Point Mutations in Pma1 H+-ATPase of Saccharomyces cerevisiae: Influence on Its Expression and Activity
V. V. Petrov
Skryabin Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, pr. Nauki 5, 142290 Pushchino, Moscow Region, Russia; E-mail: vpetrov06@mail.ru
Received March 31, 2010
Yeast Pma1 H+-ATPase is a key enzyme of cell metabolism generating electrochemical proton gradient across the plasma membrane, thus playing an important role in the maintenance of ion homeostasis in the cell. Using site-directed mutagenesis, we have previously replaced all 21 amino acid residues in the transmembrane segment M8 with Ala (Guerra et al. (2007) Biochim. Biophys. Acta, 1768, 2383-2392). In this work, we present new data on the role of these amino acid residues in the structure–function relationship in the enzyme and cell tolerance to heat shock. Mutations Q798A and I799A are lethal for cells regardless of expression of the enzyme in secretory vesicles or plasma membrane. The F796A mutation causes enzyme and cell sensitivity to heat shock when expressed in secretory vesicles. The I794A mutation increases temperature sensitivity of cells when the enzyme is expressed either in secretory vesicles or, to a lesser extent, in plasma membrane. The E803A mutation has no significant influence on the ATPase and cell sensitivity to heat shock; however, it causes a shift in the equilibrium between E1 and E2 conformations of the enzyme towards E1.
KEY WORDS: yeast, plasma membrane, secretory vesicles, ATPase, transmembrane segment, heat shock, site-directed mutagenesisDOI: 10.1134/S000629791008016X
P2-ATPases, including the yeast plasma membrane H+-ATPase, are widespread in pro- and eukaryotes [1]. They use the energy of ATP hydrolysis for transporting different cations across biological membranes [2]. Pma1 ATPase encoded by the PMA1 gene [3] is a vital enzyme generating and maintaining transmembrane electrochemical proton gradient (ΔµH+), thus supporting secondary solute transport systems. This proton pump plays an active role in maintaining yeast cell ion homeostasis and intracellular pH.
All P2-ATPases have one major catalytic subunit anchored into lipid bilayer by 10 hydrophobic segments; they also have common reaction mechanisms, when the β-phosphoryl group of ATP is bound to a conserved Asp residue to form an essential β-aspartyl phosphate reaction intermediate [1, 2]. The biochemical and molecular biological data on P2-ATPase structure and function have been recently confirmed by cryoelectronic structures obtained for mammalian Ca2+- [4-7] and Na+,K+-ATPases [8, 9] and H+-ATPases of fungi [10] and plants [11]. These ion pump structures are similar and include a large cytosolic catalytic center formed by three domains. It faces the cytosol and is connected by a stalk to the membrane domain consisting of 10 hydrophobic α-helices of different length and inclination [4-11]. Cations are transported through this membrane domain; the most important transmembrane segments are M4, M5, M6, and M8, which have amino acid residues participating in cation binding and transport. Transmembrane segments interact with each other and with the lipid bilayer. During the reaction cycle, the P2-ATPase membrane and catalytic domains undergo significant conformational changes; when ATP and transported cation(s) bind to the enzyme, it reversibly shifts between conformations E1 and E2 [5, 6].
Segment M8 is the least studied of the four transmembrane segments with the amino acid residues involved in proton transport. Previously, we have substituted the amino acid residues forming this segment, one at a time, with Ala. These ATPases with single point substitutions were then expressed in secretory vesicles [12]. The I794A, F796A, Q798A, and I799A mutant ATPases were virtually not expressed in secretory vesicles. The L797A, L801A, E803A, and L807A mutant enzymes had much lower expression (18-20% of the wild type level); however, ATPase activity of the L797A and E803A strains was high enough for detailed studies. The Glu803 residue of the Pma1 proton pump is homologous to Glu908 of the SERCA1a Ca2+-ATPase and is a part of the proton-binding site (in SERCA1a, Ca2+), being probably responsible for the coupling of ATP hydrolysis and transport of protons (hydroxonium ions) [12, 13]. Since heat shock, which was the necessary condition for isolation of secretory vesicles and, therefore, could influence the expression of ATPase causing its block in biogenesis, the aim of this work was to look further at their role in the structure–function relationship by expressing these substitutions at the level of plasma membrane.
MATERIALS AND METHODS
Yeast strains. Two Saccharomyces cerevisiae strains were used throughout the work: SY4 (MATa; ura3-52; leu2-3, 112; his4-619; sec6-4; GAL; pma1::YIpGAL-PMA1) with the chromosomal and plasmid copies of the PMA1 gene being under the control of different promoters [14] and NY13 (MATa ura3-52) with the PMA1 gene being under the control of native promoter PPMA1-PMA1 and linked to a selective marker URA3 [15].
In SY4, the chromosomal copy of the wild-type ATPase gene was placed under the control of the GAL1 promoter (PGAL-PMA1) and the plasmid allele (on the centromeric plasmid YCp2HSE [14]) was under the heat shock-inducible HSE promoter (PHSE-pma1) [14]. The plasmid-carried pma1 gene encoded either wild type or mutant enzyme [12, 13, 16]. The SY4 strain also carries a temperature-sensitive mutation in the SEC6 gene, which blocks the fusion of secretory vesicles to the plasma membrane under heat shock and leads to accumulation of secretory vesicles [17].
Site-directed mutagenesis. The polymerase chain reaction [18] was used to introduce single point mutations into a 519 bp BglII-SalI fragment of the PMA1 gene that has been previously subcloned into the modified version of Bluescript plasmid (Stratagene, USA). Each fragment was then sequenced to verify the presence of the mutation and the absence of unwanted base changes. Then, the corresponding fragments of plasmid pPMA1.2 [14] or pVP3 [12] carrying the entire coding sequence of the gene were replaced by restriction endonucleases BamHI, BglII, EcoRI, HindIII, SacI, SalI, and StyI and T4 DNA ligase (New England Biolabs, USA) with the same fragments carrying replacements. To express the Pma1 H+-ATPase in secretory vesicles, a 3.7-kb HindIII-SacI piece of pPMA1.2 containing the entire coding sequence of the gene was transferred by restriction endonucleases HindIII and SacI and T4 DNA ligase from pPMA1.2 under the control of HSE promoter into centromeric plasmid YCp2HSE, which was then used to transform the SY4 cells [12-14, 16].
To express the enzyme in plasma membrane, a 6.1 kb HindIII-HindIII fragment containing the entire coding sequence of the gene linked to the URA3 marker was excised by restriction endonuclease HindIII; then the Alkali-Cation Yeast transformation kit (Bio101, USA) was used to transform the NY13 strain, where this linear piece integrated onto the chromosome [13, 15].
Isolation of secretory vesicles and plasma membrane. To isolate secretory vesicles, the SY4 yeast cells were grown at 23oC in a liquid medium (2% bactopeptone, 1% yeast extract, 6.7 g/liter yeast nitrogen base (YNB), 20 mg/liter histidine) containing 2% galactose under continuous agitation. In the mid-exponential phase, the cells were washed free from galactose and transferred to the same medium containing 2% glucose, followed by a heat shock in 2.5 h (37oC for 2 h). Ten minutes before the heat shock termination, 10 mM NaN3 (which blocks metabolism thereby preventing the fusion of vesicles and plasma membrane [14]) was added to the cells; then the cells were chilled in ice for 10 min, sedimented, and washed twice with 10 mM NaN3; then spheroplasts were obtained as described [16]. The spheroplasts were treated with concanavalin A to make plasma membranes heavier in order to remove them during low-speed centrifugation [12-14, 16]. After spheroplasting, all procedures were performed at 0-4oC. Secretory vesicles containing newly synthesized ATPase were isolated by differential centrifugation and centrifugation in sucrose density gradient and resuspended in 0.8 M sorbitol, 1 mM EDTA, 10 mM triethanolamine-acetic acid, pH 7.2, with protease inhibitors (2 µg/ml chymostatin and leupeptin, pepstatin, and aprotinin, 1 µg/ml each) as described earlier [12, 13, 16].
To isolate plasma membranes, the S. cerevisiae strain NY13 was grown at 30°C in a liquid medium containing 6.7 g/liter YNB, 20 mg/liter histidine, and 4% glucose under continuous agitation to mid-logarithmic phase. The membranes were obtained by differential centrifugation and centrifugation in sucrose density gradient [19]. The plasma membrane-enriched fraction was washed with 1 mM EGTA-Tris, pH 7.5, containing the above protease inhibitors and was resuspended in the same buffer [12]. All preparative procedures were performed at 0-4°C.
Quantitation of expressed ATPase. The amount of ATPase in secretory vesicles and plasma membrane was estimated by SDS-PAGE and immunoblotting as described earlier [12, 13, 16]. The expression level of mutant Pma1 ATPase in secretory vesicles and plasma membrane was calculated relative to a wild-type control run in parallel.
ATPase hydrolytic activity was measured at 30oC in 0.5 ml of incubation mixture containing 5 mM MgSO4, 5 mM Na2ATP, 50 mM Mes-Tris, pH 5.70 (secretory vesicles and plasma membrane) and pH 6.25 (plasma membrane), 5 mM KN3, and an ATP-regenerating system (5 mM phosphoenolpyruvate and 50 µg/ml pyruvate kinase) in the absence and presence of 100 µM sodium orthovanadate [12, 13].
To obtain Ki for ATPase inhibition by orthovanadate ions, the stock solution of Na3VO4 was boiled for 3-5 min in a water bath to break the orthovanadate complexes formed during storage.
To obtain Km, the true concentration of MgATP was calculated according to Fabiato and Fabiato [20]. Inorganic phosphate was measured by the method of Fiske and Subbarow [21].
ATP-dependent H+-transport in secretory vesicles. ΔpH generated by ATPase was registered at 29oC by fluorescence quenching of pH-sensitive dye acridine orange using a Hitachi F2000 (Japan) spectrofluorimeter (excitation, 430 nm; emission, 530 nm) [12, 13]. Secretory vesicles (40-50 µg protein) were suspended in 1.5 ml of 0.6 M sorbitol, 100 mM KCl, 20 mM KNO3, 0.2-2.0 mM Na2ATP, 2 µM acridine orange, 20 mM Hepes-NaOH, pH 6.7; after stabilization of baseline fluorescence (120-150 sec), the reaction was started by adding 5.2-7.0 mM MgCl2 [12].
To estimate the coupling ratio between H+ transport and ATP hydrolysis, ATPase activity was assayed in parallel with the H+ transport at different concentrations of MgATP. The activity was measured under the same conditions in 100 µl of 0.6 M sorbitol, 20 mM Hepes-KOH, pH 6.7, 100 mM KCl, 20 mM KNO3, containing 0.2-2.0 mM Na2ATP and 5.2-7.0 mM MgCl2 at 29oC [12]. The reaction was stopped with 1 ml of 1.25% trichloroacetic acid, and inorganic phosphate was measured as above [21].
Protein assay. Protein was determined by the original [22] or modified Lowry method [23] using plasma membrane or secretory vesicles, respectively, with bovine serum albumin used as a standard. Aliquots of membrane resuspending buffers were added to the standard to compensate absorbance changes.
RESULTS AND DISCUSSION
Pma1 H+-ATPase expression and activity in secretory vesicles. Amino acid substitutions in transmembrane segment M8 of the yeast S. cerevisiae Pma1 H+-ATPase were constructed previously by means of alanine-scanning mutagenesis (Fig. 1) to explore the role of this segment in the structure–function relationship in this enzyme [12]. The wild type or mutated pma1 gene carrying Ala substitutions and controlled by the HSE promoter was subcloned into expression centromeric plasmid YCp2HSE [14], which then was used to transform S. cerevisiae strain SY4, where the chromosomal parental PMA1 gene was under the control of the GAL1 promoter [12-14]. The yeast was grown at 23°C in medium containing 2% galactose; under these conditions, the wild type enzyme was synthesized from the chromosomal gene PMA1. When the cells were shifted to the glucose-containing medium, the synthesis from the chromosomal gene was interrupted; when the temperature was elevated to 37°C, the Pma1 synthesis began from the plasmid pma1 gene (encoding either the wild-type ATPase used as a positive control or a mutant one). The strain SY4 also carried a temperature-sensitive mutation sec6-4, which blocked the fusion of secretory vesicles to the plasma membrane [17]. Therefore, heat shock resulted in accumulation of secretory vesicles containing the enzyme that was newly synthesized from the plasmid pma1 gene. Secretory vesicles isolated from the strain carrying plasmid YCp2HSE without the pma1 gene (Δpma1; Table 1) were used as a negative control. In this case, secretory vesicles did not contain ATPase, thus allowing us to estimate the level of contamination with plasma membranes carrying the wild type ATPase synthesized from the chromosomal PMA1 gene. Secretory vesicles practically free from plasma membrane contamination (2%, Δpma1; Table 1) were isolated using differential centrifugation and centrifugation in sucrose density gradient and used to measure the enzyme expression and activity [12, 13].
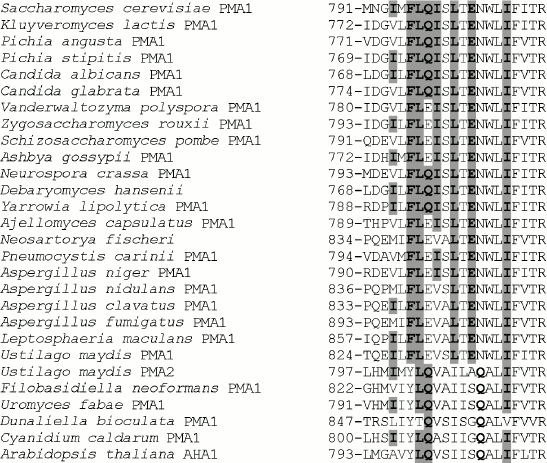
Table 1. Influence of mutations on the ATPase expression and activity in the secretory vesicles of the yeast S. cerevisiae strain SY4Fig. 1. Alignment of transmembrane segment M8 sequences from ATPases of fungi, algae, and plants. Accession numbers (top to bottom from S. cerevisiae to A. thaliana): P05030, P49380, AAD19960, ABN64423, P28877, M444794, EDO17118, P24545, P09627, AAS554405, P07038, CAG84667, CAG83458, Q07421, AW21780, AAB06958, CAK43734, AAC27991, EAW09357, AAK94755, CAP70082, AJ315590, L603947, AF077766, AY149918, P54211, E13998, P20649. Ile794, Phe796, Leu797, Gln798, Ile799, Leu801, Glu803, Ile807, and corresponding amino acid residues in other ATPases are highlighted in bold and gray.
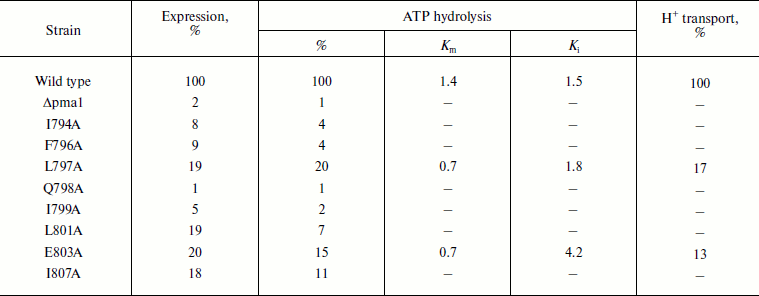
Note: Specific expression of the 100-kDa ATPase subunit was assayed by quantitative immunoblotting as described in “Materials and Methods” and represented as percentage of the wild type expression. Δpma1, secretory vesicles isolated from the strain carrying plasmid YCp2HSE [12-14] without the pma1 gene. ATPase hydrolysis was measured at pH 5.70. Proton transport was expressed as percentage of acridine orange fluorescence quenching (% F). 100% corresponds to 4.00 µmol Pi/min per mg of protein (ATP hydrolysis) and 758% F/mg of protein (H+ transport). The data are given as an average of 13 experiments for the wild type and 2-5 experiments for the mutants.
Choice of residues to be studied. There are several steps of ATPase maturation during biogenesis: it is synthesized in the endoplasmic reticulum and reaches the plasma membrane via the Golgi apparatus and secretory vesicles. Each of these steps has quality control points; if the newly synthesized enzyme has serious misfolding due to a mutation, its biogenesis is blocked [24, 25].
While isolating secretory vesicles, we found that expression of eight out of the 21 mutated enzymes was practically absent or very low (1 to 20% of the wild-type level); ATPase activity was also 1-20% of the wild type control (Table 1). These mutant enzymes could be divided into 3 groups: 1) Q798A and I799A ATPases (Fig. 1 and Table 1), the expression of which was practically equal to the level of expression of the Δpma1 strain, i.e. absent; accordingly, these enzymes were inactive. Maturation of these mutant proteins was possibly blocked due to misfolding at the early steps of biogenesis [12, 24, 25]; 2) I794A and F796A ATPases, the expression of which (8-9%; Table 1) was insignificantly higher than that of the Δpma1 strain; the ATPase activity was virtually absent (4%; Table 1). These proteins were evidently blocked at later steps of biogenesis; 3) a group of four mutant ATPases with the expression of 18-20%, which had either very low activity (7-11% in L801A and I807A; Table 1), insufficient for a detailed study due to sensitivity of methods, or high enough (L797A and E803A) even to study the proton transport and its coupling to ATP hydrolysis [12]. The E803A enzyme had the most different kinetics of ATP hydrolysis: the affinity to ATP was twofold higher and the sensitivity to orthovanadate (specific inhibitor of P-ATPases) was 3-fold lower than in the wild type (Table 1). Proton transport in this mutant appeared to be partly uncoupled to ATP hydrolysis (16% of the wild type level). Decrease in the ATPase sensitivity to orthovanadate could be explained by the shift of equilibrium between conformations E1 and E2 towards E1, which is characterized by the lower affinity to orthovanadate [25]. The kinetics of other earlier studied M8 mutant ATPases expressed in secretory vesicles were insignificantly different from the wild type control [12].
Therefore, these mutant enzymes, with the exception of I807A ATPase whose characteristics in secretory vesicles were close to L801A (and, evidently, to L797A), were included in the primary step of this study.
Conservation of the studied amino acid residues. Phe796, Leu797, Leu801, and Glu803 are the most conservative amino acid residues in M8 among those selected for this study. They are present in all Pma1 ATPases of ascomycetous fungi (from S. cerevisiae to L. maculans; Fig. 1); their substitutions in other fungal, algal, and plant enzymes are rather conservative and stereochemically similar: Phe→Tyr; Leu→Ile, Val; Glu→Gln (Fig. 1). Gln798 and Ile799, even though they are not present in all Pma1 ATPases of ascomycetes, have conservative substitutions: Gln798 is represented by Glu (Q798E mutant enzyme was almost completely identical to the wild type [12]) and Ile799 is represented by Val and Ala, which are relatively close to Ile and insignificantly disturb the enzyme stereochemistry (Fig. 1). Ile794 is the least conservative: in other proton pumps, it is replaced both by relatively conservative Leu, Val, or Ala, and by Met, which is rather different from Ile (Fig. 1).
It is worth mentioning that conservation of a residue is not always evidence for this residue being of structure–function importance. Thus, Leu797 and Ile807 (not selected for this study) are the most conservative residues, which are present in all P2-ATPase alignments of fungi, algae, and plants, with the only exception of the alga Dunaliella bioculata ATPase in both cases (Fig. 1). Nevertheless, Leu797 substitutions for Ala do not lead to significant disturbance of the enzyme structure–function relationship (Tables 1 and 3, [12]). Leu806 and Phe808 surrounding Ile807 are found in all P2 H+-ATPases; however, when they were replaced with Ala in secretory vesicles, the mutant enzymes retained rather high expression (47 and 69%) and activity (58 and 53%, respectively), and the proton transport coupling was very similar to that of the wild type [12].
Expression and activity of the Pma1 H+-ATPase at the plasma membrane. Since the 2-h incubation at 37oC was a necessary step during isolation of secretory vesicles, it could influence the Pma1 ATPase biogenesis and expression, because the enzyme passes several quality control points while maturating [24, 25]. Therefore, we decided to check heat shock tolerance of the wild type and I794A, F796A, L797A, Q798A, I799A, L801A, and E803A strains enabling these mutant proteins to reach the plasma membrane. The L797A, L801A, and E803A strains were additional controls to the wild type, since they were expressed in secretory vesicles, even though much lower, under elevated temperature. If all of these mutant ATPases were expressed at the plasma membrane, it would allow us to understand better the role of the corresponding residues in the ATPase structure–function relationship, especially of the enzyme biogenesis and functioning under heat shock.
To study the influence of different temperatures on the wild type enzyme expression and activity, S. cerevisiae NY13 cells were transformed with a linear fragment HindIII-HindIII, which contained the wild type PMA1 gene linked to the URA3 marker; the transformed cells were then plated onto Petri dishes with the selected medium and incubated at 23, 30, and 37oC. DNA was extracted from grown colonies and sequenced for the absence of unwanted mutations. The selected colonies were then grown in the liquid medium to check growth rate; plasma membranes were also isolated to measure expression and ATPase activity. The wild type appeared to be quite tolerant to temperature changes, although the growth rates and ATPase expression and activity were optimal at 30oC (Tables 2-4).
Saccharomyces cerevisiae NY13 cells were also transformed by the fragment HindIII-HindIII containing the PMA1 gene carrying mutations I794A, L796A, L797A, Q798A, I799A, L801A, or E803A. As in the case of the wild type, DNA was extracted from the grown colonies and sequenced for the presence of these mutations and absence of unwanted base changes. Only five out of the seven mutant strains grew on the selective medium: Q798A and I799A were unable to support growth even at non-restrictive 23 and 30oC. The remaining five mutants were checked for the growth rate at 30oC, and in the case of I794A and F796A also at 23 and 37oC (Tables 2-4). Plasma membranes were then isolated to check the ATPase expression and specific activity in vitro.
As is seen from Tables 2 and 3, the wild type strain grew well at 30oC and its ATPase was well expressed and highly active. The L797A and L801A strains were not very different from the wild type and therefore were not very interesting objects for further study (their ATPases were also expressed in secretory vesicles, and the L797A enzyme maintained an activity high enough for a detailed study: the kinetics of ATP hydrolysis and its coupling to proton transport was not very different from the wild type but differed from E803A [12]). The I794A and F796A strains were the most different both in activity (I794A and F796A) and growth rates and expression (I794A). These two strains were chosen for further study of the heat shock effect on ATPase biogenesis and functioning.
Role of Glu803. This residue is one of those participating in cation transport, at least in SERCA1a Ca2+-ATPase, and apparently dictating stoichiometry of ATP hydrolysis coupling to the transport of protons (hydroxonium ions) in Pma1 H+-ATPase [12, 13]. Indeed, Glu908 corresponding to Glu803 in Pma1 has been shown to be a part of the Ca2+ binding site in SERCA1a Ca2+-ATPase [5]. In Pma1 H+-ATPase, replacement of Glu803 leads either to the block of expression of the mutant enzymes in secretory vesicles (E803S and E803L with expression of 8% of the wild type level), or to the appearance of its inactive forms (E803D, E803C, and E803R with the expression of 24, 45, and 93%, respectively), or to miscoupling of proton transport and ATP hydrolysis (E803Q, E803N, and E803A with the expression of 102, 82, and 20%, activity of 123, 37, and 15%, and the coupling ratio of 2.65, 0.20, and 0.16, respectively; the wild type coupling ratio is 1.00) ([12, 13] and present study).
Our data confirm the functional importance of this amino acid residue: Glu803 replacement with Ala decreased growth rate 1.5-fold and absorbance in the stationary phase of growth at 30oC by a quarter (Table 3). In plasma membranes, E803A ATPase is 10-fold less sensitive to the enzyme specific inhibitor orthovanadate: Ki is 3.2 µM in the wild type compared with 33.9 µM in the mutant. In secretory vesicles, ATPase sensitivity to orthovanadate dropped only 3-fold (Table 1). The enzyme affinity to ATP did not change, and Km was 0.4 mM for both wild type and mutant. Mutations of amino acid residues Ile794, Phe796, Leu797, and Leu801 expressed in plasma membrane did not lead to such changes.
It is worth mentioning that Glu803, a quite conservative residue present in all Pma1 ATPases of ascomycetes and Ustilago maydis, in other proton pumps is represented by Gln, which, however, is shifted by one position towards the enzyme C-end (Fig. 1).
Role of Ile794 and Phe796. As already mentioned, Phe796 is a rather conservative residue present in all Pma1 ATPases of ascomycetes (in other ATPases, it is replaced by stereochemically close Tyr; Fig. 1). On the contrary, Ile794 is the most variable of the studied residues. Even within a single genus (e.g. Aspergillus) it is represented by Ile, Val, and Met (Fig. 1). Unlike Q798A and I799A, the I794A and F796A mutations were expressed at the plasma membrane. Their expression was the highest at 23°C and the lowest at 37°C: the drop was almost twofold. Wild type ATPase expression and activity were the highest at 30°C (Table 2). Thus, one can suggest that these mutations influence the enzyme and cell sensitivity to heat shock. However, the I794A and F796A ATPase specific activities were the highest at 30°C as in the case of the wild type (Table 2).
Table 2. Expression and activity of wild
type and mutant I794A and F796A ATPases in the S. cerevisiae
NY13 plasma membrane during growth at 23, 30, and 37°C
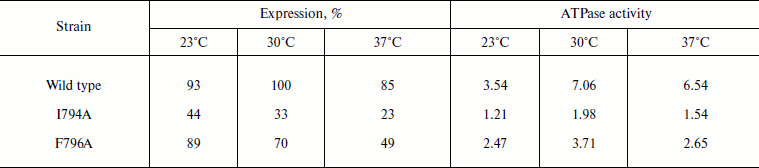
Note: The data are given as an average of 3-6 experiments. ATPase
activity was measured at pH 6.25 and expressed in µmol
Pi/min per mg of protein. 100% corresponds to the wild
type ATPase expression at 30°C. See also the note to Table 1.
More detailed study showed that even at 30°C the I794A mutation affected yeast growth and ATPase functioning more than F796A or, moreover, than the wild type: the growth rate was 1.5-fold slower, with simultaneous decrease in the maximum optical density of the culture by almost twofold. The expression of I794A ATPase dropped 3-fold and its activity decreased 7-fold (Table 3). The F796A replacement had less pronounced effect on yeast growth and ATPase expression; however, the activity of this mutant dropped 4-fold (Table 3).
Table 3. Expression and activity of ATPases
from the wild type and mutant strains in the plasma membrane of S.
cerevisiae NY13 grown at 30°C
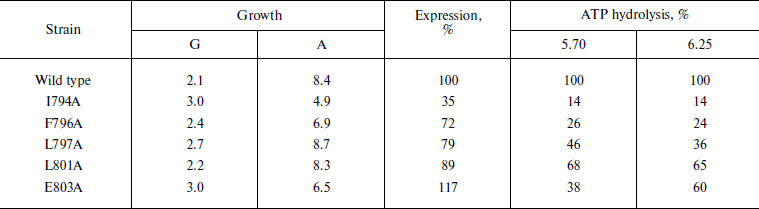
Note: G, doubling time; A, steady state optical density at
600 nm. ATP hydrolysis was measured at pH 5.70 and 6.25. 100%
corresponds to 5.72 (pH 5.70) and 6.95
(pH 6.25) µmol Pi/min per mg of
protein. See also the note to Table 1. The data
are given as an average of four experiments for the wild type and 2-3
experiments for the mutants.
Still more pronounced effect of these mutations was shown at 37oC: the growth of the I794A strain was almost twice slower, steady state optical density of the culture (biomass) dropped twice, and ATPase expression decreased 5-fold (Table 4). The relatively high percentage of the mutant activity compared to the wild type is probably explained by the low specific activity of both wild type and mutant strains. Just as at 30oC, the effect of the F796A mutation was much less pronounced; however, the expression of this mutation was twice lower compared to the less affected strain growth and enzyme activity (Table 4). Thus, it can be concluded that the I794A replacement leads to a significant but not complete temperature sensitivity of the mutant.
Table 4. Expression and activity of ATPases
from the wild type and mutant strains I794A and F796A in the plasma
membrane of S. cerevisiae NY13 grown at 37°C
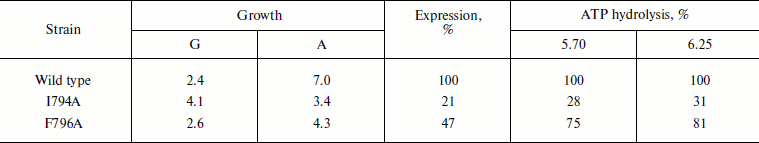
Note: G, doubling time; A, steady state optical density at
600 nm. ATP hydrolysis was measured at pH 5.70 and 6.25. 100%
corresponds to 2.15 (pH 5.70) and 2.23
(pH 6.25) µmol Pi/min per mg of
protein. See also the note to Table 1. The data
are given as an average of two experiments.
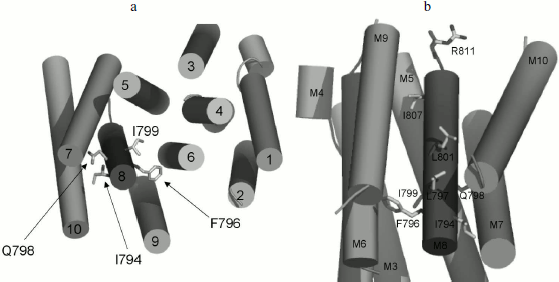
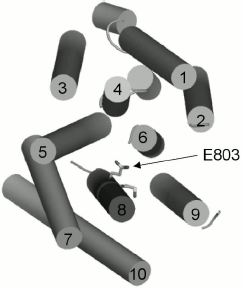
Three-dimensional structure of Pma1 H+-ATPase. Distinctive features of the I794A, F796A, and E803A mutations. A three-dimensional homological model of Pma1 ATPase has been constructed using crystallographic structures of SERCA1a Ca2+-ATPase in the E1 and E2 conformations [5, 6, 12]. It can be seen that residues Ile794, Phe796, Leu797, Gln798, Ile799, Leu801, Glu803, and Ile807 are oriented differently (Figs. 2 and 3). Glu803 and Ile799 are facing the space between the most important transmembrane segments M4, M5, M6, and M8 (Figs. 2 and 3). Residues Ile794 (closer to the N-terminus of M8) and Gln798 (in the middle of the segment) are facing the outer segments M7 and M10. Phe796 is oriented towards M6 and M9. Leu797 and Leu801 are protruding into the space between M9 and M10. Finally, Ile807, like Ile799 and Glu803, is also facing the space between M4, M5, M6, and M8; however, it is located quite close to the M8 C-terminus (Fig. 2).
Fig. 2. Homology model of the yeast ATPase showing the residues important for biogenesis and stability of the enzyme. The model was built using crystallographic structure of Ca2+-ATPase as described [12]. a) The membrane domain is viewed from the extracellular surface of the membrane. b) The membrane domain is viewed from extracellular side perpendicular to the membrane; residue R811 facing the cytosol. Adapted from [12].
Evidently, since Ala substitutions for these M8 residues (Fig. 1) have rather different effects, not only their conservation but also space arrangement is important; the latter influences both the interaction of the residue with the lipid bilayer and with the neighboring residues located in other transmembrane segments (Figs. 2 and 3). Apparently, substitutions for the most conservative residues Leu797, Leu801, and Ile807 do not significantly disturb the interaction of M8 with membrane lipids and other transmembrane segments and, therefore, should not change the enzyme conformation. At the same time, Ala substitutions for Gln798 and Ile799 are so significant structurally and/or conformationally that they cause the block of biogenesis of the mutant enzymes at early stages.Fig. 3. Homology model of the yeast ATPase showing residue E803, substitutions for which cause changes in apparent H+/ATP stoichiometry of the pump. The membrane domain is viewed from extracellular surface of the membrane. See also legend to Fig. 2. Adapted from [12].
Among the other three residues, Glu803 is a part of the cation-binding sites [4, 5, 12, 13] apparently regulating transport stoichiometry [12, 13]. The data presented here confirm the importance of this residue both for the yeast growth and for the proton pump functioning; its replacement with Ala results in a conformational shift between E1 and E2 states of the enzyme.
Residues Ile794 and Phe796 are probably not directly involved in the proton transport. In the plasma membrane, F796A mutation does not cause serious changes in contrast to I794A, the effect of which is particularly pronounced at 37°C. Probably, this can be explained by the fact that the Ile794 residue is facing the last transmembrane segment, M10, which is followed by the enzyme C-terminus (approximately 40 amino acid residues in length). This part of the enzyme is regulatory; it undergoes significant conformational changes during the reaction cycle [26]; therefore, the replacement of I794A apparently leads to malfunction of the ATPase, which is especially evident at elevated temperatures when viscosity of the lipid bilayer is changed.
The author is grateful to Prof. C. W. Slayman (Yale University School of Medicine, USA) as most of the experiments were performed at her laboratory and with her scientific advising; to Drs. T. V. Kulakovskaya and I. G. Morgunov (IBPM RAS, Pushchino) for discussion of the results; to Dr. J. P. Pardo (UNAM, Mexico City, Mexico) for creating homological models of the ATPase; to K. E. Allen (Yale University School of Medicine, USA) for expert technical assistance; and to D. G. Stepanov (STOTU, Moscow) for inspiring the writing of the manuscript.
The work was supported by Program P-24 “Origin of Biosphere and Evolution of Geo-Biological Systems” of the Presidium of the Russian Academy of Sciences and by the Russian Foundation for Basic Research, project No. 10-04-01248-a.
REFERENCES
1.Lutsenko, S., and Kaplan, J. H. (1995)
Biochemistry, 34, 15607-15613.
2.Goffeau, A., and Slayman, C. W. (1981) Biochim.
Biophys. Acta, 639, 197-223.
3.Serrano, R., Kielland-Brandt, M. C., and Fink, G.
R. (1986) Nature, 319, 689-693.
4.Zhang, P., Toyoshima, C., Yonekura, K., Green, N.
M., and Stokes, D. L. (1998) Nature, 392,
835-839.
5.Toyoshima, C., Nakasano, M., Nomura, H., and Ogawa,
H. (2000) Nature, 405, 647-655.
6.Toyoshima, C., and Nomura, H. (2002) Nature,
418, 605-611.
7.Toyoshima, C., and Muzutani, T. (2004)
Nature, 430, 529-535.
8.Morth, J. P., Pedersen, B. P., Toustrup-Jensen, M.
S., Sorensen, T. L.-M., Petersen, J., Andersen, J. P., Vilsen, B., and
Nissen, P. (2007) Nature, 450, 1043-1050.
9.Shinoda, T., Ogawa, H., Cornelius, F., and
Toyoshima, C. (2009) Nature, 459, 446-450.
10.Auer, M., Scarborough, G. A., and Kuhlbrandt, W.
(1998) Nature, 392, 840-843.
11.Pedersen, B. P., Buch-Pedersen, M. J., Morth, J.
P., Palmgren, M. G., and Nissen, P. (2007) Nature, 450,
1111-1115.
12.Guerra, G., Petrov, V. V., Allen, K. E., Miranda,
M., Pardo, J. P., and Slayman, C. W. (2007) Biochim. Biophys.
Acta, 1768, 2383-2392.
13.Petrov, V. V., Padmanabha, K. P., Nakamoto, R.
K., Allen, K. E., and Slayman, C. W. (2000) J. Biol. Chem.,
275, 15709-15716.
14.Nakamoto, R. K., Rao, R., and Slayman, C. W.
(1991) J. Biol. Chem., 266, 7940-7949.
15.Harris, S. L., Perlin, D. S., Seto-Yong, D., and
Haber, J. E. (1991) J. Biol. Chem., 266, 24439-24445.
16.Petrov, V. V. (2009) Biochemistry
(Moscow), 74, 1155-1163.
17.Novick, P., Field, C., and Schekman, R. (1980)
Cell, 21, 205-215.
18.Sarkar, G., and Sommer, S. S. (1990)
BioTechniques, 8, 404-407.
19.Perlin, D. S., Harris, S. L., Seto-Young, D., and
Haber, J. E. (1989) J. Biol. Chem., 264, 21857-21864.
20.Fabiato, A., and Fabiato, F. (1979) J.
Physiol., 75, 463-505.
21.Fiske, C. H., and Subbarow, Y. (1925) J. Biol.
Chem., 66, 375-400.
22.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and
Randall, R. J. (1951) J. Biol. Chem., 193, 265-275.
23.Bensadoun, A., and Weinstein, D. (1976) Anal.
Biochem., 70, 241-250.
24.Ferreira, T., Mason, A. B., Pypaert, M., Allen,
K. E., and Slayman, C. W. (2002) J. Biol. Chem., 277,
21027-21040.
25.Ambesi, A., Miranda, M., Petrov, V. V., and
Slayman, C. W. (2000) J. Exp. Biol., 203, 155-160.
26.Lecchi, S., Allen, K. E., Pardo, J. P., Mason, A.
B., and Slayman, C. W. (2005) Biochemistry, 44,
16624-16632.