55-61.
32.Fisher, G., Lopez, S., Peterson, K., Goff, T.,
Philip, I., Gaviria, R., Lorenzo, N., and Tsesarskaia, M. (2007)
Amino Acids, 32, 27-30.
33.Wang, Y. X., Zhou, T., and Pang, C. C. (1991)
Eur. J. Pharmacol., 200, 77-81.
34.Wang, Y. X., Poon, C. I., and Pang, C. C. (1993)
J. Pharmacol. Exp. Ther., 265, 112-119.
35.Wang, Q., Cwik, M., Wright, C. J., Cunningham,
F., and Pelligrino, D. A. (1999) J. Pharmacol. Exp. Ther.,
288, 270-273.
36.Kuznetsova, T. Yu., Gavrilov, D. V., Dudanov, I.
P., Makarevich, P. I., Balatskii, A. V., Samokhodskaya, L. M., and
Parfenova, E. B. (2008) Kardiologiya, 48, 27-33.
37.Xin, Y. F., Zhou, X. J., Cheng, X., and Wang, Y.
X. (2005) J. Pharmacol. Exp. Ther., 312, 1090-1096.
38.Abe, H., Yoshikawa, N., Sarower, M. G., and
Okada, S. (2005) Biol. Pharm. Bull., 28, 1571-1577.
39.Fisher, G. H., D'Aniello, A., Vetere, A., Padula,
L., Cusano, G. P., and Man, E. H. (1991) Brain Res. Bull.,
26, 983-985.
40.Fisher, G., Lorenzo, N., Abe, H., Fujita, E.,
Frey, W. H., Emory, C., Fiore, M. M. D., and D'Aniello, A. (1998)
Amino Acids, 15, 263-269.
41.Hamase, K., Konno, R., Morikawa, A., and Zaitsu,
K. (2005) Biol. Pharm. Bull., 28, 1578-1584.
42.Pernot, P., Mothet, J. P., Schuvailo, O.,
Soldatkin, A., Pollegioni, L., Pilone, M., Adeline, M. T., Cespuglio,
R., and Marinesco, S. (2008) Anal. Chem., 80,
1589-1597.
43.Kampel, D., Kupferschmidt, R., and Lubec, G.
(1990) in Amino Acids: Chemistry, Biology and Medicine, Escom,
Leiden, pp. 1164-1171.
44.Schieber, A., Bruckner, H., Rupp-Classen, M.,
Specht, W., Nowitzki-Grimm, S., and Classen, H.-G. (1997) J.
Chromatogr. B, 691, l-12.
45.Hamase, K., Takagi, S., Morikawa, A., Konno, R.,
Niwa, A., and Zaitsu, K. (2006) Anal. Bioanal. Chem.,
386, 705-711.
46.Tishkov, V. I., Savin, S. S., and Khoronenkova,
S. V. (2008) Russ. Chem. Bull., 57, 1014-1022.
47.Savin, S. S., Chernyshov, I. V., Tishkov, V. I.,
and Khoronenkova, S. V. (2006) Bull. Moscow University, Ser. 2.
Chemistry, 47, 25-30.
48.Khoronenkova, S. V., Savina, L. I., and Tishkov,
V. I. RF Patent application No. 2007127821/13, decision of
27.10.2008.
49.Friedman, M. (1999) J. Agric. Food Chem.,
47, 3457-3479.
50.Wcislo, M., Compagnone, D., and Trojanowicz, M.
(2007) Bioelectrochemistry, 71, 91-98.
51.Watari, H., Isomoto, A., Oda, H., and Kuroda, M.
(1968) Biochim. Biophys. Acta, 167, 184-186.
52.Cohen, H. J. (1973) Anal. Biochem.,
53, 208-222.
53.Feinstein, R. N., and Lindahl, R. (1973) Anal.
Biochem., 56, 353-360.
54.Taylor, D. W., and Nieman, T. A. (1986) J.
Chromatogr., 368, 95-102.
55.Kitzler, J. W., and Fridovich, I. (1988) Anal.
Biochem., 174, 613-617.
56.Nagata, Y., Shimojo, T., and Akino, T. (1988)
Int. J. Biochem., 20, 1235-1238.
57.Gossrau, R., van Noorden, C. J., and Frederiks,
W. M. (1989) Histochemistry, 92, 349-353.
58.Frederiks, W. M., Patel, H. R., Marx, F.,
Gossrau, R., Kooij, A., and van Noorden, C. J. (1990) Acta
Histochem., Suppl., 40, 95-100.
59.Konno, R. (1998) Biol. Proced. Online,
1, 27-31.
60.Hamase, K., Nagayasu, R., Morikawa, A., Konno,
R., and Zaitsu, K. (2006) J. Chromatogr. A, 1106,
159-164.
61.Khoronenkova, S. V., and Tishkov, V. I. (2008)
Anal. Biochem., 374, 405-410.
62.Bruckner, H., and Hausch, M. (1993) J.
Chromatogr., 614, 7-17.
63.Kumashiro, S., Hashimoto, A., and Nishikawa, T.
(1995) Brain Res., 681, 117-125.
64.Nagata, Y., Borghi, M., Fisher, G. H., and
D'Aniello, A. (1995) Brain Res. Bull., 38, 181-183.
65.Almond, S. L., Fradley, R. L., Armstrong, E. J.,
Heavens, R. B., Rutter, A. R., Newman, R. J., Chiu, C. S., Konno, R.,
Hutson, P. H., and Brandon, N. J. (2006) Mol. Cell Neurosci.,
32, 324-334.
66.Tsai, G., Yang, P., Chung, L. C., Lange, N., and
Coyle, J. T. (1998) Biol. Psychiatry, 44, 1081-1089.
67.Hashimoto, A., and Chiba, Y. (2004) Eur. J.
Pharmachol., 495, 153-158.
68.Morikawa, A., Hamase, K., Inoue, T., Konno, R.,
and Zaitsu, K. (2007) Amino Acids, 32, 13-20.
69.Brandish, P. E., Chiu, C. S., Schneeweis, J.,
Brandon, N. J., Leech, C. L., Kornienko, O., Scolnick, E. M.,
Strulovici, B., and Zheng, W. (2006) J. Biomol. Screen.,
11, 481-487.
70.Adage, T., Trillat, A. C., Quattropani, A.,
Perrin, D., Cavarec, L., Shaw, J., Guerassimenko, O., Giachetti, C.,
Greco, B., Chumakov, I., Halazy, S., Roach, A., and Zaratin, P. (2008)
Eur. Neuropsychopharmacol., 18, 200-214.
71.Aminoff, M. J. (1994) West J. Med.,
161, 303-308.
72.Kawazoe, T., Tsuge, H., Imagawa, T., Aki, K.,
Kuramitsu, S., and Fukui, K. (2007) Biochem. Biophys. Res.
Commun., 355, 385-391.
73.Naylor, S. L., Busby, L. L., and Klebe, R. J.
(1976) Somatic Cell Genet., 2, 93-111.
74.Sasamura, T., Matsuda, A., and Kokuba, Y. (1999)
Arzneim-Forsch/Drug Res., 49, 541-543.
75.Sasamura, T., Matsuda, A., and Kokuba, Y. (2002)
Ann. Clin. Biochem., 39, 595-598.
76.Fang, J., Sawa, T., Akaike, T., and Maeda, H.
(2002) Cancer Res., 62, 3138-3143.
77.Fang, J., Sawa, T., Akaike, T., Greish, K., and
Maeda, H. (2004) Int. J. Cancer, 109, 1-8.
78.Fang, J., Deng, D., Nakamura, H., Akuta, T., Qin,
H., Iyer, A. K., Greish, K., and Maeda, H. (2008) Int. J.
Cancer, 122, 1135-1144.
79.Rodwell, W. V. (1971) Meth. Enzymol.,
17B, 174-188.
80.Tranchant, C., Aubourg, P., Mohr, M.,
Rocchiccioli, F., Zaenker, Ch., and Warter, J. M. (1993)
Neurology, 43, 2044-2048.
81.Plecko, B., Hikel, C., Korenke, G. C., Schmitt,
B., Baumgartner, M., Baumeister, F., Jakobs, C., Struys, E., Erwa, W.,
and Stockler-Ipsiroglu, S. (2005) Neuropediatrics, 36,
200-205.
82.Maddess, M. L., Tackett, M. N., and Ley, S. V.
(2008) in Progress in Drug Research, Vol. 66 (Petersen, F., and
Amstutz, R., eds.) Birkhäuser Verlag, Basel, pp. 15-186.
83.Berg, C. P., and Rodden, F. A. (1976) Anal.
Biochem., 71, 214-222.
84.Huh, J. W., Yokoigawa, K., Esaki, N., and Soda,
K. (1992) J. Ferment. Bioeng., 74, 189-190.
85.Buto, S., Pollegioni, L., D'Angiuro, L., and
Pilone, M. S. (1994) Biotechnol. Bioeng., 44,
1288-1294.
86.Findrik, Z., and Vasic-Racki, D. (2007)
Biotechnol. Bioeng., 98, 956-967.
87.Tan, Q., Song, Q., Zhang, Y., and Wei, D. (2007)
Appl. Biochem. Biotechnol., 136, 279-289.
88.Upadhya, R., Nagajyothi, S., and Bhat, S. G.
(1999) Process Biochem., 35, 7-13.
89.Lutz-Wahl, S., Trost, E. M., Wagner, B., Manns,
A., and Fischer, L. (2006) J. Biotechnol., 124,
163-171.
90.Garcia-Garcia, M., Martinez-Martinez, I.,
Sanchez-Ferrer, A., and Garcia-Carmona, F. (2008) Biotechnol.
Prog., 24, 187-191.
91.Patel, R. N. (2001) Biomol. Eng.,
17, 167-182.
92.Patel, R. N. (2001) Adv. Synth. Catal.,
343, 6-7.
93.Taylor, P. P., Pantaleone, D. P., Senkpeil, R.
F., and Fotheringham, I. G. (1998) Trends Biotechnol.,
16, 412-418.
94.Caligiuri, A., D'Arrigo, P., Gefflaut, T., Molla,
G., Pollegioni, L., Rosini, E., Rossi, C., and Servi, S. (2006)
Biocatal. Biotrans., 24, 409-413.
95.Caligiuri, A., D'Arrigo, P., Rosini, E., Tessaro,
D., Molla, G., Servi, S., and Pollegioni, L. (2006) Adv. Synth.
Catal., 348, 2183-2190.
96.Sacchi, S., Rosini, E., Molla, G., Pilone, M. S.,
and Pollegioni, L. (2004) Protein Eng. Des. Sel., 17,
517-525.
97.Sidorenko, S. V., and Tishkov, V. I. (2004)
Uspekhi Biol. Khim., 44, 263-306.
98.Sonawane, V. C. (2006) Crit. Rev.
Biotechnol., 26, 95-120.
99.Khoronenkova, S. V., Shabalin, I. G., Polyakov,
K. M., and Tishkov, V. I. (2008) Biochimie, 90, in
press.
100.Mizutani, H., Miyahara, I., Hirotsu, K.,
Nishina, Y., Shiga, K., Setoyama, C., and Miura, R. (1996) J.
Biochem., 120, 14-17.
101.Umhau, S., Pollegioni, L., Molla, G.,
Diederichs, K., Welte, W., Pilone, M. S., and Ghisla, S. (2000)
Proc. Natl. Acad. Sci. USA, 97, 12463-12468.
102.Kawazoe, T., Tsuge, H., Pilone, M. S., and
Fukui, K. (2006) Protein Sci., 15, 2708-2717.