REVIEW: Biochemical and Molecular Characterization of Plant MYB Transcription Factor Family
Hai Du1,2, Li Zhang2, Lei Liu1,2, Xiao-Feng Tang1,2, Wen-Jie Yang2, Yan-Min Wu2, Yu-Bi Huang1*, and Yi-Xiong Tang2*
1Maize Research Institute, Sichuan Agricultural University, Yaan Sichuan 625014, China; E-mail: yubihuang@sohu.com2Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing 100081, China; E-mail: tangyx@mail.caas.net.cn
* To whom correspondence should be addressed.
Received November 16, 2007; Revision received February 22, 2008
MYB genes are widely distributed in higher plants and comprise one of the largest transcription factors, which are characterized by the presence of a highly conserved MYB domain at their N-termini. Over recent decades, biochemical and molecular characterizations of MYB have been extensively studied and reported to be involved in many physiological and biochemical processes. This review describes current knowledge of their structure characteristic, classification, multi-functionality, mechanism of combinational control, evolution, and function redundancy. It shows that the MYB transcription factors play a key role in plant development, such as secondary metabolism, hormone signal transduction, disease resistance, cell shape, organ development, etc. Furthermore, the expression of some members of the MYB family shows tissue-specificity.
KEY WORDS: MYB transcription factors, classification, secondary metabolism, multi-functionality, signal transductionDOI: 10.1134/S0006297909010015
Abbreviations: ABA, abscisic acid; ABRE, ABA responsive element; AMV, avian myeloblastosis virus; ANS, anthocyanidin synthase; CAD, cinnamyl alcohol dehydrogenase; C4H, cinnamate-4-hydroxylase; CHI, chalcone isomerase; CHS, chalcone synthase; 4CL, p-coumaroyl-4-CoA ligase; DFR, dihydroflavonol reductase; GA, gibberellins; HTH, helix–turn–helix; NLS, nuclear localization signal; PAL, phenylalanine ammonium lyase; TF(s), transcription factor(s).
Transcription factors are important regulators of gene expression that
are composed of at least four discrete domains, DNA binding domain,
nuclear localization signal (NLS), transcription activation domain, and
oligomerization site, which operate together to regulate many
physiological and biochemical processes by modulating the rate of
transcription initiation of target genes. In general, it has been
categorized into different families or super-families through its
conserved DNA binding domain. For instance,
helix–loop–helix, zinc finger,
helix–turn–helix, leucine zipper, scissors, MADS cassette,
etc. And the myb genes form one of the largest families, which
have the most numbers and functions in plants. The first myb
gene identified was the v-MYB gene of avian myeloblastosis
virus (AMV) [1]. Subsequently, three
v-MYB-related genes – C-MYB, A-MYB, and
B-MYB – were found in many vertebrates and are thought to
be involved in the regulation of cell proliferation, differentiation,
and apoptosis [2]. The first plant MYB gene
C1 was isolated from Zea mays, which encodes a c-MYB-like
transcription factor that is involved in anthocyanin biosynthesis [3]. The large size of the MYB family in plants
indicates their importance in the control of plant-specific processes.
In the past decade, the R2R3-MYB genes have been extensively
studied and members of the MYB family have been found to be involved in
diverse physiological and biochemical processes including the
regulation of secondary metabolism [4-7], control of cell morphogenesis [8-10], regulation of meristem
formation, floral and seed development [11-14], and the control of the cell cycle [15, 16]. Some were also involved
in various defense and stress responses [17-20] and in light and hormone signaling pathways [21-23].
CHARACTERISTIC AND CLASSIFICATION
The MYB transcription factors (TFs) family is one of the most abundant classes of transcription factors in plants, and its subfamily containing the two-repeat R2R3 DNA binding domain is the largest in higher plants [24]. A common feature of MYB proteins is the presence of a functional DNA binding domain that is conserved amongst animals, plants, and yeasts [25] that typically consists of one to three imperfect repeats (R1, R2, and R3). Each repeat is about 50-53 amino acids long and encodes three α-helices, with the second and third helices forming a helix–turn–helix (HTH) structure which intercalates in the major groove of DNA when bound to it (Scheme 1).
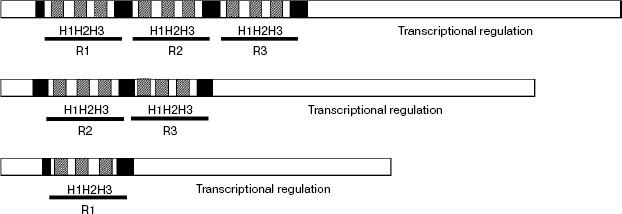
Scheme 1
For the special characteristic of its structure, the MYB gene family has been classified into several groups on different views. As showing little sequence similarity outside the MYB domain, the MYB proteins were classified into three major groups based on the number of adjacent repeats in the binding domain: R1R2R3-MYB, R2R3-MYB, and R1-MYB [24, 26, 27] (Scheme 1). Furthermore, MYB repeats typically contain regularly spaced tryptophan residues, which build a central tryptophan cluster in the three-dimensional helix–turn–helix fold. The tryptophan residues form a cluster in a hydrophobic core in each repeat and stabilize the structure of the DNA binding domain. Moreover, as flexibility in recognition, operating through a variety of mechanisms, may mean that proteins varying similarly in their DNA binding domains control quite different target genes and therefore have quite distinct physiological functions. Therefore, the plant R2R3-MYB family is categorized into three major subdivisions on the basis of the sequence of the DNA binding domain: subgroup A whose members are most similar to c-MYB and other animal MYB proteins; subgroup B, which is a relatively small group (four members in Arabidopsis); and subgroup C, which encompasses 70 members in Arabidopsis from a total of 83 defined so far, several members of which have been shown to recognize the MBSIIG binding site (T/CACCA/TAC/AC) preferentially [28]. Furthermore, based on the C-terminal conserved amino acid sequence motifs, which may facilitate the identification of functional domains, the plant myb genes are divided into 22 subgroups [24], and members of the same subgroups are suggested to have similar function. Moreover, most myb genes are reported as positive regulators of transcription. For example, ODORANT1 as a key regulator regulate floral scent biosynthesis in Petunia flowers [29]. AtMYB33 and AtMYB65 genes facilitate anther development [30]. The AtMYB103 gene regulates anther development in Arabidopsis [31]. However, not all MYB-related genes act as transcriptional activators. For example, AmMYB308 down-regulates cinnamate-4-hydroxylase (C4H), p-coumaroyl-4-CoA ligase (4CL), and cinnamyl alcohol dehydrogenase (CAD) genes [32]. Antirrhinum AmMyb305 and its Arabidopsis ortholog AtMYB4 regulate the accumulation of UV protective sinapate esters by repressing the expression of the gene encoding the key enzyme cinnamate-4-hydroxylase (C4H) [5]. Moreover, AtMYB32 was reported to repress the COMT gene in Arabidopsis [33]. Recently, two R2R3-MYB factors from maize, ZmMYB31 and ZmMYB42, were proposed to down-regulate both the Arabidopsis and the maize COMT genes as well [34].
At present, these repressors have only been attributed to R2R3-MYB factors belonging to subgroup 4. The common characteristic of these gene are that there exists a conserved repressing motif pdLNLD/ELxiG/S and a putative zinc-finger domain in the C-termini of these MYB proteins. Alternatively, these regions of sequence conservation might represent repression domains or domains for interaction with other transcription factors. Furthermore, our laboratory also isolated a new myb gene, GmMYBZ2 (DQ902861), from soybean by RT-PCR and RACE methods recently, which has the same motif and a putative zinc-finger domain in its C-terminus. It represses the expression of some flavonoid biosynthetic genes as well. To determine the role of the motif and the putative zinc-finger domain of it, we have been making some knockout mutants on it now. It is obvious that transcriptional regulation of gene expression is not mediated solely by activators, but also by the action of repressors.
MULTIFUNCTION OF PLANT MYB TRANSCRIPTION FACTORS
In higher plants, the myb-related genes constitute a rather large family of genes and play a wide variety of roles in the regulation of gene expression. For instance, WEREWOLF [35], AtMYB23 [36], and GaMYB2 [37] control epidermal cell differentiation in different plant species, whereas the cotton myb gene GhMYB109 plays a direct role in the initiation and elongation of cotton fiber cells [38] and the maize ROUGH SHEATH2 [39] gene exerts an effect on the development of lateral leaf primordia. Similarly, AtMYB21 and AtMYB24 are indicated to be key regulators of stamen and pollen maturation [40].
Control of cell morphogenesis. One of the clearly known functions of the plant MYB transcription factors is the control of cell morphogenesis and pattern formation, and the enormous improvement in this study is attributed to abundant researches of Arabidopsis root hair and epidermis.
The plant epidermis is the first point of contact between the sessile plant and its changing environment. As such, the epidermis fulfils several distinct roles, and this multi-functionality makes it an ideal system for the study of morphological diversity. The plant epidermis primarily provides an impermeable barrier to water, allowing organisms that evolved in the sea to survive in a dehydrating terrestrial atmosphere. In Arabidopsis, trichome development is a well-characterized model for the study of plant cell differentiation. Extensive molecular studies have revealed that a complex of transcription factors is involved in the determination of trichome fate. Among them, several MYB transcription factors play a key role in trichome initiation. Such as TTG1, GL1, and GL3 [41, 42] positively regulate Arabidopsis trichome cell-fate determination. The GL1 gene encodes a protein that contains a MYB domain, which is essential for trichome formation and is expressed in fields of the initiation of single cell-based trichome [43]. The TTG protein regulates R homologs the product of which could interact with the GL1 protein in trichome cells. Furthermore, several other genes have been implicated in the establishment of the root epidermal pattern, including GLABRA2 (GL2), WEREWOLF (WER), and CAPRICE (CPC) as well. The GL2 gene encodes a homeodomain transcription factor protein that is required to specify the non-hair cell type [44]. Accordingly, GL2 is expressed preferentially in the N cell position during epidermis development, beginning in the early embryo [8, 9, 45]. The WER gene encodes a MYB transcription factor required for the specification of the non-hair cell type, and it is expressed in the non-hair cells of root. Furthermore, WER is a positive regulator of GL2 expression [46]. The CPC gene encodes a small MYB protein without a putative transcriptional activation domain that is required for hair cell specification [47]. The opposing effects of the WER and CPC genes on root epidermal cell fate have been proposed to be responsible for the establishment of the cell-type pattern, perhaps as a result of competition between these MYB proteins for access to partner proteins or promoter targets [8]. Furthermore, there are some evidences that there is a transcriptional feedback loops exist between the WER, CPC, and GL2 genes, and that they are used as part of a lateral inhibition pathway to define the cell-type pattern in the root epidermis as well. All together, it has been proposed that the root and shoot epidermis employ a transcription factor complex that includes a MYB (WER or GL1), a bHLH (GL3 and/or an unknown bHLH), and a WD-repeat protein (TTG) to induce GL2 expression and the non-hair or trichome fate [48].
Responses to environmental stress. Plants respond to environmental changes with a number of physiological and developmental changes to tolerate stresses. Functional analyses of plant R2R3-MYB TFs indicate that they regulate numerous processes, including responses to environmental stress. Drought stress is one such condition, and it affects almost all plant functions including growth and development. Up to now, a number of myb genes have been described to respond to drought stress. For instance, the Arabidopsis R2R3-MYB transcription factor AtMYB2 is transiently induced by dehydration. Further analyses indicate that AtMYB2 functions as transcriptional activator in ABA (abscisic acid)-inducible gene expression during drought [18]. AtMYB68 is modulated by temperature, and loss of AtMYB68 reduces the ability of myb68 plants to compensate their growth at higher temperatures [49]. Moreover, the BcMYB1 isolated from Boea crassifolia was strongly induced by drought stress as well [50]. Furthermore, AtMYB60 was specifically expressed in guard cells, and its expression was negatively modulated during drought, which indicated that it was a transcriptional modulator of physiological responses in guard cells and opened new possibilities to engineering stomatal activity to help plants survive desiccation [51], etc.
Further studies found that light can activate transient expression in various tissues, including protoplasts, developing and germinating embryos, and root and leaf tissues. And many MYB proteins have been reported to display light-inducible expression. For example, the expression of C1 was inhibited by far-red light, suggesting a possible involvement of phytochrome [3]. The ATM4 and AtMYB21 genes isolated from Arabidopsis were induced by light [52, 53]. Expression of MYB-p1 increased 10-fold in the red relative to the green form of Perilla frutescens, which indicates that this gene also is induced by light [54]. Recently, several cold-inducible genes were reported as well. For example, the rice OsMYB4 gene encodes a MYB transcription factor involved in cold acclimation whose constitutive expression in Arabidopsis resulted in improved cold and freezing tolerance [55]. And the Arabidopsis HOS10 gene also is reported to be essential for cold acclimation and may affect dehydration stress tolerance in plants by controlling stress induced ABA biosynthesis [56]. Further studies indicate that the R2R3-MYB genes are involved not only in the signal transduction pathways of drought, low-temperature, and light but also in the signal transduction pathways of nutritional deficiency [57, 58], UV-B [5, 50, 59], low oxygen [59], etc. The same myb gene may activate expression of different functional genes to response to different environmental factors [50, 55, 59]. Its mechanism may be that the different functional genes have the same cis-elements in their promoters, which could be bound by the same MYB protein. Hence, over-expression of some MYB TFs could produce transgenic plants carrying resistance to multiple environmental stresses [55, 59].
Based on these studies, a large number of transgenic plants could be produced that could increase the transgenic plants’ resistance to numerous unfavorable environmental factors through plant breeding.
Response to phytohormone. R2R3-MYB genes are involved in the signal transduction pathways of salicylic acid [60], abscisic acid [18], gibberellic acid [61, 62], and jasmonic acid [63] as well. The phytohormone ABA is produced under water deficit conditions, which cause stomata closure and play an important role in the adaptation of vegetative tissues to abiotic environmental stresses, such as drought and high salinity [55, 59, 64]. AtMYB2 was the first gene that was induced by water stress and high-salt conditions and ABA induction [65]. The maize C1 gene is regulated by ABA during seed development as well [66]. HOS10 is essential for cold acclimation and affects dehydration stress tolerance in plants by controlling stress-induced ABA biosynthesis [56]. Further studies support that most of the drought-inducible genes are induced by the plant hormone ABA [50, 59, 67]. Analyses of drought-induced genes indicate the existence of ABA-independent as well as ABA-dependent signal transduction cascades between the initial signal of water deficit and the expression of specific genes [68]. For instance, in Arabidopsis the induction of a dehydration-responsive gene, rd22, is mediated by ABA and requires protein biosynthesis for ABA-dependent gene expression. It appears that dehydration triggers the production of ABA, which in turn induces various cis- and trans-acting factors involved in ABA induced gene expression. A conserved sequence, PyACGTGGC, has been reported to function as an ABA-responsive element (ABRE) in the promoters of many ABA-responsive genes [69, 70]. Recently, several groups have isolated genes for the ABRE binding proteins that interact with ABRE and regulate gene expression [71, 72]. These ABRE binding proteins contain a similar DNA binding motif of basic domain/Leu zipper (bZIP) structure and three conserved regions in their N termini.
Gibberellins (GA) control many aspects of plant growth and development, such as seed germination, seedling growth, elongation of leaf and stem, flowering, anther development, and fruit set. However, only a few downstream genes have been identified. GAMYB is the first GA signaling protein to have been identified in barley aleurone cells [73]. Orthologous proteins have since been identified in other grasses, such as rice, wheat, and Lolium temulentum [66, 74, 75]. The GAMYB protein induces the expression of GA-inducible genes by interacting directly with the GA-responsive cis-acting elements of these genes in aleurone tissue [62, 76, 77]. It was demonstrated that HvGAMyb from barley and OsGAMyb from rice, which are required for the expression of the α-amylase in aleurone, are both regulated by GA signal [74, 78].
Phenylpropanoid biosynthetic pathway. Phenylpropanoid metabolism is one of the three main types of secondary metabolism involving modification of compounds derived initially from phenylalanine, which is now well understood. As the first step, phenylalanine is deaminated to yield cinnamic acid by the action of phenylalanine ammonia lyase (PAL). Cinnamic acid is hydroxylated by cinnamate-4-hydroxylase (C4H) to 4-coumaric acid, which is then activated to 4-coumaroyl-coenzyme A (CoA) by the action of 4-coumarate-CoA ligase (4CL). Then it is divided into two major pathways—the flavonoid biosynthesis pathway and the lignin biosynthetic pathway (Scheme 2). To date, most R2R3-MYB proteins have been reported to play a major role in the regulation of secondary metabolism, such as the phenylpropanoid biosynthetic pathway [4, 79, 80].
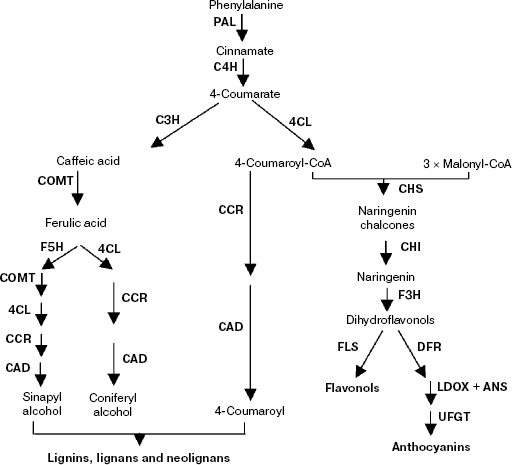
Scheme 2
Flavonoid biosynthesis is a branch of the large phenylpropanoid pathway, and virtually all genes encoding enzymes for the biosynthesis of flavones have been identified in plants (Scheme 2). The flavonoids, a group of polyphenolic plant secondary metabolites, are important for plant biology and human nutrition; they form a large group of polyphenolic compounds that occur naturally in plants. The biological importance of flavonoids has been implicated in plant growth and developmental processes, including pollen tube germination, resistance to insect feeding, formation of pollinator attractant pigments, UV light protection, pathogen resistance, and acting as signal molecules in plant–microbe interactions. To date more than 6000 different flavonoids have been identified, and the number is still increasing [81, 82]. There is increasing evidence to suggest that flavonoids, in particular those belonging to the class of flavonols, are potentially health-protecting components in the human diet as a result of their high antioxidant capacity [82-84], and their dietary intake is correlated with a reduced risk of cardiovascular diseases [85, 86] and osteoporosis [87, 88]. Based on these findings, it is obvious that flavonoids might offer protection against major diseases such as coronary heart diseases, certain cancers, and other age-related diseases.The first enzyme of the pathway is chalcone synthase (CHS) that uses malonyl-CoA and 4-coumaroyl-CoA as substrates to form tetrahydroxy chalcone, which are derived from carbohydrate metabolism and the phenylpropanoid pathway, respectively. Then the chalcone is isomerized by chalcone isomerase (CHI) to form the flavonone naringenin, which is converted to dihydrokaempferol by flavonone 3-hydroxylase (F3H). Subsequently, the dihydrokaempferol is further hydroxylated by flavonoid 3′-hydroxylase (F3′H) or flavonoid 3′,5′-hydroxylase (F3′5′H) to form the dihydroflavonols DHQ and DHM, which are required for the production of delphinidin-based anthocyanins [89]. Furthermore, these colorless molecules are converted into anthocyanins by at least three steps involving the enzymes dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and UDP-glucose:flavonoid 3-O-glucosyl-transferase (UFGT) [90] (Scheme 2).
The flavonoid biosynthetic pathway has now been almost completely elucidated. Many R2R3-MYB TFs have been identified from several model plants, such as maize, Antirrhinum, tobacco, Petunia, and Arabidopsis, which are involved in the regulation of different branches of flavonoid biosynthesis metabolism. For instance, the Antirrhinum majus genes AmMYB305 and AmMYB340 both can activate the gene encoding the first enzyme of phenylpropanoid metabolism, PAL, and also could activate other two enzymes of flavonol synthesis, CHI and F3H [75]. The sorghum MYB protein y1 gene could regulates the expression of CHS, CHI, and DFR genes that are required for biosynthesis of 3-deoxyflavonoids [82]. Expression of the maize myb C1 gene in tomato leads to the induction of all of the genes required for the production of flavonols and anthocyanins, except for CHI and the two B-ring hydroxylases F3′H and F3′5′H [100]. The seed-specific R2R3-MYB factor TRANSPARENT TESTA2 (TT2) regulates proanthocyanidin accumulation in developing seeds in Arabidopsis [6]. Moreover, Arabidopsis AtMYB12 was recently reported as a flavonol-specific activator of flavonoid biosynthesis. Further study using mutation showed AtMYB12 to be a transcriptional regulator of CHS and FLS in plants, the gene products of which are indispensable for the biosynthesis of flavonols [92]. Furthermore, we cloned two R2R3-MYB factors GmMYBJ6 and GmMYBJ7 from soybean, which are proved to regulate the synthesis of flavonoids as well.
Anthocyanins are pigments derived from a specialized branch of phenylpropanoid metabolism (Scheme 2). At least three enzymes are required for converting the dihydroflavonols to anthocyanins. The first of these enzymatic conversions is the reduction of dihydroflavonols to flavon-3, 4-cis-diols by DFR. These leucoanthocyanidins are the immediate precursors for the synthesis of anthocyanins. Then the leucoanthocyanidins are converted into anthocyandins by ANS. The enzyme UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT) was regarded as an indispensable enzyme of the main biosynthetic pathway to anthocyanins [93]. The first characterized plant R2R3-MYB was C1 from maize, which regulates genes encoding enzymes of the anthocyanin biosynthetic pathway [3]. Similar to C1, some R2R3-MYB family members control anthocyanin biosynthesis, albeit in a different developmental context [94]. For instance, overexpression the Kyoho grape MYBA gene leads to the induction of reddish-purple spots and UFGT gene expression in non-colored embryos, which indicated that this gene was involved in the regulation of anthocyanin biosynthesis in the grape via expression of the UFGT gene [95]. The strong correlation between the expression of the apple MdMYB10 gene and apple anthocyanin levels during fruit development suggests that this transcription factor is responsible for controlling anthocyanin biosynthesis in apple fruit in the red-fleshed cultivar and in the skin of other varieties. And there is an induction of MdMYB10 expression concurrent with color formation during development [96]. However, biochemical studies have suggested that the involvement of MYB-related transcription factors may extend beyond the control of flavonoid metabolism and include other branches of phenylpropanoid metabolism.
Lignin is one of the major components of the secondary walls of xylem cells, allowing mechanical support and efficient conduction of water and solutes over long distances within the vascular system (Scheme 2). In woody plant species, a large proportion of photosynthetically assimilated carbon is channeled to lignin synthesis and, as a consequence, lignified cell walls represent a major proportion of plant biomass and a huge reservoir of carbon stored within the polymers of lignocelluloses [97]. Lignin biosynthesis involves the phenylpropanoid pathway, which converts phenylalanine to p-coumaroyl-CoA, the precursor of a wide range of phenolic compounds. The subsequent hydroxylation and methylation steps have recently been shown to occur at the level of hydroxyl-cinnamic acid esters and their corresponding aldehydes and/or alcohols [98].
Over the last few years, the regulation of some genes of the lignin biosynthetic pathway has begun to be elucidated by the isolation and characterization of R2R3-MYB factors, whose belonging to different subgroups has been described as regulators of lignifications. For example, the PAP1 gene from Arabidopsis encodes an R2R3-MYB which, when over-expressed in Arabidopsis, alters lignin biosynthesis. The Pinus taeda R2R3-MYB TFs PtMYB1 and PtMYB4 that can bind to DNA motifs known as AC elements, which are ubiquitous in the promoters encoding lignin biosynthetic enzymes, can alter the accumulation of transcripts corresponding to genes encoding lignin biosynthetic enzymes in transgenic plants [99, 100]. And the eucalyptus EgMYB2 is also able to regulate transcription of two lignin biosynthetic genes, CCR and CAD, in both transient and stable expression assays [101]. Recently two new maize R2R3-MYB transcription factors, ZmMYB31 and ZmMYB42, have been reported to down-regulate both the Arabidopsis and the maize COMT genes. Furthermore, over-expression of the two genes also affects the expression of other genes of the lignin pathway and produces a decrease in lignin content of transgenic plants [34]. These examples illustrate the potential for the involvement of R2R3-MYB proteins in the regulation of lignification in xylem.
TISSUE-SPECIFIC REGULATION
Based on genome sequencing and annotation, more than 125 members of MYB TFs are present in Arabidopsis, scattered throughout different chromosomes, and some display gene-specific expression patterns. For example, AtMYB7 and AtMYB44 are expressed in most tissues, while AtMYB46 was only detected in siliques and AtMYB21 in flower buds [13]. The C1 gene regulates the expression of structural genes for enzymes involved in anthocyanin biosynthesis during seed development. Several MYB-related genes expressed in anthers have been identified, namely AtMYB26 and AtMYB103 in Arabidopsis [102, 103] and NtMYBAS1 in tobacco [104]. For the Arabidopsis AtMYB103 gene, whose expression is restricted to the tapetum of developing anthers and to trichomes, down-regulation results in early tapetal degeneration and aberrant pollen, possibly by interfering with polyploidization. Recently the new MYB gene AtMYB32 was reported, and it is expressed in most tissues, but it is most strongly expressed in the anther tapetum, stigma papillae, and lateral root primordia. Mutation of AtMYB32 leads to aberrant pollen and partial male sterility [33]. In other plants evidence of tissue-specific regulation are reported as well, such as HbMYB1 being expressed in leaves, bark, and latex of rubber trees, but its expression is significantly decreased in bark of TPD (tapping panel dryness) trees [105]. GhMYB109 is specifically expressed in cotton fiber initial cells as well as elongating fibers [106]. GhMYB7/9 are expressed in flowers and fibers, and their expression in fibers is developmentally regulated [107]. Furthermore, some recent studies have suggested that GAMYB may be involved in floral initiation, stem elongation, anther development, and seed development [108]. GAMYB is expressed at a high level in the floral meristem at the double-ridge stage and in stamen primordia of the grass L. temulentum. Moreover, the two soybean R2R3-MYB genes GmMYBJ6 (DQ902863) and GmMYBJ7 (DQ902864) are expressed only in leaf and stem according to our recent study. Taken together, it may be a common characteristic of the MYB TFs having different expression patterns in higher plants. But its detailed molecular mechanism remains largely unknown.
COMBINATIONAL REGULATION
Gene-specific regulation of transcription is of fundamental importance for virtually every aspect of cellular functions. Specificity is provided by the action of transcription factors, modular proteins typically composed of a DNA binding domain and effector domains responsible for activator or repressor activity. In eukaryotes, gene expression is frequently mediated by multi-protein complexes. The formation of these complexes involves the combinatorial action of transcription factors that bind conserved promoter elements in precise spatial orientation and on the basis of both specific protein–DNA and protein–protein interactions. This type of transcriptional regulation, termed combinatorial control, is thought to facilitate the complex regulatory networks found in higher eukaryotes. Both genetic and direct physical interactions suggest an intimate functional relationship between MYB proteins and bHLH proteins. The cooperative action of MYB and bHLH proteins has been most extensively studied with respect to the phenylpropanoid biosynthetic pathways.
The regulation of the phenylpropanoid biosynthetic pathways by MYB proteins in combination with bHLH proteins seems to be conserved throughout the plant kingdom, as exemplified by the MYB protein PhAN2 and the bHLH proteins PhJAF13 and PhAN1 from Petunia hybrida, or strawberry FaMYB1 that is able to interact with the maize bHLH protein ZmR [109, 110]. The regulation of flavonoid biosynthetic gene expression by the cooperation of R2R3-MYB and basic helix–loop–helix (bHLH) transcription factors provides one of the best-described examples of combinatorial gene regulation in plants. The interplay between R2R3-MYB TFs and bHLH families was first described in Zea mays with the interaction of C1 and R for the activation of anthocyanin biosynthesis. In Zea mays, the transcriptional activation of anthocyanin biosynthesis genes by the R2R3-MYB proteins ZmC1 or ZmPl1 was reported to require a member of the R/B gene family, bHLH. And a direct interaction between the MYB domain of ZmC1 and the N-terminal domain of the bHLH protein ZmB has been described as well [111].
Moreover, extensive studies of flavonoid regulation in different plant species have demonstrated the conservation of MYB–bHLH interactions in the control of flavonoid biosynthesis [112] as well. Molecular analysis of the TRANSPARENT TESTA mutants of A. thaliana revealed that several steps of the flavonoid biosynthetic pathway are also controlled by the combinatorial action of MYB and bHLH proteins. For example, the chalcone synthase (CHS), chalcone isomerase (CHI), flavonone 3-hydroxylase (F3H), and flavonol synthase (FLS), which catalyze successive steps of the flavonol biosynthetic pathway, are coordinately expressed in response to light and are spatially co-expressed in siliques, flowers, and leaves in Arabidopsis. These data suggested that a bHLH and a R2R3-MYB factor cooperate in directing tissue-specific production of flavonoids, while an ACE binding factor, potentially a BZIP, and a R2R3-MYB factor work together in conferring light responsiveness [113]. Analyses of the CHI, F3H, and FLS promoters functionally identified the cis-acting elements involved in conferring light responsiveness in all three promoters. Further study demonstrated the differential combinatorial interaction of MRE with either ACE or RRE, respectively, in response to different activating stimuli [113]. Moreover, in the anthocyanins pathway, the PAP1/MYB75 and PAP2/MYB90, which regulate genes required for the production of anthocyanins including those of PAL, CHS, DFR, and glutathione-S-transferase, also depend on a bHLH partner [4, 114].
Furthermore, the cooperation between bHLH and R2R3-MYB transcription factors not only control of the regulation of flavonoid biosynthesis, but also the regulation of epidermal cell differentiation and cell patterning in root hair and trichome development. For instance, the GL1 R2R3-MYB protein interacts with the GL3 and EGL3 bHLH factors to regulate the accumulation of trichomes in Arabidopsis [115]. Similarly, the bHLH rd22BP1 and R2R3-MYB AtMYB2 Arabidopsis proteins cooperate for drought and abscisic acid-regulated gene expression [116]. Recent results suggest that in Arabidopsis certain MYB proteins and R/B-like bHLH proteins work together with the WD40 protein TRANSPARENT TESTA GLABRA1 (TTG1) in a regulatory network, which underlies not only the control of phenylpropanoid biosynthesis, but also the regulation of epidermal cell differentiation and cell patterning in root hair and trichome development [79, 116]. These findings suggest a general mechanism of cooperation between R2R3-MYB proteins and other factors in transcriptional regulation in the higher plant.
EVOLUTION OF MYB DNA BINDING DOMAINS
Evolutionary studies based on the sequences of MYB domains from several organisms indicate that plant MYB ancestors may have had three MYB repeats and that the first repeat was lost. A model for evolution of MYB proteins has been presented by Lipsick [25]. This mode reports that the MYB domain first originated over one billion years ago, shortly after the divergence of eubacteria and eukaryotes. According to this model, R1R2R3-MYBs were generated by successive intragenic duplications or triplications in the primitive eukaryotes, and these evolved into today’s two repeat (R2R3-MYB) and three repeat (R1R2R3-MYB) genes in plants and animals. Furthermore, duplication of the MYB domain to give multiple-repeat MYB proteins followed by later expansion of MYB proteins through duplication of entire genes. This expansion was considerable in plants, and there are estimated to be over 125 members of this subfamily in Arabidopsis [24]. However, it is limited for the three-repeat MYB proteins in animals and epiphytes. So, there are more R2R3-MYB and R1R2R3-MYB in higher plants and animal, respectively.
From studies of amino acid homologies, the model suggested that the R2R3-MYB-related proteins arose after loss of the sequences encoding R1 in an ancestral three-repeat MYB gene. It is likely that, upon loss of R1, several subgroups of genes encoding R2R3-MYB proteins went through selective amplification and subgroup expansion during plant evolution [79].
In contrast, some reports [118] argued that typical MYB proteins might have a polyphyletic origin, with only the MYB DNA binding motif being derived from a common origin. With substantial sequence data available [119] we can investigate the origin of two major types of MYB genes, R1R2R3-MYB and R2R3-MYB. Although the evolutionary relationship of the atypical myb genes (one-repeat and partial myb genes) remains unclear, the topology of their study suggests that the R2R3 and R1R2R3 MYB genes were generated by successive gain of repeat units. This “gain” model is illustrated schematically as first, the ancestral R2R3-MYB was produced by an intragenic domain duplication; subsequently, the ancestral R1R2R3-MYB was formed by a further intragenic domain duplication.
Both R2R3 and R1R2R3 MYB co-existed in primitive eukaryotes, and they gave rise to the currently extant myb genes. Furthermore, when these two models were applied to the topology indicated by our results, the gain model can be accommodated with two changes, whereas the loss model requires five changes. Thus, the gain model provides a more parsimonious explanation for myb gene evolution and hence is favored over the loss model. Both of the models about the evolution of the MYB family have their advantage and disadvantage to explain the origin of MYB TFs family. The discussion of it also means that evolution needs further studies in the future.
DEGREE OF GENETIC REDUNDANCY
Many R2R3-MYB proteins share an extended degree of sequence similarity, especially within the highly conserved MYB domain. However, it remains unclear if in general this apparent structural redundancy also accounts for functional similarity. For example, the structurally closely related proteins AtMYB68 and AtMYB84 from Arabidopsis [49], PhMYBAN2 from Petunia hybrida [120], and AmMYBROSEA from Antirrhinum majus [121] belong to a subfamily of R2R3-MYB proteins controlling anthocyanin biosynthesis. Although the loss of function phenotype is the same in all these cases, the factors display slightly different, though overlapping target gene specificities [122], indicating no complete functional homology between these closely related MYB proteins. An example for the existence of true functional homology between closely related MYB proteins are the two Arabidopsis factors AtMYB66/WER and AtMYB0/GL1, which are involved in the regulation of trichome and root hair development, respectively. By reciprocal complementation experiments, it was shown that AtMYB66/WER and AtMYB0/GL1, although expressed in different tissues, encode functionally fully equivalent proteins. Hence, their unique roles in plant development are entirely due to differences in their expression patterns [9]. Moreover, AmMYB308 and AmMYB330 are particularly similar in their DNA binding domains (94% identical), suggesting that they may bind the same target DNA motifs, whereas these two proteins show no conservation of sequences in their C-termini [32]. These structural features suggest that AmMYB308 and AmMYB330 can recognize the same or very similar target motifs. However, because of the divergence in their C-termini, they may have distinct functions in regulating transcription. And AmMYB308 is expressed throughout Antirrhinum plants, whereas AmMYB330 is expressed primarily in mature flowers.
This work was supported by the China–The Netherlands cooperation fund.
REFERENCES
1.Klempnauer, K. H., Gonda, T. J., and Bishop, J. M.
(1982) Cell, 31, 453-463.
2.Weston, K. (1998) Curr. Opin. Genet.,
8, 76-81.
3.Paz-Ares, J., Ghosal, D., Wienand, U., Peterson,
P., and Saedler, H. (1987) EMBO J., 6, 3553-3558.
4.Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A.,
and Lamb, C. (2000) Plant Cell, 12, 2383-2394.
5.Jin, H., Cominelli, E., Bailey, P., Parr, A.,
Mehrtens, F., Jones, J., Tonelli, C., Weisshaar, B., and Martin, C.
(2000) EMBO J., 19, 6150-6161.
6.Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and
Lepiniec, L. (2001) Plant Cell, 13, 2099-2114.
7.Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M.,
Weisshaar, B., and Lepiniec, L. (2004) Plant J., 39,
366-380.
8.Lee, M. M., and Schiefelbein, J. (1999)
Cell, 99, 473-483.
9.Lee, M. M., and Schiefelbein, J. (2001)
Development, 128, 1539-1546.
10.Higginson, T., Li, S. F., and Parish, R. W.
(2003) Plant J., 35, 177-192.
11.Penfield, S., Meissner, R. C., Shoue, D. A.,
Carpita, N. C., and Bevan, M. W. (2001) Plant Cell, 13,
2777-2791.
12.Schmitz, G., Tillmann, E., Carriero, F., Fiore,
C., and Theres, K. (2002) Proc. Natl. Acad. Sci. USA, 99,
1064-1069.
13.Shin, B., Choi, G., Yi, H., Yang, S., Cho, I.,
Kim, J., Lee, S., Paek, N. C., Kim, J. H., Song, P. S., and Choi, G.
(2002) Plant J., 30, 23-32.
14.Steiner-Lange, S., Unte, U. S., Eckstein, L.,
Yang, C., Wilson, Z. A., Schmelzer, E., Dekker, K., and Saedler, H.
(2003) Plant J., 34, 519-528.
15.Ito, M., Araki, S., Matsunaga, S., Itoh, T.,
Nishihama, R., Machida, Y., Doonan, J. H., and Watanabe, A. (2001)
Plant Cell, 13, 1891-1905.
16.Araki, S., Ito, M., Soyano, T., Nishihama, R.,
and Machida, Y. (2004) J. Biol. Chem., 279,
32979-32988.
17.Vailleau, F., Daniel, X., Tronchet, M.,
Montillet, J. L., Triantaphylides, C., and Roby, D. (2002) Proc.
Natl. Acad. Sci. USA, 99, 10179-10184.
18.Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki,
K., and Yamaguchi-Shinozaki, K. (2003) Plant Cell, 15,
63-78.
19.Denekamp, M., and Smeekens, S. C. (2003) Plant
Physiol., 132, 1415-1423.
20.Nagaoka, S., and Takano, T. (2003) J. Exp.
Bot., 54, 2231-2237.
21.Gocal, G. F., Sheldon, C. C., Gubler, F., Moritz,
T., Bagnall, D. J., MacMillan, C. P., Li, S. F., Parish, R. W., Dennis,
E. S., Weigel, D., and King, R. W. (2001) Plant Physiol.,
1270, 1682-1693.
22.Seo, H. S., Yang, J. Y., Ishikawa, M., Bolle, C.,
Ballesteros, M. L., and Chua, N. H. (2003) Nature, 424,
995-999.
23.Newman, L. J., Perazza, D. E., Juda, L., and
Campbell, M. M. (2004) Plant J., 37, 239-250.
24.Stracke, R., Werber, M., and Weisshaar, B. (2001)
Curr. Opin. Plant Biol., 4, 447-456.
25.Lipsick, J. S. (1996) Oncogene, 13,
223-235.
26.Braun, E. L., and Grotewold, E. (1999) Plant
Physiol., 121, 21-24.
27.Kranz, H., Scholz, K., and Weisshaar, B. (2000)
Plant J. Jan., 21, 231-235.
28.Romero, I., Fuertes, A., Benito, M. J., Malpical,
J. M., Leyva, A., and Paz-Ares, J. (1998) Plant J., 14,
273-284.
29.Verdonk, J. C., Haring, M. A., van Tunen, A. J.,
and Schuurink, R. C. (2005) Plant Cell, 17,
1612-1624.
30.Millar, A., and Frank, G. (2005) Plant
Cell, 17, 705-721.
31.Zhang, Z. B., Zhu, J., Gao, J. F., Wang, C., Li,
H., Zhang, H. Q., Zhang, S., Wang, D. M., Wang, Q. X., Huang, H., Xia,
H. J., and Yang, Z. N. (2007) Plant J., 52, 528-538.
32.Tamagnone, L., Merida, A., Parr, A., Mackay, S.,
Culianez-Macia, F. A., Roberts, K., and Martin, C. (1998) Plant
Cell, 10, 135-154.
33.Preston, J., Wheeler, J., Heazlewood, J., Li, S.
F., and Parish, R. W. (2004) Plant J., 40, 979-995.
34.Fornale, S., Sonbol, F. M., Maes, T., Capellades,
M., Puigdomenech, P., Rigau, J., and Caparros-Ruiz, D. (2006) Plant
Mol. Biol., 62, 809-823.
35.Ishida, T., Hattori, S., Sano, R., Inoue, K.,
Shirano, Y., Hayashi, H., Shibata, D., Sato, S., Kato, T., Tabata, S.,
Okada, K., and Wada, T. (2007) Plant Cell, 19,
2531-2543.
36.Matsui, K., Hiratsu, K., Koyama, T., Tanaka, H.,
and Ohme-Takagi, M. (2005) Plant Cell Physiol., 46,
147-155.
37.Wang, S., Wang, J. W., Yu, N., Li, C. H., Luo,
B., Gou, J. Y., Wang, L. J., and Chen, X. Y. (2004) Plant Cell,
16, 2323-2334.
38.Suo, J., Liang, X., Pu, L., Zhang, Y., and Xue,
Y. (2003) Biochim. Biophys. Acta, 1630, 25-34.
39.Timmermans, M. C., Hudson, A., Becraft, P.
W., and Nelson, T. (1999) Science, 284, 151-153.
40.Mandaokar, A., Thines, B., Shin, B., Lange,
B. M., Choi, G., Koo, Y. J., Yoo, Y. J., Choi, Y. D., Choi, G.,
and Browse, J. (2006) Plant J., 46, 984-1008.
41.Herman, P. L., and Marks, M. D. (1989) Plant
Cell, 1, 1051-1055.
42.Koornneef, M., Dellaert, L., and van der Veen, J.
(1982) Mutat. Res., 93, 109-123.
43.Oppenheimer, D. G., Herman, P. L., Sivakumaran,
S., Esch, J., and Marks, M. D. (1991) Cell, 67,
483-493.
44.DiCristina, M. D., Sessa, G., Dolan, L.,
Linstead, P., Baima, S., Ruberti, I., and Morelli, G. (1996) Plant
J., 10, 393-402.
45.Masucci, J. D., Rerie, W. G., Foreman, D. R.,
Zhang, M., Galway, M. E., Marks, M. D., and Schiefelbein, J. W. (1996)
Development, 122, 1253-1260.
46.Lin, Y., and Schiefelbein, J. (2001)
Development, 128, 3697-3705.
47.Wada, T., Tachibana, T., Shimura, Y., and Okada,
K. (1997) Science, 277, 1113-1116.
48.Schiefelbein, J. (2003) Plant Biol.,
6, 74-78.
49.Feng, C. P., Andreasson, E., Maslak, A., Mock, H.
P., Mattsson, O., and Mundy, J. (2004) Plant Sci., 167,
1099-1107.
50.Chen, B. J., Wang, Y., Hu, Y. L., Wu, Q., and
Lin, Z. P. (2005) Plant Sci., 168, 493-500.
51.Cominelli, E., Galbiati, M., Vavasseur, A.,
Conti, L., Sala, T., Vuylsteke, M., Leonhardt, N., Dellaporta, S. L.,
and Tonelli, C. (2005) Curr. Biol., 15, 1196-1200.
52.Quaedvlieg, N., Dockx, J., Keultjes, G., Kock,
P., Wilmering, J., Weisbeek, P., and Smeekens, S. (1996) Plant Mol.
Biol., 32, 987-993.
53.Li, J., Yang, X., Wang, Y., Li, X., Gao, Z.,
Pei, M., Chen, Z., Qu, L. J., and Gu, H. (2006) Biochem.
Biophys. Res. Commun., 341, 1155-1163.
54.Gong, Z. Z., Yamazaki, M., and Saito, K. (1999)
Mol. Gen. Genet., 262, 65-72.
55.Vannini, C., Locatelli, F., Bracale, M., Magnani,
E., Marsoni, M., Osnato, M., Mattana, M., Baldoni, E., and Coraggio, I.
(2004) Plant J., 37, 115-127.
56.Zhu, J., Verslues, P. E., Zheng, X., Lee, B. H.,
Zhan, X., Manabe, Y., Sokolchik, I., Zhu, Y., Dong, C. H., Zhu, J. K.,
Hasegawa, P. M., and Bressan, R. A. (2005) Proc. Natl. Acad. Sci.
USA, 102, 9966-9971.
57.Miyake, K., Ito, T., and Sends, M. (2003)
Plant Mol. Biol., 53, 237- 245.
58.Hernandez, G., Ramirez, M., Valdes-Lopez,
O., Tesfaye, M., Graham, M. A., Czechowski, T., Schlereth, A.,
Wandrey, M., Erban, A., Cheung, F., Wu, H. C., Lara, M.,
Town, C. D., Kopka, J., Udvardi, M. K., and Vance, C. P. (2007)
Plant Physiol., 144, 752-767.
59.Kazuhiro, M., Soichi, K., Taku, D., Junko, T.,
and Yoshihiro, O. (2005) Plant Mol. Biol., 59,
739-752.
60.Raffaele, S., Rivas, S., and Roby, D. (2006)
FEBS Lett., 580, 3498-3504.
61.Murray, F., Kalla, R., Jacobsen, J., and Gubler,
F. (2003) Plant J., 33, 481-491.
62.Gocal, G. F., Poole, A. T., Gubler, F., and King,
R. W. (1999) Plant Physiol., 119, 1271-1278.
63.Lee, M. W., Qi, M., and Yang, Y. (2001) Mol.
Plant–Microbe Interact., 14, 527-535.
64.Maeda, K., Kimura, S., Demura, T., Takeda J., and
Ozeki, Y. (2005) Plant Mol. Biol., 59, 739-752.
65.Abe, H., Yamaguchi-Shinozaki, K., Urao, T.,
Iwasaki, T., and Shinozaki, K. (1997) Plant Cell, 9,
1859-1868.
66.Hattori, T., Vasil, V., Rosenkrans, L., Hannah,
L. C., McCarty, D. R., and Vasil, I. K. (1992) Genes Dev.,
6, 609-618.
67.Jeong, S. T., and Goto-Yamamoto, N. (2004)
Plant Sci., 167, 247-252.
68.Bray, E. A. (1997) Trends Plant Sci.,
2, 48-54.
69.Guiltinan, M. J., Marcotte, W. R., and Quatrano,
R. S. (1990) Science, 250, 267-271.
70.Mundy, J., Yamaguchi-Shinozaki, K., and Chua, N.
H. (1990) Proc. Natl. Acad. Sci. USA, 87, 406-410.
71.Choi, H. I., Hong, J. H., Ha, J. O., Kang, J. Y.,
and Kim, S. Y. (2000) J. Biol. Chem., 275, 1723-1730.
72.Lopez-Molina, L., and Chua, N. H. (2000) Plant
Cell Physiol., 41, 541-547.
73.Gubler, F., Kalla, R., Roberts, J. K., and
Jacobsen, J. V. (1995) Plant Cell, 7, 1879-1891.
74.Gubler, F., Watts, R. J., Kalla, R., Matthews,
P., Keys, M., and Jacobsen, J. V. (1997) Plant Cell Physiol.,
38, 362-365.
75.Moyano, E., Martinez-Garcia, J. F., and Martin,
C. (1996) Plant Cell, 8, 1519-1532.
76.Gubler, F., Raventos, D., Keys, M., Watts, R.,
Mundy, J., and Jacobsen, J. V. (1999) Plant J., 17,
1-9.
77.Piazza, P., Procissi, A., Jenkins, G. I., and
Tonelli, C. (2002) Plant Physiol., 128, 1077-1086.
78.Diaz, I., Vicente-Carbajosa, J., Abraham, Z.,
Martinez, M., Isabel-La, M. I., and Carbonero, P. (2002) Plant
J., 29, 453-464.
79.Jin, H., and Martin, C. (1999) Plant Mol.
Biol., 41, 577-585.
80.Boddu, J., Jiang, C., Sangar, V., Olson, T.,
Peterson, T., and Chopra, S. (2006) Plant Mol. Biol.,
60, 185-199.
81.Harborne, J. B., and Williams, C. A. (2001)
Nat. Prod. Rep., 18, 310-333.
82.Duthie, G., and Crozier, A. (2000) Curr. Opin.
Lipidol., 11, 43-47.
83.Sugihara, N., Arakawa, T., Ohnishi, M., and
Furuno, K. (1999) Free Radic. Biol. Med., 27,
1313-1323.
84.Dugas, A. J. J., Castaneda-Acosta, J., Bonin, G.
C., Price, K. L., Fischer, N. H., and Winston, G. W. (2000) J. Nat.
Prod., 63, 327-331.
85.Choi, D., and Mallet, J. (1999) Neurobiol.
Dis., 6, 309.
86.Hollman, P. C., and Katan, M. B. (1999) Free
Radic. Res., 31, 75-80.
87.Toda, T. (2006) Clin. Calcium, 16,
1693-1699.
88.Wu, J., Oka, J., Higuchi, M., Tabata, I., Toda,
T., Fujioka, M., Fuku, N., Teramoto, T., Okuhira, T., Ueno, T.,
Uchiyama, S., Urata, K., Yamada, K., and Ishimi, Y. (2006)
Metabolism, 55, 423-433.
89.Toda, K., Yang, D., Yamanaka, N., Watanabe, S.,
Harada, K., and Takahashi, R. (2002) Plant Mol. Biol.,
50, 187-196.
90.Pelletier, M. K., Burbulis, I. E., and
Winkel-Shirley, B. (1999) Plant Mol. Biol., 40,
45-54.
91.Arnaud, B., Ricde, V., Mark, K., Elio, S., Maria,
A. P., Shelagh, M., Geoff, C., Sue, R., Martine, V., Steve, H.,
Celestino, S. B., and Arjenvan, T. (2002) The Plant Cell,
14, 2509-2526.
92.Mehrtens, F., Kranz, H., Bednarek, P., and
Weisshaar, B. (2005) Plant Physiol., 138, 1083-1096.
93.Schijlen, E. G., Ricde, V. C. H., van Tunen, A.
J., and Bovy, A. G. (2004) Phytochemistry, 65,
2631-2648.
94.Cocciolone, S. M., Chopra, S., Flint-Garcia, S.
A., McMullen, M. D., and Peterson, T. (2001) Plant J.,
27, 467-478.
95.Kobayashi, S., Ishimaru, M., Hiraoka, K., and
Honda, C. (2002) Planta, 215, 924-933.
96.Espley, R. V., Hellens, R. P., Putterill,
J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007) Plant
J., 49, 414-427.
97.Boudet, A. M., Kajita, S., Grima-Pettenati, J.,
and Goffner, D. (2003) Trends Plant Sci., 8, 576-581.
98.Humphreys, J. M., and Chapple, C. (2002) Curr.
Opin. Plant Biol., 5, 224-229.
99.Patzlaff, A., Newman, L. J., Dubos, C., Whetten,
R. W., Smith, C., McInnis, S., Bevan, M. W., Sederoff, R. R., and
Campbell, M. M. (2003) Plant Mol. Biol., 53, 597-608.
100.Gomez-Maldonado, J., Avila, C., de la Torre,
F., Canas, R., Canovas, F. M., and Campbell, M. M. (2004) Plant
J., 39, 513-526.
101.Goicoechea, M., Lacombe, E., Legay, S.,
Mihaljevic, S., Rech, P., Jauneau, A., Lapierre, C., Pollet, B.,
Verhaegen, D., Chaubet-Gigot, N., and Grima-Pettenati, J. (2005)
Plant J., 43, 553-567.
102.Steiner-Lange, S., Unte, U. S., Eckstein, L.,
Yang, C., Wilson, Z. A., Schmelzer, E., Dekker, K., and Saedler, H.
(2003) Plant J., 34, 519-528.
103.Higginson, T., Li, S. F., and Parish, R. W.
(2003) Plant J., 35, 177-179.
104.Yang, S., Sweetman, J. P., Amirsadeghi, S.,
Barghchi, M., Huttly, A. K., Chung, W. I., and Well, T. D. (2001)
Plant Physiol., 126, 1738-1753.
105.Chen, S., Peng, S., Huang, G., Wu, K., Fu, X.,
and Chen, Z. (2003) Plant Mol. Biol., 51, 51-58.
106.Suo, J., Liang, X., Pu, L., Zhang, Y., and Xue,
Y. (2003) Biochim. Biophys. Acta, 1630, 25-34.
107.Chuan, Y. H., Johnie, N., Jenkins, A., Sukumar,
S. H., and Din, P. M. (2005) Plant Sci., 168,
167-181.
108.Woodger, F. J., Gubler, F., Pogson, B. J., and
Jacobsen, J. V. (2003) Plant J., 33, 707-717.
109.Aharoni, A., de Vos, C. H., Wein, M., Sun, Z.,
Greco, R., Kroon, A., Mol, J. N., and O’Connell, A. P. (2001)
Plant J., 28, 319-332.
110.Quattrocchio, F., Wing, J., Woude, K., Souer,
E., Vetten, N., Mol, J., and Koes, R. (1999) Plant Cell,
11, 1433-1444.
111.Goff, S. A., Cone, K. C., and Chandler, V. L.
(1992) Genes Dev., 6, 864-875.
112.Quattrocchio, F., Verweij, W., Kroon, A.,
Spelt, C., Mol, J., and Koes, R. (2006) Plant Cell, 18,
1274-1291.
113.Hartmann, U., Sagasser, M., Mehrtens, F.,
Stracke, R., and Weisshaar, B. (2005) Plant Mol. Biol.,
57, 155-171.
114.Zimmermann, I. M., Heim, M. A., Weisshaar, B.,
and Uhrig, J. F. (2004) Plant J., 40, 22-34.
115.Zhang, F., Gonzalez, A., Zhao, M., Paynek, C.
T., and Lloyd, A. (2003) Development, 130, 4859-4869.
116.Schiefelbein, J. (2003) Curr. Opin. Plant
Biol., 6, 74-78.
117.Johnson, C. S., Kolevski, B., and Smyth, D. R.
(2002) Plant Cell, 14, 1359-1375.
118.Rosinski, J. A., and Atchley, W. R. (1998)
J. Mol. Evol., 46, 74-83.
119.Jiang, C., Gu, J., Chopra S., Gu, X., and
Peterson, T. (2004) Gene, 326, 13-22.
120.Quattrocchio, F., Wing, J. F., Leppen, H., Mol,
J., and Koes, R. E. (1993) Plant Cell, 5, 1497-1512.
121.Schwinn, K., Venail, J., Shang, Y., Mackay, S.,
Alm V., Butelli, E., Oyama, R., Bailey, P., Davies, K., and Martin, C.
(2006) Plant Cell, 18, 831-851.
122.Martin, C., and Paz-Ares, J. (1997) Trends
Genet., 13, 67-73.C4H