Biogenic Polyamines Spermine and Spermidine Activate RNA Polymerase and Inhibit RNA Helicase of Hepatitis C Virus
A. N. Korovina#, V. L. Tunitskaya#, M. A. Khomutov, A. R. Simonian, A. R. Khomutov, A. V. Ivanov*, and S. N. Kochetkov
Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, ul. Vavilova 32, 119991 Moscow, Russia; fax: (499) 135-1405; E-mail: aivanov@yandex.ru; aivanov@eimb.ru# Contributed equally to this work.
* To whom correspondence should be addressed.
Received April 25, 2012
Influence of the biogenic polyamines spermine, spermidine, and putrescine as well as their derivatives on the replication enzymes of hepatitis C virus (HCV) was investigated. It was found that spermine and spermidine activate HCV RNA-dependent RNA polymerase (NS5B protein). This effect was not caused by the stabilization of the enzyme or by competition with template–primer complex, but rather it was due to achievement of true maximum velocity Vmax. Natural polyamines and their derivatives effectively inhibited the helicase reaction catalyzed by another enzyme of HCV replication – helicase/NTPase (NS3 protein). However, these compounds affected neither the NTPase reaction nor its activation by polynucleotides. Activation of the HCV RNA polymerase and inhibition of the viral helicase were shown at physiological concentrations of the polyamines. These data suggest that biogenic polyamines may cause differently directed effects on the replication of the HCV genome in an infected cell.
KEY WORDS: polyamines, spermine, spermidine, analogs, hepatitis C virus, RNA polymerase, helicaseDOI: 10.1134/S0006297912100094
Biogenic polyamines—spermine and spermidine and their precursor putrescine—are present in cells in millimolar and submillimolar concentrations. Their interactions with nucleic acids and nucleoprotein complexes are involved in the regulation of diverse cellular processes, including DNA replication and transcription and mRNA processing and translation [1]. Polyamines are also involved in protecting cells from oxidative stress by neutralizing reactive oxygen species and activating gene expression for protective proteins [2-4].
A rate-limiting step of the biosynthesis of spermidine and spermine is catalyzed by ornithine decarboxylase (ODC) forming putrescine from L-ornithine [5]. The degradation of polyamines occurs via two alternative pathways. One of the most studied is the N1-monoacetylation of spermine and spermidine by the spermidine/spermine-N1-acetyltransferase (SSAT) and the subsequent oxidative cleavage of the N1-acetyl derivatives to putrescine and spermidine, respectively, which is catalyzed by polyamine oxidase (APAO) [6]. An alternative pathway of degradation of spermine, which was discovered in 2001, is a one-step reaction catalyzed by spermine oxidase (SMO) [7]. Maintenance of the desired concentration of each polyamine is achieved via regulation of expression of ODC, SSAT, and SMO at the levels of transcription of their genes, splicing, and translation of mRNA and degradation of the protein in cells [5, 6].
It has been found that disturbances in the metabolism of spermine and spermidine, which lead to a change in their concentration in the cell, usually accompany the development of a number of diseases. It was established that cancer cells and tumor tissues contain high concentrations of biogenic polyamines, which is primarily due to their increased biosynthesis. The level of expression of ODC correlates with the stage of tumor development and its invasiveness [8-10]. In this case we cannot exclude that the overexpression of both ODC and SSAT contributes to the transformation of normal cells into tumor cells [11-13]. In addition to carcinogenesis, a connection between defects in the metabolism of polyamines with the development of metabolic and parasitic diseases has been found [14, 15].
Despite the significant progress in biomedical research on spermine and spermidine, their role in the development of viral diseases remains rather unclear. The main work in this area dates back to 1970-1980 and relates mainly to the viruses of plants and pox, herpes, and retroviruses [16]. Polyamines were also found in viral particles, forming complexes with genomic DNA or RNA and partially neutralizing its charge. The viruses themselves can cause increased levels of polyamines in the host cell. In addition, inhibition of the biosynthesis of the polyamine putrescine revealed the importance of polyamines for the effective reproduction of a number of viruses. The analysis of expression of individual enzymes of polyamine metabolism, as well as the influence of polyamines on the functioning of individual viral proteins and the occurrence of certain stages of the viral life cycle remained beyond the scope of that study. One of the few exceptions was work [17], which reported an increase in enzymatic activity of DNA polymerase of herpes simplex virus type 1 (HSV-1) in activated DNA assay and an inhibition of DNA polymerase in the system of homopolymer primer–template complexes.
The object of research in our work is the hepatitis C virus (HCV), which belongs to the family Flaviviridae [18]. HCV is widespread and causes chronic liver disease, often accompanied by the development of fibrosis and cirrhosis, impaired metabolism of lipids (steatosis), carbohydrates, iron and other substances, as well as the occurrence of liver cancer (hepatocellular carcinoma) and blood (non-Hodgkin’s lymphoma) [18, 19]. Since the discovery of HCV in 1989, the structure of its genome and basic functions of ten virus encoded proteins were studied, and many aspects of the viral life cycle and the basic mechanisms of a number of metabolic abnormalities in patients with hepatitis C were revealed [18]. However, there have been no studies of possible links between HCV infection and polyamine metabolism.
The goal of the current study was to investigate the regulation by biogenic polyamines and analogs of HCV replication enzymes: RNA-dependent RNA polymerase (NS5B protein) and RNA-helicase/NTPase (NS3 protein).
MATERIALS AND METHODS
Reagents and materials. The following reagents and materials were used in our study: Escherichia coli strain Rosetta (DE3) and Ni-NTA-agarose (Novagen, USA), bacto-tryptone, yeast extract, bacto-agar, and ammonium persulfate (Amresco, USA), Tris, EDTA, and 2-mercaptoethanol (Merck, Germany), glycerol, dithiothreitol, imidazole, Triton X-100, phenylmethylsulfonyl fluoride (PMSF) and a cocktail of proteinase inhibitors (Sigma, USA), acrylamide and methylene-bis-acrylamide (Roth, Germany), Coomassie Brilliant Blue R-250 (Bio-Rad, USA), and tetramethylethylenediamine (Reanal, Hungary). Other reagents of analytical or chemically pure grade were from Reakhim (Russia). Oligonucleotides for determination of helicase activity were obtained from Lytech (Russia).
Spermintrien (SmpTrien, 1,12-diamino-3,6,9-triazadodecane) [20], 5-oxaspermine (5-oxaSpm, 1,12-diamino-4,9-diaza-5-oxadodecane) [21], α,α′-dimethylspermine (α,α′-Me2Spm, 2,13-diamino-5,10-diazatetradecane) [22, 23], α-methylspermidine (α-MeSpd, 1,8-diamino-5-azanonane) [23, 24], β-methylspermidine (β-MeSpd, 1,8-diamino-2-methyl-4-asaoctane), γ-methylspermidine (γ-MeSpd, 1,8-diamino-3-methyl-4-azaoctane), and β,β-dimethylspermidine (β,β-Me2Spd, 1,8-diamino-2,2-dimethyl-4-azaoctane) [25] were obtained as previously described in the literature. Synthesis of β,β′-dimethylspermine (β,β′-Me2Spm, 1,12-diamino-2,11-dimethyl-4,9-diazadodecane) and γ,γ′-dimethylspermine (γ,γ′-Me2Spm, 1,12-diamino-3,10-dimethyl-4,9-diazadodecane) will be the subject of a separate publication. The formulas of these compounds are shown in Fig. 1.
Fig. 1. Biogenic polyamines and their analogs used in this work.
Isolation of recombinant HCV RNA-dependent RNA polymerase and helicase/NTPase. The recombinant RNA-dependent RNA polymerase (protein NS5B) and helicase/NTPase domain of NS3 protein were obtained using plasmids pET-21d-2c-NS5BΔ55 and pET-21d-2c-NS3hel previously constructed by us [26-28].
Investigation of effect of polyamines on activity of HCV RNA-dependent RNA polymerase. The activity of HCV RNA polymerase was determined in the poly(A)–oligo(U) system as previously described [29]. Spermine and spermidine were added to the reaction mixture to achieve final concentrations of 0.03-3 mM.
Investigation of effect of polyamines on helicase and NTPase activity of HCV NS3 protein. Determination of NTPase and helicase enzyme activities is described in detail in the Appendix to our previous paper [30]. To obtain a duplex, the following deoxyoligonucleotides were used:
5′-CGTCGACACTGACACGACTCACCACAGGACTCCA-3′;
5′-GTCGTGTCAGTGTCGACG-3′.
In addition, to prevent reannealing of unwound oligonucleotides, an oligonucleotide-capture, 5′- cgtcgacactgacacgac-3′, was introduced into the reaction mixture. Radioactive label to the 5′-end of one of the oligonucleotides was introduced using phage T4 polynucleotide kinase and [γ-32P]ATP in accordance with the standard protocol.
During the study of the inhibitory effect of biogenic polyamines, they were added to the tested sample in aqueous solution. The concentrations of polyamines and their analogs in preliminary experiments were 0.05-10 mM, then for each compound an adequate range of concentrations to determine IC50 (concentration at which the enzyme helicase activity is inhibited by 50%) was selected.
In the study of complex formation of the enzyme with the duplex, 5′-32P-labeled 34-mer oligonucleotide was used. The standard reaction mixture was similar to that used for determination of helicase activity, but it did not contain ATP and oligonucleotide-capture. Samples were incubated for 10 min at 20°C. Upon completion of the reaction, the samples were supplemented (instead of the stop buffer) with 2 µl (10%) of glycerol and immediately applied on 8% polyacrylamide gel (gel buffer and electrode buffer was 0.5× TBE). After preliminary electrophoresis for 30 min, the separation was performed at 4°C, as in the case of determination of helicase activity. Radioactive bands corresponding to duplex and DNA–protein complex were visualized by the Packard Cyclone Storage Phosphor System, the intensity of the bands being calculated by the TotalLab 2.01 program.
Statistical analysis of results. The data were statistically analyzed using BioStat 2008 software (AnalystSoft, Canada). All data represent the mean ± standard deviation (SE, N = 3). Significance of differences between the two groups of values was determined by the paired t-test. Comparison of several groups of values was performed using analysis of variance (ANOVA) and Tukey’s test. Statistically significant differences between the two groups of values were considered for p < 0.05.
RESULTS
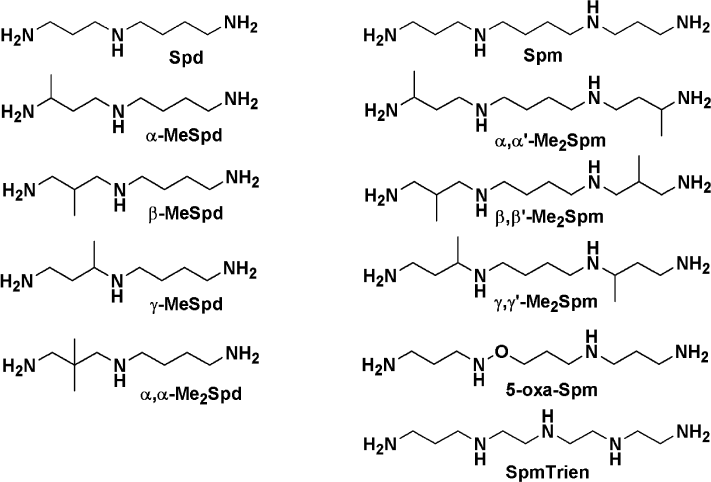
Spermine and spermidine are activators of HCV RNA-dependent RNA polymerase. The activity of HCV RNA-dependent RNA polymerase was determined in the poly(A)–oligo(U) primer–template complex evaluating the incorporation of radioactively-labeled [α-32P]UTP [26]. The influence of biogenic polyamines was studied in the range of concentrations from 30 µM to 3 mM. Spermine and spermidine were found to be activators of RNA polymerase activity of protein NS5B, while their metabolic precursor putrescine had no appreciable effect on the functioning of the enzyme (Fig. 2a). The effect of activation was observed at the concentrations of polyamines below 2 mM and was more pronounced for spermine than for spermidine. At higher concentrations of polyamines, a partial reduction of the effect was observed.
Fig. 2. Effect of polyamines on the activity of HCV RNA-dependent RNA polymerase. a) Dependence of enzymatic activity on the concentration of polyamines. b) Activation by spermine of enzyme preparations stored in the absence and presence of 2.5 mM spermine. c) Dependence of activation of RNA polymerase by spermine on the ratio of primer–template complex and the enzyme: 1) the standard ratio (see “Materials and Methods”); 2) 3-fold excess of primer–template complex; 3) 4.3-fold excess of primer–template complex. d) Absolute activity of HCV RNA polymerase (counts/min) under conditions similar to those in panel (c).
HCV RNA polymerase has a very low stability—the enzyme activity decreases even during the enzymatic reaction [31]. Since biogenic polyamines are often used to stabilize protein solutions in storage, it was necessary to investigate the effect of spermine on the stability of the enzyme. The introduction of 2.5 mM spermine in the storage buffer of the RNA polymerase stored for six weeks did not affect the specific activity of the enzyme (Fig. 2b).
Activation of HCV RNA polymerase by spermine was studied at different concentrations of the substrate, poly(A)–oligo(U) primer–template complex. It was found that an increase in the amount of the complex is accompanied by a decrease in the maximum degree of activation of the enzyme (Fig. 2c), which could indicate a possible competition between polyamine and primer–template complex. However, we considered the dependence of the concentration of the incorporated nucleotide spermine. In the absence of polyamine, the RNA polymerase exhibits higher activity with increasing concentration of primer–template complex, whereas increase in the concentration of spermine leads to a leveling of the differences (Fig. 2d). This fact may indicate that the enzyme reaches the real maximum reaction rate (Vmax).
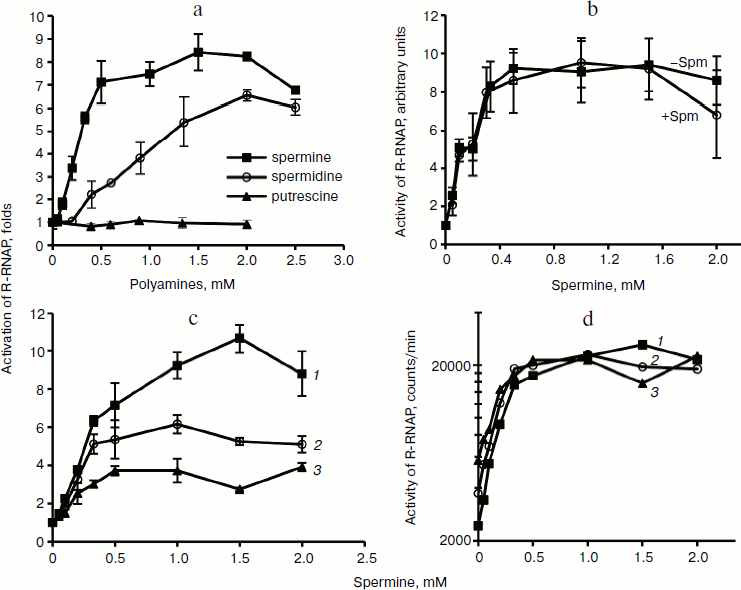
Biogenic polyamines inhibit HCV helicase. The nonstructural protein HCV NS3, along with viral RNA polymerase, is a component of the virus replicase. It exhibits three types of activities: protease (N-terminal domain), helicase, and NTPase (C-terminal domain). In this work we studied the effect of biogenic polyamines on helicase and NTPase activities of the protein using the helicase C-terminal domain of NS3. NTPase activity was measured by hydrolysis of radioactively-labeled [γ-32P]ATP and analysis of the product by TLC. Helicase activity of the protein was measured based on the unwinding of DNA duplex with a long 3′-unpaired end of one of the chains and the separation of reaction products by electrophoresis in polyacrylamide gel under nondenaturing conditions. It was found that spermine, spermidine, and putrescine had no noticeable effect on NTPase activity and its activation by polynucleotides (Fig. 3a) and that spermine, spermidine, and putrescine inhibited the helicase activity of NS3 (Fig. 3b). As in the case of HCV RNA polymerase, the effect of spermine was observed at lower concentrations (IC50 = 0.30 mM) compared with spermidine (IC50 = 0.75 mM) (table). Putrescine inhibited helicase from HCV only at millimolar concentrations (table). The study of the effects of several analogs of spermine and spermidine on the helicase activity of NS3 showed that mono- and dimethylated derivatives of spermidine and spermine are effective at the same concentrations as those that were determined for spermidine and spermine, respectively (table). It can be noted that the introduction of methyl substituents in the spermine molecule somewhat (~30%) increased the effect of suppressing the helicase reaction by the respective compounds, while the introduction of a substituent into the spermidine molecule, on the contrary, led to a deterioration of inhibitory properties in comparison to the parent compound. Spermintrien and 5-oxaspermine, charge-deficient spermine analogs, appeared to be much less effective inhibitors of the enzyme. Diethyl-nor-spermine showed inhibitory activity similar to that of spermidine (table).
Fig. 3. Influence of biogenic polyamines on ATPase and helicase activity of NS3 protein. a) Detection of ATPase activity by TLC. Lanes: 1) control without enzyme; 2, 3) in absence of polyamines; 4, 5) in presence of 0.5 mM spermine; 6, 7) in presence of 1 mM spermidine; 2, 4, 6) without addition of poly(U); 3, 5, 7) in presence of 1 mg/ml poly(U). b) Determination of helicase activity in the absence of polyamines and in the presence of increasing concentrations of spermine and spermidine.
Inhibition of helicase reaction catalyzed by NS3 by polyamines
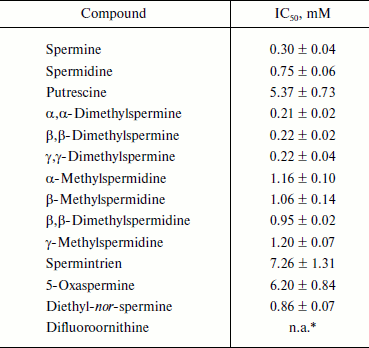
Note: IC50 values presented in the table are means from three
independent experiments ± SE.
* n.a., not active at concentrations below 6 mM.
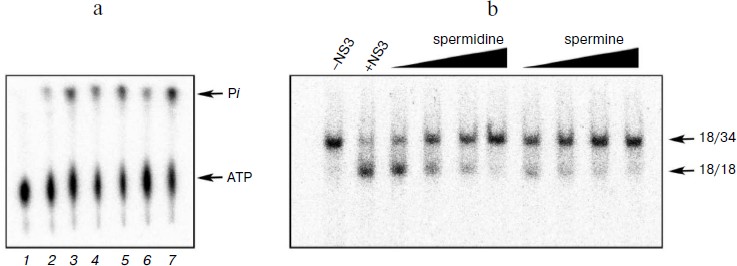
Biogenic polyamines inhibit helicase reaction catalyzed by HCV NS3 via their interaction with the DNA duplex, thus preventing its binding to the enzyme. To elucidate the mechanism of inhibition of the helicase reaction by biogenic polyamines, the effect of spermine (as the most active inhibitor) on the binding of the enzyme to the substrate – DNA duplex was investigated. The effect of spermine on the formation of DNA–protein complex was assessed by their inhibition in the gel. It was shown that spermine inhibits the formation of the corresponding complex, where IC50 value (for the inhibition of binding) was 0.13 ± 0.03 mM (Fig. 4a). Thus, the inhibition is apparently due to the inhibition of the binding of the enzyme with its oligonucleotide substrate. To confirm this hypothesis, we evaluated the inhibitory effect of spermine on the binding of the duplex with the enzyme at a constant concentration (0.2 mM) and its dependence on the order of adding components to the reaction mixture. As expected, the greatest inhibition of binding was achieved for the preincubation of DNA duplex with spermine (Fig. 4b).
Fig. 4. a) Formation of duplex complexes with NS3 in the absence and presence of spermine; b) dependence of inhibition of complex formation on the order of addition of components of the reaction mixture. 1) Control (in the absence of spermine); 2) the order of addition of the reagents (duplex + spermine); addition of the enzyme after 10 min at room temperature; 3) (enzyme + spermine); addition of the duplex after 10 min; 4) (enzyme + duplex); addition of spermine (0.2 mM) after 10 min.
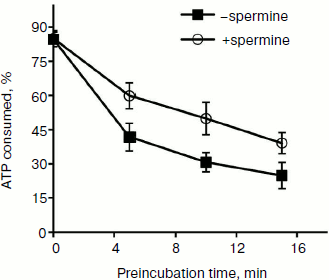
It might be assumed that preincubation of the enzyme with the duplex before the addition of spermine would minimize the inhibition of binding. However, it was found that the lowest inhibitory effect was observed for preincubation of the enzyme with spermine (Fig. 4b, columns 3 and 4). A possible explanation for this could be a partial stabilization of the enzyme in the presence of polyamine during the preincubation. The presence of two opposite effects (inhibition and stabilization) for the helicase reaction makes it difficult to assess the effect of stabilization by this technique. Therefore, this hypothesis was tested for the NTPase reaction catalyzed by NS3, since biogenic polyamines do not inhibit NTPase activity of the protein. Analysis of the dependence of the rate of hydrolysis of ATP on the time of preincubation of the protein in the absence/presence of spermine showed that in the presence of spermine in dilute solution at room temperature a slow inactivation of the enzyme was observed (Fig. 5).
Fig. 5. ATPase activity of helicase/NTPase NS3 depending on time of preincubation of the enzyme at room temperature in the absence or presence of 0.2 mM spermine.
Thus, we can assume that the biological effect of biogenic polyamines on NS3 helicase is largely confined to inhibition of the reaction due to the interaction of polyamine with the duplex, which hinders its binding to the enzyme. On the other hand, the observed inhibitory effect is probably somewhat lower than the actual one because the native structure of the enzyme during the reaction in some way could be stabilizes by the same biogenic amine.
DISCUSSION
There are several examples in the literature of regulation of activity of DNA polymerases by polyamines. For example, Wallace et al. showed that spermine alters the activity of DNA-dependent DNA polymerase of HSV-1 [17]. Thus, when activated DNA was used as a substrate, activation of the enzyme (maximal in the presence of 2.5 mM spermine) was observed, whereas in systems with a homopolymer poly(dA)–oligo(dT) or poly(dC)–oligo(dG) primer–template complexes a significant inhibition of DNA polymerase activity due to the reduction of its processivity was observed. Inhibition of enzyme activity has also been described for eukaryotic DNA polymerases α and β [32, 33], and the activation of enzyme activity has also been described for the DNA polymerase of the malaria parasite Plasmodium falciparum [34] and DNA-dependent RNA polymerases I, II, and III [35]. Furthermore, there are data on the regulation of the activity and specificity of other DNA-binding enzymes including topoisomerases and restriction endonucleases [36, 37]. This work is the first to report that spermine and spermidine are activators of HCV RNA polymerase, and this effect is not related to a possible stabilization of this rather labile enzyme.
For HSV DNA polymerase, the observed effect of spermine is due to its interaction with the primer–template complex [17]. RNA polymerase, however, interacts with polyamines directly, this being supported by the following facts. First, putrescine, which is doubly protonated at physiological pH, showed no catalytic activity, as opposed to spermine and spermidine, which are four- and three-fold protonated. Second, the RNA polymerase showed a similar level of activity in the presence of spermine at various concentrations of the primer–template complex, indicating the achievement of the real maximum reaction rate (Vmax). Third, it has been previously shown that HCV RNA polymerase binds a single stranded template followed by annealing of the primer, which does not allow the observed activation of HCV RNA polymerase to be interpreted in terms of stabilizing of primer–template duplex by polyamines.
The exact mechanism of activation of HCV RNA polymerase by polyamines is unknown. The polymerase reaction can be represented as two separate stages: binding of the enzyme to the template–primer complex and primer elongation. The standard approach to the elucidation of the influence of substances on the elongation stage is to perform enzymatic reactions in the presence of heparin, which has a high affinity for the free enzyme and thus prevents re-initiation of the synthesis [29, 31]. Unfortunately, the use of heparin in this case is impossible because of its binding to the positively charged polyamines [38].
It can be assumed that the activation of HCV RNA polymerase is due to the influence of polyamines on the oligomerization of the protein. It is known that RNA polymerase from HCV is active in the monomeric and homodimeric forms, and a change in the degree of oligomerization affects the specific activity of the protein [39, 40]. Recently it was shown that the oligomerization state of NS5B protein could be disrupted by nonionic and zwitterionic detergents and thereby could activate polymerase reaction [41]. If such a mechanism is realized also in the case of biogenic polyamines, this indicates existence of the differences in the activation by spermine between variants of HCV RNA polymerase of different genotypes. It was established that the NS5B protein of genotype 2a (isolate JFH) does not form dimers and, consequently, its activity is not increased in the presence of detergents [41].
For NS3 protein (HCV helicase), we showed that the inhibitory effect of biogenic polyamines and their methylated derivatives is due to the binding of the compounds to the DNA duplex. Interaction of polyamines with the protein does not appear to have a significant impact on the functioning of the enzyme. The observed stabilization of the enzyme in the presence of spermine is only partial and cannot completely prevent the loss of activity. Finally, there was no significant difference between the activity of spermine and spermidine compared to their corresponding mono- and dimethyl derivatives. In the case of methylated analogs of spermidine, it was previously shown that their interaction with enzymes of polyamines metabolism and their ability to influence the regulatory pathways, which are responsible for maintenance of the homeostasis of polyamines in the cell, depends on the methyl group position and the configuration of the chiral center [25, 42]. In our case, the change in substituents along the chain of the polyamine did not change their properties. Another confirmation that the effect is due to simple electrostatic interaction of polyamines with DNA is reduced activity of spermintrien and 5-oxaspermine, because these are charge-deficient analogs of spermine.
Activation of HCV RNA polymerase can lead to a change in its functions in the cell. In the case of activation of the enzyme by detergent, it was shown that these compounds enhance the binding of protein to RNA, as well as alter the processivity of the enzyme [41]. We can assume that similar changes in the cell can facilitate the initiation of RNA replication, which is the rate-limiting step of the enzymatic reaction. We have previously shown that in vitro nonstructural phosphoprotein NS5A is a regulator of the initiation of RNA biosynthesis catalyzed by HCV RNA polymerase, and the phosphorylation status of NS5A protein determines its ability to act as the regulator [43]. It was also found that that in a cellular system, this mechanism does not occur for unknown reasons [44]. Perhaps this could be explained by changes in the properties of HCV RNA polymerase in the presence of biogenic polyamines.
These data evidently show that biogenic polyamines can cause differently directed effects on the replication of the HCV genome in an infected cell. Thus, activation of RNA polymerase can be balanced by the inhibition of the viral helicase. Both processes are regulated by physiological concentrations of spermine and spermidine, which can reach millimolar values in cells [45]. Further elucidation of the role of biogenic polyamines in the replication of the HCV genome can be performed using analogs of putrescine, spermidine, and spermine, which are polyamine regulators of metabolic enzymes. A significant decrease in the intracellular concentration of individual polyamines with difluoromethylornithine (an inhibitor of ODC) or diethyl-nor-spermine (inductor of SSAT [46]) may allow investigation of the efficiency of viral replication in various systems including HCV replicons, infection system (HCVcc) [47], or by using a reporter plasmid [48]. In this case the obtained effects are not due to the interaction of these regulators of metabolism of polyamines with viral enzymes as at the concentrations used in cell models (10-30 µM for diethyl-nor-spermine and 2-4 mM for difluoroornithine) they do not affect the viral helicase activity.
This work was supported by the Russian Foundation for Basic Research grants 10-04-00047-a (AVI) and 12-04-01487-a (ARKh), a grant of the President of the Russian Federation (MK-5035.2011.4) (AVI), the State Contract No. 11.519.11.2017 (SNK) and Russian Academy of Sciences Presidium Program “Molecular and cellular biology”. Some of the experiments were performed on the equipment of NBI “Genome” (Institute of Molecular Biology, Russian Academy of Sciences).
REFERENCES
1.Pegg, A. E. (2009) IUBMB Life, 61,
880-894.
2.Ha, H. C., Sirisoma, N. S., Kuppusamy, P., Zweier,
J. L., Woster, P. M., and Casero, R. A., Jr. (1998) Proc. Natl.
Acad. Sci. USA, 95, 11140-11145.
3.Tkachenko, A. G., and Fedotova, M. V. (2007)
Biochemistry (Moscow), 72, 109-116.
4.Rider, J. E., Hacker, A., Mackintosh, C. A., Pegg,
A. E., Woster, P. M., and Casero, R. A. (2007) Amino Acids,
33, 231-240.
5.Pegg, A. E. (2006) J. Biol. Chem.,
281, 14529-14532.
6.Pegg, A. E. (2008) Am. J. Physiol. Endocrinol.
Metab., 294, 995-1010.
7.Wang, Y., Devereux, W., Woster, P. M., Stewart, T.
M., Hacker, A., and Casero, R. A., Jr. (2001) Cancer Res.,
61, 5370-5373.
8.Milovic, V., and Turchanowa, L. (2003) Biochem.
Soc. Trans., 31, 381-383.
9.Linsalata, M., Giannini, R., Notarnicola, M., and
Cavallini, A. (2006) BMC Cancer, 6, 191.
10.Deng, W., Jiang, X., Mei, Y., Sun, J., Ma, R.,
Liu, X., Sun, H., Tian, H., and Sun, X. (2008) Acta. Biochim.
Biophys. Sin. (Shanghai), 40, 235-243.
11.Coleman, C. S., Pegg, A. E., Megosh, L. C., Guo,
Y., Sawicki, J. A., and O’Brien, T. G. (2002)
Carcinogenesis, 23, 359-364.
12.O’Brien, T. G., Megosh, L. C., Gilliard,
G., and Soler, A. P. (1997) Cancer Res., 57,
2630-2637.
13.Megosh, L., Gilmour, S. K., Rosson, D., Soler, A.
P., Blessing, M., Sawicki, J. A., and O’Brien, T. G. (1995)
Cancer Res., 55, 4205-4209.
14.Alhonen, L., Parkkinen, J. J., Keinanen, T.,
Sinervirta, R., Herzig, K. H., and Janne, J. (2000) Proc. Natl.
Acad. Sci. USA, 97, 8290-8295.
15.Birkholtz, L. M., Williams, M., Niemand, J.,
Louw, A. I., Persson, L., and Heby, O. (2011) Biochem. J.,
438, 229-244.
16.Tyms, A. S., and Williamson, J. D. (1987) in
Inhibition of Polyamine Metabolism (McCann, P. P., Pegg, A. E.,
and Sjoerdsma, A., eds.) Academic Press, Inc., Orlando, pp.
277-304.
17.Wallace, H. M., Baybutt, H. N., Pearson, C. K.,
and Keir, H. M. (1981) FEBS Lett., 126, 157-160.
18.Lemon, S. M., Walker, C. M., Alter, M. J., and
Yi, M.-K. (2007) in Fields Virology (Knipe, D. M., and Howley,
P. M., eds.) Lippincott, Williams & Wilkins, Philadelphia, pp.
1253-1304.
19.Libra, M., Gasparotto, D., Gloghini, A.,
Navolanic, P. M., de Re, V., and Carbone, A. (2005) Front.
Biosci., 10, 2460-2471.
20.Weisell, J., Hyvonen, M. T., Hakkinen, M. R.,
Grigorenko, N. A., Pietila, M., Lampinen, A., Kochetkov, S. N.,
Alhonen, L., Vepsalainen, J., Keinanen, T. A., and Khomutov, A. R.
(2010) J. Med. Chem., 53, 5738-5748.
21.Khomutov, A. R., Simonian, A. R., Vepsalianen,
J., Keinanen, T. A., Alhonen, L., and Janne, Ju. (2005) Bioorg.
Khim., 31, 206-212.
22.Grigorenko, N. A., Vepsalainen, J., Jarvinen, A.,
Keinanen, T. A., Alhonen, L., Janne, Ju., and Khomutov, A. R. (2005)
Bioorg. Khim., 31, 200-205.
23.Jarvinen, A. J., Cerrada-Gimenez, M., Grigorenko,
N. A., Khomutov, A. R., Vepsalainen, J. J., Sinervirta, R. M.,
Keinanen, T. A., Alhonen, L. I., and Janne, J. E. (2006) J. Med.
Chem., 49, 399-406.
24.Grigorenko, N. A., Vepsalainen, J., Jarvinen, A.,
Keinanen, T. A., Alhonen, L., Janne, Ju., Krizin, A. M., and Khomutov,
A. R. (2004) Bioorg. Khim., 30, 441-445.
25.Hyvonen, M. T., Keinanen, T. A., Khomutov, M.,
Simonian, A., Weisell, J., Kochetkov, S. N., Vepsalainen, J., Alhonen,
L., and Khomutov, A. R. (2011) J. Med. Chem., 54,
4611-4618.
26.Ivanov, A. V., Korovina, A. N., Tunitskaya, V.
L., Kostyuk, D. A., Rechinsky, V. O., Kukhanova, M. K., and Kochetkov,
S. N. (2006) Protein Expr. Purif., 48, 14-23.
27.Mukovnya, A. V., Komissarov, V. V., Kritsyn, A.
M., Mit’kevich, V. A., Tunitskaya, V. L., and Kochetkov, S. N.
(2011) Mol. Biol. (Moscow), 44, 1045-1053.
28.Mukovnya, A. V., Tunitskaya, V. L.,
Khandazhinskaya, A. L., Golubeva, N. A., Zakirova, N. F., Ivanov, A.
V., Kukhanova, M. K., and Kochetkov, S. N. (2008) Biochemistry
(Moscow), 73, 660-668.
29.Ivanov, A. V., Kozlov, M. V., Kuzyakin, A. O.,
Kostyuk, D. A., Tunitskaya, V. L., and Kochetkov, S. N. (2004)
Biochemistry (Moscow), 69, 782-788.
30.Tunitskaya, V. L., Mukovnya, A. V., Ivanov, A.
A., Gromyko, A. V., Ivanov, A. V., Streltsov, S. A., Zhuze, A. L., and
Kochetkov, S. N. (2011) Bioorg. Med. Chem. Lett., 21,
5331-5335.
31.Tomei, L., Vitale, R. L., Incitti, I., Serafini,
S., Altamura, S., Vitelli, A., and De Francesco, R. (2000) J. Gen.
Virol., 81, 759-767.
32.Wallace, H. M., Duff, P. M., Pearson, C. K., and
Keir, H. M. (1981) Biochim. Biophys. Acta, 652,
354-357.
33.Kim, D. G., Du, J., Miao, C., Jung, J. H., Park,
S. C., and Kim, D. K. (2008) Oncol. Rep., 19,
535-539.
34.Bachrach, U., and Abu-Elheiga, L. (1990) Eur.
J. Biochem., 191, 633-637.
35.Eichler, W., and Corr, R. (1989) Biol. Chem.
Hoppe Seyler., 370, 451-466.
36.Pommier, Y., Kerrigan, D., and Kohn, K. (1989)
Biochemistry, 28, 995-1002.
37.Kuosmanen, M., and Poso, H. (1985) FEBS
Lett., 179, 17-20.
38.Belting, M., Havsmark, B., Jonsson, M., Persson,
S., and Fransson, L. A. (1996) Glycobiology, 6,
121-129.
39.Wang, Q. M., Hockman, M. A., Staschke, K.,
Johnson, R. B., Case, K. A., Lu, J., Parsons, S., Zhang, F.,
Rathnachalam, R., Kirkegaard, K., and Colacino, J. M. (2002) J.
Virol., 76, 3865-3872.
40.Qin, W., Luo, H., Nomura, T., Hayashi, N.,
Yamashita, T., and Murakami, S. (2002) J. Biol. Chem.,
277, 2132-2137.
41.Weng, L., Kohara, M., Wakita, T., Shimotohno, K.,
and Toyoda, T. (2012) Gene, 496, 79-87.
42.Hyvonen, M. T., Howard, M. T., Anderson, C. B.,
Grigorenko, N., Khomutov, A. R., Vepsalainen, J., Alhonen, L., Janne,
J., and Keinanen, T. A. (2009) Biochem. J., 422,
321-328.
43.Ivanov, A. V., Tunitskaya, V. L., Ivanova, O. N.,
Mitkevich, V. A., Prassolov, V. S., Makarov, A. A., Kukhanova, M. K.,
and Kochetkov, S. N. (2009) FEBS Lett., 583, 277-280.
44.Tellinghuisen, T. L., Foss, K. L., and Treadaway,
J. (2008) PLoS Pathog., 4, e1000032.
45.Tabor, C. W., and Tabor, H. (1984) Annu. Rev.
Biochem., 53, 749-790.
46.Wallace, H. M., and Niiranen, K. (2007) Amino
Acids, 33, 261-265.
47.Duverlie, G., and Wychowski, C. (2007) World
J. Gastroenterol., 13, 2442-2445.
48.Lee, J. C., Tseng, C. K., Chen, K. J., Huang, K.
J., Lin, C. K., and Lin, Y. T. (2010) Anal. Biochem.,
403, 52-62.