New Data on Biochemical Mechanism of Programmed Senescence of Organisms and Antioxidant Defense of Mitochondria
V. P. Skulachev
Belozersky Institute of Physico-Chemical Biology and Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University, 119991 Moscow, Russia; fax: (495) 939-0338; E-mail: skulach@belozersky.msu.ru
Received October 19, 2009
Much evidence has recently been reported suggesting that reactive oxygen species (ROS) produced in mitochondria play a crucial role in the programmed senescence of organisms. In particular, it has been shown that antioxidants addressed to mitochondria slow down the appearance of symptoms of senescence and development of senile diseases and increase the median lifespan of various organisms from fungi to mammals. At the biochemical level, the mechanism of action of such rechargeable antioxidants as plastoquinonyldecyltriphenyl phosphonium (SkQ1) includes, in particular, prevention of oxidation of mitochondrial cardiolipin by ROS. The hormone melatonin also exhibits a number of such effects, and decrease in its level with age could explain the weakening of antioxidant protection upon aging. According to Moosmann et al., there exists a natural mechanism of antioxidant protection that, like SkQ1, is localized in the internal mitochondrial membrane and is rechargeable. It involves methionine residues in the surface regions of proteins encoded by mitochondrial DNA. It appears that in organisms with high respiratory metabolism the genetic code in the mitochondrial system of protein biosynthesis has changed. In these organisms (including some yeasts, insects, crustaceans, and vertebrates), the AUA codon codes for methionine rather than isoleucine, as in the case of synthesis of proteins encoded either in the nucleus or in mitochondria of organisms with lower rates of metabolism (other yeast species, sponges, and echinoderms). Methionine quenches ROS, being converted to methionine sulfoxide, which is re-reduced to the initial methionine by NADPH.
KEY WORDS: programmed senescence, reactive oxygen species, mitochondria, antioxidants, methionine, mitochondrial genetic codeDOI: 10.1134/S0006297909120165
In recent times, our journal has repeatedly reported on the problem of programmed senescence of organisms [1-6]. However, this field has developed so quickly and is so promising in a practical sense that new data of great significance appear every month.
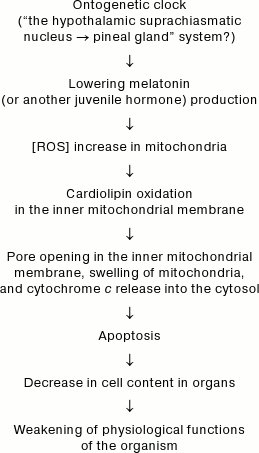
In 2003-2007, I published a hypothetical scheme for the main events of programmed senescence [1, 7, 8]; the scheme postulated the key role in this process of reactive oxygen species (ROS) generated in mitochondria. According to this scheme (see below), the signal for switching on the mammalian senescence program is the decrease with age of the level of melatonin (or another juvenile hormone) formed in pineal gland under control of the “master biological clock”, probably localized in suprachiasmatic nucleus of the hypothalamus and responsible for ontogenesis [7]. The decrease in melatonin level (according to Karasek [9], reaching 7-fold value during the period from 5- to >70-year-olds in humans) causes an increase in ROS level because melatonin itself is an antioxidant and it serves as an inducer of a number of antioxidant enzymes (glutathione peroxidase, both superoxide dismutases, etc.) [10, 11].
Hypothetical mechanism of programmed aging of organisms
As applied to mitochondria, the increase in ROS means increased probability of oxidation of cardiolipin [2], which in turn results in opening of pores in the inner mitochondrial membrane, swelling of mitochondria, rupture of the outer mitochondrial membrane, cytochrome c release into the cytosol, and activation of apoptosis [12, 13]. The enhanced apoptosis will decrease the number of cells in organs and tissues (decreases in their “cellularity” [14]), thus lowering functional resources and finally causing senescence of the organism [8].
In accordance with this scheme, it has been recently shown that melatonin eliminates the decrease in cardiolipin in mitochondria upon oxidative stress in vitro [13, 15] and upon aging in vivo [16]. However, in vitro experiments required very high (non-physiological) melatonin concentrations, namely 10 µM [13] and 100 µM [15]. The in vivo dose was about 0.1 µmol/kg per day [16].
The involvement of mitochondrial ROS in senescence was confirmed in our experiments showing that cationic derivatives of antioxidant plastoquinones (SkQs), selectively accumulated in mitochondria, prevent oxidation of cardiolipin upon oxidative stress [2], lengthen the lifespan of a wide circle of eukaryotes (from fungi to mammals) [6], and retard the development of approximately 30 typical symptoms of senescence and senile diseases [3-6, 17, 18].
The most important feature of the SkQs is the extremely low concentrations required for their effects to be seen: in experiments on cells this is the range of 10–12-10–9 M; in the case of treatment of senile ophthalmic diseases, one drop of 2.5·10–7 M solution daily; in therapy of heart arrhythmia, 1·10–10 mol/kg per day; in experiments on lifespan elongation, 5·10–10-5·10–9 mol/kg per day. This becomes understandable when it is taken into account that the coefficient of SkQ1 accumulation on the way from the intercellular space in vivo (or from the cell growth medium in vitro) to the inner half-membrane leaflet of inner mitochondrial membrane can in principle achieve such huge values as 108 times. This effect is composed of SkQ1 accumulation (i) in the cytosol (about 10 times, because Δψ value on the cell plasma membrane is about 60 mV), (ii) in the above-mentioned layer of mitochondrial membrane (about 1000 times, because Δψ in mitochondria is about 180 mV), and (iii) in any membrane (about 13,000 times because the distribution coefficient of SkQ1 in the water–octanol system is 13,000). Another factor providing for the high efficiency of SkQ1 is its ability to return from its oxidized form to the initial reduced (working) form. According to our data [2, 17], SkQ1 receives electrons from the respiratory chain complex III bH heme, which in turn is reduced by NADH:
NADH → complex I → CoQ → bL → bH → SkQ1.
All the above-said allows us to conclude that SkQ1 is an artificial rechargeable antioxidant addressed to the inner mitochondrial membrane. In 2008, a report from Moosmann’s group [19] appeared describing a natural rechargeable antioxidant localized in the same membrane. It is remarkable that to achieve this, mitochondria changed their genetic code. As shown by authors of the cited work, proteins encoded by mitochondrial DNA in many eukaryote species contain much more methionine compared to their analogs encoded by nuclear DNA in different eukaryote species. It was also found that in species with low methionine content this amino acid is encoded in mitochondria by a single codon (AUG), while in methionine-rich species not only AUG but additionally AUA codon is competent in encoding methionine (in species with low methionine content, AUA is one of three isoleucine codons). As a rule, the AUA codon encodes methionine instead of isoleucine in mitochondria of species with high aerobic metabolism (vertebrates, insects, crustaceans, and nematodes), whereas in species with low aerobic metabolism (Echinodermata, sponges, platyhelminthes, Cnidaria) AUA encodes isoleucine in mitochondria just as in the case of proteins encoded by nuclear DNA. Remarkable variety is demonstrated by fungi. Within the Saccharomycotina group, some members (in particular, Saccharomyces cerevisiae) use AUA for encoding methionine, while others use it to encode isoleucine (the latter include also Schizosaccharomyces pombe).
It is important that both isoleucines and methionines encoded by AUA are usually located on the surface of proteins of the inner mitochondrial membrane, and some are turned to the membrane–water interphase, while others are in the membrane core, i.e. in its hydrophobic region. The following example is demonstrative. The bee Melipona bicolor 10-fold exceeds feather star Florometra serratissima in the number of surface methionines in mitochondrial cytochrome b (33 residues against three). As a result, the whole bee cytochrome b molecule is in fact covered with methionines.
According to Moosmann and his colleagues [19], antioxidant activity of methionine gives the key to understanding a similar situation. It is a unique amino acid able to react with practically all natural ROS, including even the relatively inert H2O2. In this case, ROS are quenched harmlessly and methionine is converted into the stable and harmless methionine sulfoxide (compared with the other sulfur-containing amino acid cysteine that also reacts with ROS but forms a highly aggressive radical1). Besides, it is very important that methionine sulfoxide can be regenerated to the initial methionine by reduction with the following electron transport chain:
NADPH → thioredoxin reductase → thioredoxin → methionine sulfoxide reductase → methionine.
1 It is not surprising that the reverse correlation exists between the longevity of organisms and the number of cysteine residues in proteins coded in mitochondria [20, 21]. Thus, methionines of proteins encoded by mitochondrial DNA appear to be an analog of SkQ1 as a rechargeable antioxidant localized in the inner mitochondrial membrane. It is important that additional methionines that emerged in mitochondria due to changes in the genetic code are localized at the periphery of the protein molecule, encountering ROS on the way to active centers of the key proteins of oxidative phosphorylation. Such distribution of methionines is due to the fact that earlier the same loci were occupied by residues of the hydrophobic amino acid isoleucine. These proteins are inlaid into the membrane like bricks in a wall, while surface hydrophobic amino acids cement them with their partners inside the membrane. Probably a second change in genetic code in mitochondria of certain fungi, resulting in replacement of leucine by threonine, forming hydrogen bonds between α-helical regions of membrane proteins, strengthens such bonds [21].
The antioxidant role of methionine residues in proteins has been quite recently confirmed by Luo and Levine [22] in experiments on E. coli. The bacteria were grown in a medium with a mixture of amino acids in which methionine was replaced by norleucine. Since the protein biosynthesis system of E. coli can use norleucine instead of methionine, an increasing number of cells with proteins in which norleucine residues replace methionines accumulated with time. When the percentage of such replacements reached 40%, the cells were washed free of norleucine and methionine. Then the concentrations of soluble methionines and S-adenosylmethionines in the E. coli cells were normalized but the methionine content in proteins remained low. It was found that such cells were much more sensitive than normally to the toxic effect of H2O2.
Despite high similarity between antioxidant effects of protein methionines and SkQ1, there is a very substantial distinction between them concerning the applicability of these two mechanisms for aging therapy. We cannot enhance the methionine system by increasing the amount of methionine in mitochondrial proteins without interfering with the mitochondrial genome. However, it is not difficult to vary the intensity of the SkQ1 effect by simply changing its content in the food or drinking water. It is also important that methionine is the most toxic amino acid, and its excess in the diet results in numerous pathological consequences for the organism [23].
REFERENCES
1.Skulachev, V. P. (2007) Biochemistry
(Moscow), 72, 1385-1396.
2.Antonenko, Yu. N., Avetisyan, A. V., Bakeeva, L.
E., Chernyak, B. V., Chertkov, V. A., Domnina, L. V., Ivanova, O. Yu.,
Izyumov, D. S., Khailova, L. S., Klishin, S. S., Korshunova, G. A.,
Lyamzaev, K. G., Muntyan, M. S., Nepryakhina, O. K., Pashkovskaya, A.
A., Pletyushkina, O. Yu., Pustovidko, A. V., Roginsky, V. A.,
Rokitskaya, T. I., Ruuge, E. K., Saprunova, V. B., Severina, I. I.,
Simonyan, R. A., Skulachev, I. V., Skulachev, M. V., Sumbatyan, N. V.,
Sviryaeva, I. V., Tashlitsky, V. N., Vassiliev, J. M., Vyssokikh, M.
Yu., Yaguzhinsky, L. S., Zamyatnin, A. A., Jr., and Skulachev, V. P.
(2008) Biochemistry (Moscow), 73, 1273-1287.
3.Bakeeva, L. E., Barskov, I. V., Egorov, M. V.,
Isaev, N. K., Kapel’ko, V. I., Kazachenko, A. V., Kirpatovsky, V.
I., Kozlovsky, S. V., Lakomkin, V. L., Levina, S. V., Pisarenko, O. I.,
Plotnikov, E. Yu., Saprunova, V. B., Serebryakova, L. I., Skulachev, M.
V., Stelmashuk, E. V., Studneva, I. M., Tskitishvili, O. V., Vasilieva,
A. K., Viktorov, I. V., Zorov, D. B., and Skulachev, V. P. (2008)
Biochemistry (Moscow), 73, 1288-1299.
4.Agapova, L. S., Chernyak, B. V., Domnina, L. V.,
Dugina, V. B., Efimenko, A. Yu., Fetisova, E. K., Ivanova, O. Yu.,
Kalinina, N. I., Khromova, N. V., Kopnin, B. P., Kopnin, P. B.,
Korotetskaya, M. V., Lichinitser, M. R., Lukashev, A. N., Pletyushkina,
O. Yu., Popova, E. N., Skulachev, M. V., Shagieva, G. S., Stepanova, E.
V., Titova, E. V., Tkachuk, V. A., Vasiliev, Yu. M., and Skulachev, V.
P. (2008) Biochemistry (Moscow), 73, 1300-1316.
5.Neroev, V. V., Arkhipova, M. M., Bakeeva, L. E.,
Fursova, A. Zh., Grigoryan, E. N., Grishanova, A. Yu., Iomdina, E. N.,
Ivashchenko, Zh. N., Katargina, L. A., Khoroshilova-Maslova, I. P.,
Kilina, O. V., Kolosova, N. G., Kopenkin, E. P., Korshunov, S. S.,
Kovaleva, N. A., Novikova, Yu. P., Filippov, P. P., Pilipenko, D. I.,
Robustova, O. V., Saprunova, V. B., Senin, I. I., Skulachev, M. B.,
Sotnikova, L. F., Stefanova, N. A., Tikhomirova, N. K., Tsapenko, I.
V., Shchipanova, A. I., Zinovkin, R. A., and Skulachev, V. P. (2008)
Biochemistry (Moscow), 73, 1317-1328.
6.Anisimov, V. N., Bakeeva, L. E., Egormin, P. A.,
Filenko, O. F., Isakova, E. F., Manskikh, V. N., Mikhelson, V. M.,
Panteleeva, A. A., Pasyukova, E. G., Pilipenko, D. I., Piskunova, T.
S., Popovich, I. G., Roshchina, N. V., Rybina, O. Yu., Saprunova, V.
B., Samoilova, T. A., Semenchenko, A. V., Skulachev, M. V., Spivak, I.
M., Tsibulko, E. A., Tyndyk, M. L., Vyssokikh, M. Yu., Yurova, M. N.,
Zabezhinsky, M. A., and Skulachev, V. P. (2008) Biochemistry
(Moscow), 73, 1329-1342.
7.Skulachev, V. P. (2003) in Topics in Current
Genetics (Nystrom, T., and Osiewacz, H. D., eds.) Model Systems
in Ageing, Vol. 3, Springer-Verlag, Berlin-Heidelberg, pp.
191-238.
8.Skulachev, V. P. (2009) Ros. Khim. Zh.,
3, 125-140.
9.Karasek, M. (2004) Exp. Gerontol.,
39, 1723-1729.
10.Anisimov, S. V., and Popovic, N. (2004) Rev.
Neurosci., 15, 209-230.
11.Anisimov, V. N. (2008) Molecular and
Physiological Mechanisms of Aging [in Russian], Nauka, St.
Petersburg.
12.Petrosillo, G., Casanova, G., Matera, M.,
Ruggiero, F. M., and Paradies, G. (2006) FEBS Lett., 580,
6311-6316.
13.Petrosillo, G., Moro, N., Ruggiero, F. M., and
Paradies, G. (2009) Free Rad. Biol. Med., 47,
969-974.
14.Szilard, L. (1959) Proc. Natl. Acad. Sci.
USA, 45, 30-45.
15.Jou, M. J., Peng, T. I., Yu, P. Z., Jou, S. B.,
Reiter, R. J., Chen, J. Y., Wu, H. Y., Chen, C. C., and Hsu, L. F.
(2007) J. Pineal Res., 43, 389-403.
16.Petrosillo, G., Moro, N., Ruggiero, F. M., and
Paradies, G. (2009) (in press).
17.Skulachev, V. P. Anisimov, V. N., Antonenko, Yu.
N., Bakeeva, L. E., Chernyak, B. V., Erichev, V. P., Filenko, O. F.,
Kalinina, N. I., Kapelko, V. I., Kolosova, N. G., Kopnin, B. P.,
Korshunova, G. A., Lichinitser, M. R., Obukhova, L. A., Pasyukova, E.
G., Pisarenko, O. I., Roginsky, V. A., Ruuge, E. K., Senin, I. I.,
Severina, I. I., Skulachev, M. V., Spivak, I. M., Tashlitsky, V. N.,
Tkachuk, V. A., Vyssokikh, M. Yu., Yaguzhinsky, L. S., and Zorov, D. B.
(2009) Biochim. Biophys. Acta, 1787, 437-461.
18.Obukhova, L. A., Skulachev, V. P., and Kolosova,
N. G. (2009) Aging, 1, 389-401.
19.Bender, A., Hajieva, P., and Moosmann, B. (2008)
Proc. Natl. Acad. Sci. USA, 105, 16496-16501.
20.Moosmann, B., and Behl, C. (2008) Aging
Cell, 7, 32-46.
21.Kitazoe, Y., Kishino, H., Hasegava, M., Nakajima,
N., Thorne, J. L., and Tanaka, M. (2008) PloS ONE,
3, e3343.
22.Luo, S., and Levine, R. (2009) FASEB J.,
23, 464-472.
23.Gomez, J., Caro, P., Sanchez, I., Naudi, A.,
Jove, M., Portero-Otin, M., Lopez-Torres, M., Pamplona, R., and Barja,
G. (2009) J. Bioenerg. Biomembr., 41, 309-321.