Characteristics of σ-Dependent Pausing by RNA Polymerases from Escherichia coli and Thermus aquaticus
E. V. Zhilina1, N. A. Miropolskaya1, I. A. Bass1, K. L. Brodolin2, and A. V. Kulbachinskiy1*
1Institute of Molecular Genetics, Russian Academy of Sciences, pl. Kurchatova 2, 123182 Moscow, Russia; fax: (499) 196-0221; E-mail: akulb@img.ras.ru2Université Montpellier 1, Université Montpellier 2, CNRS UMR 5236, Centre d’études d’agents Pathogènes et Biotechnologies pour la Santé, Montpellier 34293, France
* To whom correspondence should be addressed.
Received May 27, 2011
The σ70 subunit of RNA polymerase (RNAP) is the major transcription initiation factor in Escherichia coli. During transcription initiation, conserved region 2 of the σ70 subunit interacts with the –10 promoter element and plays a key role in DNA melting around the starting point of transcription. During transcription elongation, the σ70 subunit can induce pauses in RNA synthesis owing to interactions of region 2 with DNA regions similar to the –10 promoter element. We demonstrated that the major σ subunit from Thermus aquaticus (σA) is also able to induce transcription pausing by T. aquaticus RNAP. However, hybrid RNAP containing the σA subunit and E. coli core RNAP is unable to form pauses during elongation, while being able to recognize promoters and initiate transcription. Inability of the σA subunit to induce pausing by E. coli RNAP is explained by the substitutions of non-conserved amino acids in region 2, in the subregions interacting with the RNAP core enzyme. Thus, changes in the structure of region 2 of the σ70 subunit have stronger effects on transcription pausing than on promoter recognition, likely by weakening the interactions of the σ subunit with the core RNAP during transcription elongation.
KEY WORDS: RNA polymerase, σ-dependent pausing, transcription regulationDOI: 10.1134/S0006297911100038
Abbreviations: nt, nucleotides; RNAP, RNA polymerase.
Specific recognition of promoters during transcription initiation by
bacterial RNA polymerase (RNAP) occurs with the participation of the
initiation factor – the σ subunit. Interaction of the
σ subunit with core enzyme of RNAP (subunit composition:
α2ββ′ω) results in formation of
RNAP holoenzyme, which is able to initiate RNA synthesis without
participation of additional factors. During initiation the holoenzyme
recognizes promoters, causes local melting of DNA around the
transcription start point, binds initiating nucleotides, and starts RNA
synthesis. When RNA reaches a length of about 10 nt, contacts of RNAP
with the promoter are disrupted and the transition to transcription
elongation occurs [1-3].
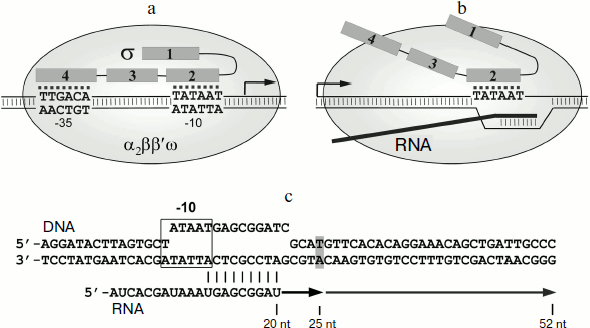
Рис.1Bacterial cells generally contain several different σ subunits responsible for transcription of different gene groups. Transcription of most genes is mediated by a so-called major σ subunit (σ70 in Escherichia coli, σA in other bacteria). Promoters recognized by the σ70 subunit contain several conservative elements, the main being the –10 and the –35 elements (consensus sequences TATAAT and TTGACA, respectively) (Fig. 1a) [1]. The σ subunit in the holoenzyme directly recognizes promoter elements and participates in DNA melting in the starting point of transcription. All major σ subunits include several conserved regions, which are divided into subregions (1.1-1.2, 2.1-2.4, 3.0-3.2, 4.1-4.2) and perform different functions during transcription initiation. In particular, the 2.3/2.4 and the 4.2 regions participate in the recognition of the –10 and –35 promoter elements, respectively, and the 2.1 and the 2.2 regions play a major role in interactions with the core RNAP enzyme. It should be noted that the remaining regions of the σ subunit are also involved in interactions with the core enzyme (Fig. 1a) [1, 4-7]. The melting of DNA is initiated by the 2.3 region of the σ70 subunit, which contains several aromatic and positively charged residues and interacts with the non-template DNA chain at the –10 promoter element. Mutations of these residues lead to an increase in the temperature of promoter DNA melting during transcription initiation, which confirms their important role in separation of DNA strands [8-12].
It was initially thought that during the transition to elongation the σ subunit dissociates from the transcription complex. However, the σ70 subunit in E. coli was found to interact with the elongation complex (EC) and to induce pauses during transcription. Such pauses were initially found in the initially-transcribed promoter regions of lambdoid bacteriophages (phages λ, 82, and 21) [13, 14]. Later the σ70-dependent pauses were found in the promoter-proximal region of the lac gene in vitro [15-17] and also in a number of E. coli genes in vivo [18]. In the case of lambdoid phage promoters, the σ-dependent pausing plays a regulatory role and is required for interaction of the EC with the Q protein, which acts as a transcription antiterminator [14]. However, the functional role of σ-dependent pauses in the transcription of other genes remains unknown.Fig. 1. Participation of the RNAP σ-subunit in promoter recognition and in formation of transcription pauses. a) Scheme of promoter recognition by the σ-subunit. Conserved regions of the σ-subunit are numbered. The sequences of –10 and –35 promoter elements are shown; the transcription start point is indicated by an arrow. b) Scheme of pause signal recognition by the σ-subunit of RNAP. c) Scheme of the synthetic template used in this study for analysis of σ-dependent transcription pause. Pause signal, corresponding to the –10 promoter element, is outlined by a frame. DNA nucleotides corresponding to the RNA 3′-end in the pause site are shown in gray. Arrows indicate the transcripts corresponding to transcription pause (25 nt) and full-sized RNA product (52 nt).
Pausing requires the presence in the transcribed DNA of a region similar to the –10 promoter element. This region interacts with region 2 in the σ70 subunit (Fig. 1b) [13, 15, 17]. The binding of the σ subunit to EC is likely to occur through interactions of region 2 σ with the β′ subunit of the RNAP core enzyme; mutations in the 2.2 region of the σ70 subunit disrupt pausing [14, 19-21]. It should be noted that the presence of RNA transcript in EC prevents the interaction of the core enzyme with regions 3 and 4 of the σ subunit, which in the holoenzyme are located in the RNA exit channel [2, 6, 22]. Binding of σ70 to the –10-like element leads to “anchoring” of the EC at the upstream edge of the transcription bubble. Continuation of the RNA synthesis leads to the formation of unstable stressed EC, which can then backtrack on the DNA template; as a result, the RNAP active site loses contact with the 3′-end of RNA, leading to stop of transcription [14, 15, 17, 23, 24].
Many details of σ70-dependent pausing as well as the EC structure during pausing remain unknown. In particular, the molecular mechanisms of interactions of the σ-subunit with the RNAP core enzyme and with the non-template DNA chain in the EC are not sufficiently studied. Additionally, it is not known whether σ-dependent pausing occurs during transcription in other bacteria and if so, whether there are any differences of this process in comparison with E. coli RNAP.
It should be noted that most data about RNAP structure was obtained in the study of T. aquaticus RNAP, as well as RNAP of the closely related bacterium Thermus thermophilus, which makes functional analysis of this RNAP during different stages of transcription very important. Escherichia coli and T. aquaticus are not evolutionarily related, and their RNAP differ greatly in their sequence, structure [25-28], and transcriptional properties [29, 30]. The major σ-subunits in E. coli and T. aquaticus (σ70 and σA, respectively) were found to exhibit significant differences at the stage of transcription initiation. In particular, the σA-subunit is not able to melt promoter DNA at the transcription start point at temperatures below 45°C, unlike the σ70-subunit [7, 30, 31]. Thus, comparison of the mechanisms of pause formation by E. coli and T. aquaticus RNAPs can give important information about the features of transcription regulation mechanisms in different groups of bacteria.
The purpose of this study was to examine the process of T. aquaticus RNAP pausing compared with E. coli RNAP and to determine how the differences in the structures of the σ-subunits from these bacteria affect the species-specific characteristics of transcription pausing.
MATERIALS AND METHODS
Enzymes and DNA. Core enzymes and RNAP σ-subunits of E. coli and T. aquaticus were isolated from E. coli cells as described previously [30]. Preparation of mosaic σ-subunits is described in [7]. DNA and RNA oligonucleotides were synthesized by Syntol (Moscow). Promoter DNA fragment containing the lacUV5-promoter (from position –57 to +58) was obtained by PCR as described previously [16].
RNAP activity measurement. The transcription reaction was performed in buffer containing 40 mM Tris-HCl, pH 7.9, 40 mM KCl, and 10 mM MgCl2. To obtain the holoenzyme, RNAP core enzyme of E. coli (in final concentration of 100 nM) was mixed with a fivefold excess of σ-subunit (500 nM), incubated for 5 min at 37°C, then promoter DNA fragment (10 nM) was added, the mixture was incubated for another 5 min, and the samples were placed on ice for 3 min. The sample was then transferred to the desired temperature, incubated for 5 min, and a mixture of nucleotides (100 μM ATP, GTP, CTP, 10 μM UTP with addition of α-[32P]UTP) was added. The reaction was stopped after 5 min by addition of an equal volume of stop buffer containing 8 M urea and 50 mM EDTA. RNA products were analyzed by electrophoresis in 15% (29 : 1.5) polyacrylamide gel.
Analysis of pausing. Artificial EC was obtained using a modified method [32, 33]. Oligonucleotide sequences used for the complex assembly are shown in Fig. 1c. RNA oligonucleotide was labeled at the 5′-end with T4 polynucleotide-kinase (New England Biolabs, USA) and γ-[32P]ATP. Assembly was carried out in buffer containing 40 mM Tris-HCl, pH 7.9, and 40 mM KCl. The oligonucleotide corresponding to the template DNA strand was incubated with RNA oligonucleotide (final concentrations of 2.5 µM and 250 nM, respectively) for 3 min at 65°C and cooled for 30 min to 20°C. The mixture was diluted three times with buffer, RNAP core enzyme was added (250 nM), and the mixture was incubated for 20 min at 40°C. The oligonucleotide corresponding to the non-template DNA strand (1 µM) was added to the sample, and the mixture was incubated for 20 min at 40°C and transferred to 20°C. The resulting EC was diluted 10 times with buffer containing 40 mM Tris-HCl, pH 7.9, and 40 mM KCl, σ-subunit was added (final concentration 500 nM), and the mixture was incubated for 5 min at 37°C. Samples were transferred to the desired temperature, incubated for 3 min, then NTP substrates were added to the reaction mixture to the final concentration of 100 µM or 1 mM, and MgCl2 solution was added (10 mM). The reaction was stopped after the required periods of time by addition of an equal volume of stop buffer. RNA products were separated by electrophoresis in 15% polyacrylamide gel and analyzed with a phosphorimager (GE Healthcare). Three or four independent experiments were conducted for each of the studied RNAPs.
RESULTS
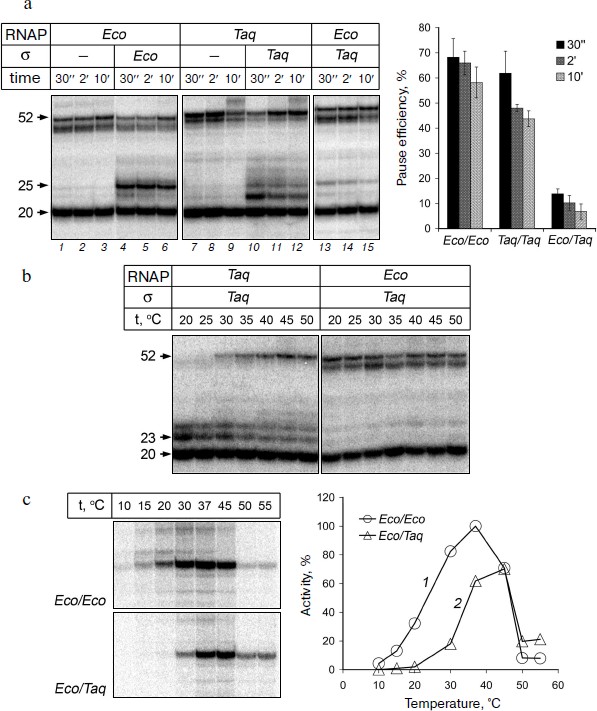
Formation of σ-dependent transcription pauses by RNAPs from E. coli and T. aquaticus. To study the formation of σ-dependent RNAP pauses, we used the method of in vitro EC assembly. This approach allows to assemble fully functional EC using synthetic DNA and RNA oligonucleotides and RNAP core enzyme, bypassing the stage of transcription initiation [32, 33]. The scheme of oligonucleotide template used in the experiments is shown in Fig. 1c. Two complementary DNA oligonucleotides (template and non-template) with a length of 60 nt and RNA oligonucleotide with length of 20 nt, complementary to the template DNA oligonucleotide, were used for the EC assembly (see “Materials and Methods”). A –10 element of the promoter was introduced into the template sequence; its position relative to the 3′-end of the RNA corresponded to its position relative to the transcription starting point in the promoter complex (Fig. 1c). We then added to the EC assembled on this template the σ-subunit and nucleotide substrates. The transcription reaction was stopped after different time intervals, RNA products were analyzed by gel electrophoresis. The results of measurements of RNAP transcriptional activity in this experiment are shown in Fig. 2a.
It was shown that the EC of E. coli RNAP obtained on this template is transcriptionally active. After addition of nucleotides to the EC obtained in the absence of σ-subunit, RNAP synthesizes an RNA product of 52 nt that corresponds to transcription up to the end of the template (Fig. 2a, lanes 1-3). A fraction of the starting RNA oligonucleotide (50-70% in our experiments) is not extended, likely because the efficiency of EC assembly does not reach 100%, and part of the RNA oligonucleotide is not included in the EC and is not involved in transcription (data not shown). When the reaction was performed in the presence of σ70-subunit we observed a transcription pause that occurs when the RNA reaches a length of 25 nt (lanes 4-6). The effectiveness of pausing is about 70% and gradually decreases over time, as a result of RNAP transition to productive elongation (Fig. 2a, plot on right). High efficiency of σ-dependent pause formation on this template allows using this approach in further studies of the mechanism of pausing.Fig. 2. Formation of transcription pauses by RNAPs from E. coli and T. aquaticus. a) Electrophoretic analysis of transcription products synthesized on the synthetic template by RNAPs from E. coli and T. aquaticus in the absence or in the presence of E. coli (Eco) or T. aquaticus (Taq) σ-subunits. The position of RNA products of different length is indicated on the left of the figure. The efficiencies of pause formation against time are shown in the plot on the right for RNAPs from E. coli (Eco/Eco), T. aquaticus (Taq/Taq), and for the hybrid RNAP consisting of E. coli core enzyme and the σA-subunit of T. aquaticus (Eco/Taq). The experiment was performed at 37°C in the presence of 100 μM NTP. b) Pausing of RNAPs from T. aquaticus and E. coli in the presence of the σA-subunit at different temperatures. The experiment was performed at 1 mM NTP concentration. c) Activities of E. coli RNAP (1) and the hybrid RNAP (2) on the template containing the lacUV5-promoter at different temperatures. The plot shows the efficiency of synthesis of full-length RNA product as a percentage of maximum activity of E. coli RNAP.
RNAP of T. aquaticus also possesses transcription activity on the synthetic template and is able to synthesize full-length RNA product (Fig. 2a, lanes 7-9). It is worth noting that RNAP from T. aquaticus has a higher temperature optimum of activity than RNAP from E. coli [29, 30]. However, under the conditions of our experiments, RNAP from T. aquaticus can synthesize full-length RNA at 37°C with the same efficiency as the RNAP from E. coli (Fig. 2a). When the reaction was performed in the presence of σA-subunit, we observed a transcription pause and the efficiency of pausing was comparable to that of RNAP from E. coli (Fig. 2a, lanes 10-12 and plot). Thus, RNAP of T. aquaticus is also capable of σ-dependent pausing during transcription elongation. It is worth noting that, in contrast to RNAP from E. coli, the pause is mainly formed when the length of the RNA is 23 nt and, to a lesser extent, 25 nt. Analysis of the temperature dependence of the reaction showed that the efficiency of the RNAP pause formation in T. aquaticus is not changed when the temperature is increased to 50°C, but it rises significantly with temperature decrease and reaches almost 100% at 20°C (Fig. 2b).
The σA-subunit of T. aquaticus cannot induce transcription pausing of RNAP from E. coli. It was shown previously that the σA-subunit of T. aquaticus forms an active holoenzyme in complex with core enzyme of E. coli [30, 31, 34]. This allows to directly compare the properties of σ-subunits of E. coli and T. aquaticus in complex with E. coli core enzyme and also to investigate to what extent the characteristics of RNAP pause formation in T. aquaticus are associated with structural features of the σA-subunit. It is worth noting that the σ70-subunit of E. coli is not capable of forming the holoenzyme with the core enzyme of T. aquaticus, which makes it impossible to compare properties of these two subunits in the complex with the core enzyme of T. aquaticus [29, 34].
We investigated whether the σA-subunit of T. aquaticus can interact with EC of E. coli RNAP and induce a transcription pause. It was found that pause formation occurs with much lower efficiency (10%) in the presence of the σA-subunit than in the presence of the σ70-subunit (Fig. 2a, lanes 13-15, and plot on right). The efficiency of pause formation does not increase when changing the reaction temperature (in the range from 20 to 50°C; Fig. 2b). The pause occurs mainly in EC containing RNA with length of 25 nt, i.e. in the same template position as in the case of pausing in the EC of E. coli RNAP in the presence of the σ70-subunit (Fig. 2a). Thus, the differences in the pause position between RNAPs from E. coli and T. aquaticus are apparently determined by structural features of the RNAP core enzyme.
To confirm that the σA-subunit of T. aquaticus is able to stimulate promoter-dependent transcription of E. coli RNAP under the conditions of our experiments, we compared the activities of E. coli RNAP and the hybrid RNAP containing the E. coli core enzyme and the σA-subunit in an in vitro transcription test sing a DNA fragment containing the lacUV5-promoter as a template. This promoter is one of the well-studied classical promoters and contains –10 and –35 elements [15]. It was shown that the hybrid RNAP is capable of transcription initiation at the lacUV5-promoter. The temperature optimum of promoter-dependent activity of the hybrid RNAP is slightly shifted towards higher temperatures, but its activity at 37-45°C is comparable to the activity of E. coli RNAP (Fig. 2c). Thus, the σA-subunit of T. aquaticus is able to interact with the core enzyme of E. coli and to recognize the –10 and –35 promoter elements.
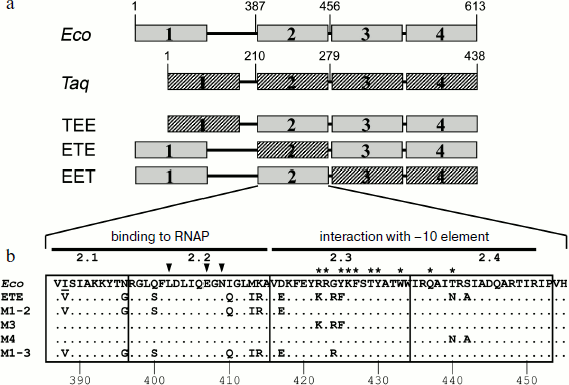
Effect of substitutions in conserved regions of the σ70-subunit on pause formation. Reduced efficiency of pause formation by the hybrid RNAP containing core enzyme of E. coli and the σA-subunit of T. aquaticus allowed us to search for regions of the σ-subunit that play a functionally important role in pausing. To locate these regions, we used an approach based on analysis of mosaic σ-subunits comprised of different parts of the E. coli and T. aquaticus σ-subunits. Since the recognition of the pause signal (the –10 element) involves conserved region 2 of the σ-subunit, the E. coli and T. aquaticus sequences were connected at the boundaries of this region. In total, we prepared and studied three mosaic σ-subunits based on the E. coli σ70-subunit, in which some parts were replaced with the corresponding sequences of the σA-subunit of T. aquaticus (Fig. 3a): 1) σTEE, σ-subunit with the replacement of conserved region 1 and a nonconserved segment between regions 1 and 2 (residues 1-386 of E. coli replaced by residues 1-209 of T. aquaticus; the difference in the lengths of fragments is explained by the different length of the nonconserved segment in these σ-subunits); 2) σETE, σ-subunit with the replacement of conserved region 2 (residues 387-455 of E. coli replaced by residues 210-278 of T. aquaticus); 3) σEET, σ-subunit with the replacement of conserved regions 3 and 4 (residues 456-613 replaced by residues 279-438).
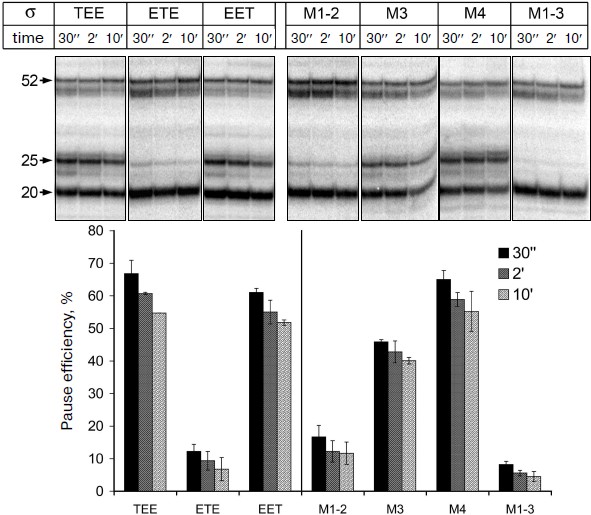
It was shown previously that all three mosaic σ-subunits form active holoenzymes with core enzyme of E. coli [7]. Mosaic subunits were expressed in E. coli cells and obtained in purified form. Analysis of E. coli RNAP transcription on a synthetic template containing the pause signal in the presence of these subunits showed that the replacement of regions 1, 3 and 4 in the σ-subunits σTEE and σEET had practically no effect on the pause formation. However, the replacement of region 2 in the σ-subunit σETE reduced the efficiency of the pause formation to the same level as observed in the case of the σA-subunit of T. aquaticus (Fig. 4). Thus, the low efficiency of E. coli RNAP transcription pausing with σA-subunit of T. aquaticus is apparently explained by the structural features of region 2 of this subunit.Fig. 3. Structure of studied σ-subunit variants. a) Schematic representation of σ-subunits from E. coli, T. aquaticus, and the mosaic subunits σTEE, σETE, and σEET. Numbers above the diagrams correspond to amino acid residues. The relative sizes of the conserved areas are shown not to scale. b) Region 2 sequence comparison of the σ70-subunit of E. coli (Eco) and of the mosaic subunits. σETE contains a sequence of region 2 from T. aquaticus. Regions 2.1, 2.2, 2.3 and 2.4 are marked by frames. The numbers under the sequences denote amino acid residue numbers in the σ70-subunit of E. coli. Lines above the sequences denote the parts of region 2 involved in interactions with the RNAP core enzyme and promoter DNA. Arrows indicate the conserved residues in subregion 2.2 whose substitutions prevent E. coli RNAP pausing [19, 21]. Asterisks denote amino acid residues whose substitutions increased the temperature of promoter melting by E. coli RNAP [8-10].
Effect of nonconserved amino acids substitutions in region 2 of the σ70-subunit on pausing. Conserved region 2 of the σ-subunit consists of four subregions: 2.1, 2.2, 2.3, and 2.4 (Fig. 3b). Sequence comparison of region 2 from σ-subunits of E. coli and T. aquaticus shows that they differ in 12 amino acid residues, and substitutions are present in all four subregions of this region (Fig. 3b). To determine which of these changes affect pause formation, we examined four additional mutant variants of the σ70-subunit that contained only some of these substitutions (Fig. 3b). The σ-subunit M1-2 contained seven substitutions in subregions 2.1 and 2.2, σ M3 contained three substitutions in subregion 2.3, σ M4 had two substitutions in subregion 2.4, and σ M1-3 combined substitutions in subregions 2.1 and 2.2 and the G424R substitution in subregion 2.3 (the most nonconserved substitution out of three substitutions present in this subregion). It was found that substitutions in σ M1-2 significantly reduce the efficiency of E. coli RNAP pause formation; the effect of these changes is almost as pronounced as in the case of σETE (Fig. 4). Substitutions in σ M3 reduce the efficiency of pause formation about 1.5-fold, and substitutions in σ M4 have practically no effect on pausing. The combination of substitutions in subregions 2.1-2.3 in the case of σ M1-3 reduces the efficiency of the pause formation to the level characteristic of the σA-subunit of T. aquaticus and mosaic subunit σETE (Fig. 4). None of the studied mutations altered pause position, i.e. transcription stop occurred at RNA length of 25 nt in all cases. Thus, the inability of the T. aquaticus σA-subunit to induce formation of E. coli RNAP pauses is mainly due to nonconserved substitutions of amino acid residues in regions 2.1 and 2.2 of the σ-subunit.Fig. 4. Transcription pausing by E. coli RNAP in the presence of mosaic σ-subunits. The experiment was performed at 37°C in the presence of 100 μM NTP substrates.
DISCUSSION
Transcription elongation is an important regulatory step in gene expression. Regulatory pauses of various types, which are formed during transcription elongation, can affect the level of gene expression by changing the efficiency of elongation and of transcription termination and by RNAP interaction with additional regulatory factors [35-37]. The σ70-dependent pauses were detected in initially-transcribed regions of lambdoid bacteriophage late genes, as well as in many genes of E. coli [14, 18, 38]. Although the functional role of these pauses is largely unknown, it is assumed that they can change the properties of EC and affect its interaction with transcription factors [38]. In this study we compared the formation of σ-dependent RNAP pauses of E. coli and T. aquaticus and investigated how substitutions of evolutionary nonconserved amino acids in different parts of the σ-subunit, including the regions involved in interactions with DNA and RNAP in the elongation complex, influence pausing.
It is worth noting that the majority of studies of σ-dependent pauses have been devoted to analysis of E. coli RNAP. At the same time, RNAP of T. aquaticus is one of the main models for structural studies of molecular mechanisms of transcription, since for this RNAP the high-resolution three-dimensional structure was first deciphered [22, 39]. This allows using the results of studies of transcription pausing by T. aquaticus RNAP for structural modeling of the pausing process. Our studies have shown that RNAP of T. aquaticus is capable of forming σA-dependent transcription pauses, and the pause formation efficiency is comparable with that of E. coli RNAP at temperatures 37°C. As the temperature decreases, the efficiency of T. aquaticus RNAP pausing increases dramatically. This is likely to be explained by a significantly lower rate of transcription elongation by RNAP of T. aquaticus at low temperatures [30, 40], which stimulates transcription stop at the pause site.
The pause occurs earlier in the case of T. aquaticus RNAP than in the case of RNAP from E. coli: preferential position of pause is shifted two nucleotides upstream on the template DNA. Analysis of E. coli RNAP pausing in the presence of σA-subunit from T. aquaticus showed that such a shift in the pause position is probably caused by the properties of RNAP core enzyme. We can propose that the position of the pause is affected by the differences in contacts between core enzymes of E. coli and T. aquaticus and transcribed DNA template. The stressed complex formed during transcription at the pause site is probably less stable in the case of T. aquaticus RNAP, which leads to an earlier stop of transcription.
We found that the σA-subunit of T. aquaticus is not able to induce transcriptional pausing of RNAP from E. coli with high efficiency, at neither low nor high temperatures. However, a hybrid holoenzyme that consists of E. coli core enzyme and the σA-subunit possesses normal transcriptional activity comparable to that of the holoenzyme of E. coli. The most likely explanation for this fact is that the σA-subunit cannot efficiently bind to the EC of E. coli RNAP.
We analyzed mosaic subunits containing various conserved regions from E. coli and T. aquaticus to find differences in the regions of the σ-subunits that affect pause formation. It was found that the inability of σA-subunit to induce a pause in the transcription by RNAP of E. coli is caused by structural features of subunit region 2. This region plays the main role in interactions of σ-subunit with core enzyme of RNAP and –10 element during transcription initiation. In particular, the substitutions of amino acid residues in subregions 2.1 and 2.2 (V387A, L402F, D403A, Q406A, E407A, N409D, M413T) disrupt the interaction of σ70 with RNAP core enzyme [5], and substitutions of the conserved aromatic and positively charged residues in subregions 2.3 and 2.4 affect the interaction of RNAP holoenzyme with promoters and increase the temperature of transcription initiation (Fig. 3b) [8-10, 12, 41].
The influence of nonconserved amino acid substitutions in region 2 of the σ70-subunit on transcription pausing could be explained both by a change in its interactions with the RNAP core enzyme and by a change in contacts with the non-template DNA chain in the transcribed region. The study of substitutions in different parts of the σ70-subunit region 2 showed that the formation of transcription pauses is most greatly influenced by substitutions in subregions 2.1 and 2.2; the introduction of corresponding substitutions from the σA-subunit to the σ70-subunit leads to a significant decrease in the efficiency of pause formation. The effect of substitutions in subregions 2.1 and 2.2 on pause formation is most probably directly caused by their influence on the interaction of the σ-subunit with the RNAP core enzyme. The amino acid substitution G424R in subregion 2.3 also leads to some decrease in pausing efficiency, and a combination of substitutions in subareas 2.1, 2.2, and 2.3 has the most substantial effect. G424R substitution is located in a protein loop that apparently does not interact with DNA and RNAP core enzyme; this substitution may affect the conformational changes of the σ-subunit that occur during its interactions with the core enzyme and DNA [7]. It was shown previously that conserved amino acid substitutions in subregion 2.2 of σ70 (L402F, E407K, N409D; Fig. 3b) disrupt pausing, probably due to changes in interactions with the RNAP core enzyme [19, 21]. Our results indicate an important role of nonconserved amino acid residues in interactions with RNAP core enzyme in the process of transcription elongation.
It was shown previously that the replacement of nonconserved amino acid residues in region 2 of σ70 for corresponding residues of the σA-subunit of T. aquaticus increases promoter melting temperature compared with the wild-type σ70-subunit [7]. As in the case of σ-dependent transcription pausing, nonconserved amino acid substitution in subregions 2.1 and 2.2 had the greatest effect on promoter recognition. Nevertheless, all the studied mutant variants of the σ70-subunit as well as the σA-subunit of T. aquaticus can form active holoenzyme with core enzyme of E. coli and are capable of promoter-dependent transcription initiation (reference [7] and this work). Thus, the substitutions in region 2 have a stronger impact on the interaction of σ-subunit with transcription complex during elongation than on the recognition and melting of promoters during initiation. This is probably due to the fact that the recognition of promoters and the interaction with the RNAP core enzyme in the promoter complex involves additional regions of σ-subunit (including regions 3 and 4), whereas the main role in the interaction of the σ-subunit with EC is played by region 2.
Our studies have allowed us to identify σ-subunit regions involved in pause formation and revealed the role of nonconserved amino acid region 2 in this process. Thus, the study of mosaic subunits is a promising approach for future studies of transcription mechanisms in different organisms. A similar approach can be used to identify functionally important regions of RNAP and to identify evolutionary changes that alter the functional properties of the protein and have adaptive value.
This work was partially supported by the Russian Foundation for Basic Research grant No. 10-04-00925, Program of the Russian Academy of Sciences Presidium “Molecular and Cellular Biology”, Russian President Grant for State Support of Young Scientists (MD-618.2011.4), and the Targeted Federal Program “Scientific and Educational Research Personnel of Innovative Russia” for the years 2009-2013 (state contracts No. 02.740.11.5132 and P335). Work of K.L.B. was supported by INSERM.
REFERENCES
1.Gross, C. A., Chan, C., Dombroski, A., Gruber, T.,
Sharp, M., Tupy, J., and Young, B. (1998) Cold Spring Harb. Symp.
Quant. Biol., 63, 141-155.
2.Murakami, K. S., and Darst, S. A. (2003) Curr.
Opin. Struct. Biol., 13, 31-39.
3.Haugen, S. P., Ross, W., and Gourse, R. L. (2008)
Nat. Rev. Microbiol., 6, 507-519.
4.Campbell, E. A., Muzzin, O., Chlenov, M., Sun, J.
L., Olson, C. A., Weinman, O., Trester-Zedlitz, M. L., and Darst, S. A.
(2002) Mol. Cell, 9, 527-539.
5.Sharp, M. M., Chan, C. L., Lu, C. Z., Marr, M. T.,
Nechaev, S., Merritt, E. W., Severinov, K., Roberts, J. W., and Gross,
C. A. (1999) Genes Dev., 13, 3015-3026.
6.Vassylyev, D. G., Sekine, S., Laptenko, O., Lee,
J., Vassylyeva, M. N., Borukhov, S., and Yokoyama, S. (2002)
Nature, 417, 712-719.
7.Barinova, N., Zhilina, E., Bass, I., Nikiforov, V.,
and Kulbachinskiy, A. (2008) J. Bacteriol., 190,
3088-3092.
8.Panaghie, G., Aiyar, S. E., Bobb, K. L., Hayward,
R. S., and de Haseth, P. L. (2000) J. Mol. Biol., 299,
1217-1230.
9.Fenton, M. S., Lee, S. J., and Gralla, J. D. (2000)
EMBO J., 19, 1130-1137.
10.Tomsic, M., Tsujikawa, L., Panaghie, G., Wang,
Y., Azok, J., and DeHaseth, P. L. (2001) J. Biol. Chem.,
276, 31891-31896.
11.Schroeder, L. A., Gries, T. J., Saecker, R. M.,
Record, M. T., Jr., Harris, M. E., and DeHaseth, P. L. (2009) J.
Mol. Biol., 385, 339-349.
12.Schroeder, L. A., Choi, A. J., and DeHaseth, P.
L. (2007) Nucleic Acids Res., 35, 4141-4153.
13.Ring, B. Z., Yarnell, W. S., and Roberts, J. W.
(1996) Cell, 86, 485-493.
14.Perdue, S. A., and Roberts, J. W. (2011) J.
Mol. Biol., in press.
15.Brodolin, K., Zenkin, N., Mustaev, A., Mamaeva,
D., and Heumann, H. (2004) Nat. Struct. Mol. Biol., 11,
551-557.
16.Zenkin, N., Kulbachinskiy, A., Yuzenkova, Y.,
Mustaev, A., Bass, I., Severinov, K., and Brodolin, K. (2007) EMBO
J., 26, 955-964.
17.Nickels, B. E., Mukhopadhyay, J., Garrity, S. J.,
Ebright, R. H., and Hochschild, A. (2004) Nat. Struct. Mol.
Biol., 11, 544-550.
18.Hatoum, A., and Roberts, J. (2008) Mol.
Microbiol., 68, 17-28.
19.Ko, D. C., Marr, M. T., Guo, J., and Roberts, J.
W. (1998) Genes Dev., 12, 3276-3285.
20.Mooney, R. A., Darst, S. A., and Landick, R.
(2005) Mol. Cell, 20, 335-345.
21.Sevostyanova, A., Svetlov, V., Vassylyev, D. G.,
and Artsimovitch, I. (2008) Proc. Natl. Acad. Sci. USA,
105, 865-870.
22.Murakami, K. S., Masuda, S., and Darst, S. A.
(2002) Science, 296, 1280-1284.
23.Marr, M. T., and Roberts, J. W. (2000) Mol.
Cell, 6, 1275-1285.
24.Perdue, S. A., and Roberts, J. W. (2010) Mol.
Microbiol., 78, 636-650.
25.Iyer, L. M., Koonin, E. V., and Aravind, L.
(2004) Gene, 335, 73-88.
26.Lane, W. J., and Darst, S. A. (2010) J. Mol.
Biol., 395, 671-685.
27.Lane, W. J., and Darst, S. A. (2010) J. Mol.
Biol., 395, 686-704.
28.Opalka, N., Brown, J., Lane, W. J., Twist, K. A.,
Landick, R., Asturias, F. J., and Darst, S. A. (2010) PLoS
Biol., 8.
29.Minakhin, L., Nechaev, S., Campbell, E. A., and
Severinov, K. (2001) J. Bacteriol., 183, 71-76.
30.Kulbachinskiy, A., Bass, I., Bogdanova, E.,
Goldfarb, A., and Nikiforov, V. (2004) J. Bacteriol.,
186, 7818-7820.
31.Kulbachinskiy, A. V., Nikiforov, V. G., and
Brodolin, K. L. (2005) Biochemistry (Moscow), 70,
1227-1230.
32.Sidorenkov, I., Komissarova, N., and Kashlev, M.
(1998) Mol. Cell, 2, 55-64.
33.Komissarova, N., Kireeva, M. L., Becker, J.,
Sidorenkov, I., and Kashlev, M. (2003) Methods Enzymol.,
371, 233-251.
34.Kuznedelov, K., Minakhin, L., and Severinov, K.
(2003) Methods Enzymol., 370, 94-108.
35.Roberts, J. W., Shankar, S., and Filter, J. J.
(2008) Annu. Rev. Microbiol., 62, 211-233.
36.Landick, R. (2006) Biochem. Soc. Trans.,
34, 1062-1066.
37.Santangelo, T. J., and Artsimovitch, I. (2011)
Nat. Rev. Microbiol., 9, 319-329.
38.Deighan, P., Pukhrambam, C., Nickels, B. E., and
Hochschild, A. (2011) Genes Dev., 25, 77-88.
39.Zhang, G., Campbell, E. A., Minakhin, L.,
Richter, C., Severinov, K., and Darst, S. A. (1999) Cell,
98, 811-824.
40.Miropolskaya, N., Artsimovitch, I., Klimasauskas,
S., Nikiforov, V., and Kulbachinskiy, A. (2009) Proc. Natl. Acad.
Sci. USA, 106, 18942-1897.
41.Schroeder, L. A., and DeHaseth, P. L. (2005)
J. Biol. Chem., 280, 17422-17429.