Initiation of Nitric Oxide (NO) Synthesis in Roots of Etiolated seedlings of Pea (Pisum sativum L.) under the Influence of Nitrogen-Containing Compounds
A. K. Glyan’ko
Siberian Institute of Plant Physiology and Biochemistry, Siberian Branch of the Russian Academy of Sciences, 664033 Irkutsk, P.O. box 317, Russia; fax: (3952) 51-0754; E-mail: akglyanko@sifibr.irk.ru
Received December 26, 2012; Revision received January 24, 2013
The level of nitric oxide (NO) in roots of 2-day-old etiolated pea (Pisum sativum L.) seedlings was investigated by fluorescence microscopy using the fluorescent probe 4,5-diaminofluorescein diacetate. Segments representing transversal (cross) cuts of the roots having thickness of 100 to 150 µm (a segment of the root located 10 to 15 mm from the apex) were analyzed. A substantial concentration of NO in the roots was registered when the seedlings were grown in water (control). Addition of 4 mM sodium nitroprusside, 20 mM KNO3, 2 mM NaNO2, 2 mM L-arginine into the growth medium increased NO concentration with respect to the control by 1.7- to 2.3-fold. Inhibitors of animal NO-synthase – 1 mM Nω-nitro-L-arginine methyl ester hydrochloride and 1 mM aminoguanidine hydrochloride – reduced the intensity of fluorescence in the root segments in the presence of all the studied compounds. In medium with KNO3, the inhibitor of nitrate reductase –150 µM sodium tungstate – lowered the fluorescence intensity by 60%. Scavengers of nitric oxide – 100 µM 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide and 4 µM hemoglobin – lowered NO concentration in all the studied variants. Potassium ferrocyanide (4 mM) as the inactive analog of sodium nitroprusside inhibited generation of NO. These results are discussed regarding possible pathways of NO synthesis in plants.
KEY WORDS: Pisum sativum L., etiolated seedlings of pea, fluorescence probe, nitric oxide (NO), inhibitors of animal NO-synthase (NOS) and plant nitrate reductase, NO scavengersDOI: 10.1134/S0006297913050052
Abbreviations: DAF-2DA, 4,5-diaminofluorescein diacetate; L-NAME, Nω-nitro-L-arginine methyl ester hydrochloride; NO, nitric oxide; NOS, nitric oxide synthase; NR, nitrate reductase; PFC, potassium ferrocyanide; PTIO, 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide; SNP, sodium nitroprusside.
Nitric oxide (NO) is a free radical – a gaseous and neutral
lipophilic molecule that easily diffuses through biological membranes.
As a molecule that is known to participate in signaling and as a toxic
substance, it is characterized by diverse biological activity [1]. Biological functions of NO are manifested in
plants in such processes as growth, development, hormonal regulation,
programmed cell death (apoptosis), protective reactions, blooming,
gravitropism, stomatal motions, etc. [2, 3]. When present in large concentrations, NO is toxic
for bacteria, fungi, tumor cells, viruses, plants, and animals [4, 5]. Toxicity of NO is related
first to its high reactivity with respect to proteins containing metals
with variable valence and with respect to oxygen, and second with its
ability to form products with amines and thiols [6]. The obviously high chemical reactivity of NO and
the multifunctional character of its effect are obstacles complicating
the development of a model representing its role in signaling [7]. Nevertheless, according to a viewpoint accepted at
present, NO forms one of the eight signal systems of organisms, i.e.
the NO-synthase (NOS) signal system [8].
One of the unresolved problems related to NO in plants is the process of its synthesis. This issue has been solved unequivocally for animal cells. This is the NADPH-dependent oxidation of L-arginine to L-citrulline and NO with the participation of three isoforms of NOS. Meanwhile, there are several mechanisms of NO synthesis in plants already discovered [9]. We will concentrate the readers’ attention on the two principal mechanisms of NO synthesis in plants: nitrate/nitrite-dependent and L-arginine-dependent pathways. The first pathway presumes reducing nitrate and nitrite in leaves and in roots down to NO with the participation of cytosolic nitrate reductase [10] and possibly nitrate reductase and nitrite-NO-reductase, which are localized on plasma membranes of roots [11, 12]. The second pathway presumes the presence of an arginine-dependent formation of NO similar to the one existing in animal cells. This is confirmed, in particular, by the fact that the reaction forming NO in plants is blocked by the activity of inhibitors of animal NOS, which leads to the mediated influence of NO on physiological processes: growth, development, blooming, hormonal signaling, and protective reactions [9]. Unfortunately, the protein homological to animal NOS has not yet been extracted from plants. At the same time, much attention has been given in recent years to the protein AtNOS1 from Arabidopsis thaliana, which is homological to the protein extracted from the snail helix pomatia (Achatina fulica). The latter catalyzes the reaction of forming NO from arginine. But, according to biochemical and structural properties, AtNOS1 belongs to the family of GTP-binding proteins and does not demonstrate NOS activity [13]. It can be presumed that this protein interacts with other proteins, which implies formation of an enzyme complex catalyzing NO synthesis [13].
Synthesis of NO in plant cells may also occur by nonenzymatic reduction of nitrite in the acidic medium of the apoplast in the presence of reductants [14] and with the participation of polyamines [15]. Therefore, it can be assumed that various pathways of NO synthesis act in plants, and the activation will probably depend on the kind of exogenous factor (biotic or abiotic). According to Flores et al. [16], there are several sources for NO formation in plants, and some of these sources may be regulated via signaling pathways.
The objective of the present investigation was, first, assessment of the endogenous NO concentration in roots of etiolated seedlings of pea (Pisum sativum L.) depending on the effect of nitrogen compounds upon the plants, these compounds in planta probably being the sources (substrates) for synthesis of NO, and, second, the study of influence of inhibitors and NO-scavengers on this synthesis.
MATERIALS AND METHODS
Etiolated seedlings of Pisum sativum L., variety Yamalsky (selected by the Siberian Agricultural Company, Ltd., Russia) were the used in the present investigation. The seeds washed with warm water and soap and sterilized on the surface for 15 min in 3% solution of hydrogen peroxide were sprouted in flasks on wet filter paper at 22°C for 48 h from the moment of initial soaking. Only homogeneous material was selected for the investigation. Root length of 25-30 mm (including the epicotyl) was the criterion of homogeneity.
For the purpose of further growth, the seedlings were placed into flasks on filter paper soaked with either distilled water or solutions of the compound to be tested. To stain the segments, we used 4,5-diaminofluorescein diacetate (DAF-2DA) as a fluorescence probe. Normally this compound penetrates through the cell membrane and deacetylates (catalyzed by intracellular esterases) into 4,5-diaminofluorescein (DAF-2), which (together with NO) forms a fluorescing compound – diamino-triazole fluorescein (DAF-2T) [17]. To obtain the labeled compound DAF-2T, the segments of roots were incubated with 10 µM DAF-2DA in 10 mM Tris-HCl (pH 7.4) for 30 min at 26°C.
From the incubated segments of roots, we obtained cross cuts having thickness of 100 to 150 µm and analyzed them with the aid of an Axio Observer Z1 fluorescence microscope (Germany) equipped with an Axio Cam MRm3 digital monochromatic camera and Axio Vision Rel.4.6 software package for capturing and analysis of images using filter block No. 10 with excitation wavelength of 450 to 490 nm and emission wavelength of 515 to 565 nm. The fluorescence intensity was expressed in relative units. In our analysis, we used 20 snapshots for each of the variants, for which the fluorescence intensity was computed. In our experiments, we tested various time exposures of the seedlings in solutions of the investigated compounds: from short-term exposure (15 to 30 min) up to long-term ones (2, 5, and 24 h). Below we discuss exposures that gave the highest observed levels of NO in the root segments.
The content of free L-arginine was determined in water extracts from the roots of etiolated pea seedlings using an AAA T339 amino acid analyzer (Czech Republic). The concentration of nitrate in pea roots was quantitatively determined by the method of Cataldo et al. [18].
The following chemical reagents were used: hemoglobin extracted from horse erythrocytes and sodium nitroprusside (SNP) (MP Biomedicals, USA); DAF-2DA (Calbiochem, Germany); 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (PTIO), Nω-nitro-L-arginine methyl ester hydrochloride (L-NAME), and aminoguanidine hydrochloride (Sigma, USA); L-arginine, sodium tungstate, potassium ferrocyanide (PFC) [K4(Fe(CN)6·3H2O], KNO3 (nitrate) and NaNO2 (nitrite) (Reakhim, Russia).
The results of the investigation are presented in the form of microphotographs and diagrams showing the fluorescence intensity in the cells. The values are given in terms of arithmetic means, with standard deviations obtained for three independent experiments conducted with three biological recurrences. The significance of differences between the mean values was assessed by Student’s t-criterion. The statistical processing of the data was conducted with the aid of the Microsoft Excel program.
RESULTS AND DISCUSSION
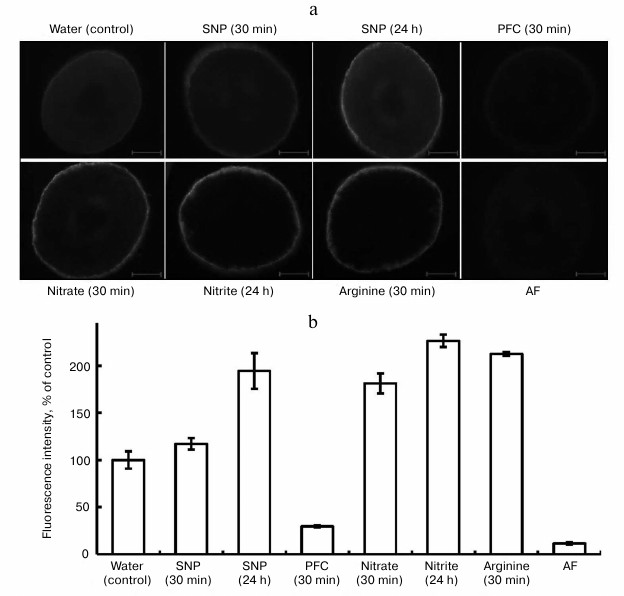
In the experiments, fluorescence was observed in epidermal cells belonging to segments of roots of pea seedlings (Fig. 1). This probably gives evidence that etiolated seedlings synthesize NO on the surface cells of the root, forming a constant “NO-field”, which may aid the transfer of signal information into the genome, especially under the influence of stresses. As is obvious from Fig. 1, fluorescence in the epidermal cells was observed after the exposure of the seedlings in distilled water (control), solutions of SNP, nitrate and nitrite salts, and L-arginine.
Fig. 1. Influence of exogenous compounds on NO level in roots of etiolated pea seedlings. SNP, 4 mM sodium nitroprusside; PFC, 4 mM potassium ferrocyanide; nitrate, 20 mM KNO3; nitrite, 2 mM NaNO2; arginine, 2 mM L-arginine; AF, autofluorescence. a) Electronic microphotographs, bar 100 µm; b) intensity of fluorescens in root cuts.
Formation of NO in plant cells on the background of SNP is due to the ability of this compound to release nitric oxide in plant tissues. The function of SNP as an exogenous donor of NO explains its wide application in investigations. Note that in a number of investigations, we employed PFC as an inactive analog of SNP [19] in the capacity of the control for SNP. In our experiments, PFC at the concentration of 4 mM reduced fluorescence after a 30 min exposure and after longer exposures (Fig. 1). However, the mechanism of its action cannot be completely explained. It is known from the literature that PFC inhibits the formation of lateral roots and lowers the accumulation of dry substance of roots of mountain ginseng (Panax ginseng) seedlings with respect to SNP [19]. According to Tewari et al. [19], unlike for SNP, PFC activates the cross oxidation of lipids, and it does not induce synthesis of non-protein thiols (ascorbate, glutathione), which play a clear role in modulation of NO concentration. Probably, at a high concentration of PFC (4 mM) the synthesis of compounds that oxide endogenous NO in pea roots makes NO unavailable for the reaction with DAF-2DA. This issue needs further investigation.
The generation of NO in roots of seedlings grown on water is, in our opinion, related to the presence of a sufficient quantity of free L-arginine in the cells. Indeed, in our cases, attempts to determine free L-arginine in roots of 48-h pea seedlings elucidated its high concentration: 7.73 ± 1.17 µmol/g dry substance (n = 5). According to the data of other authors, the arginine pool is rather large (5-10 µmol/g dry substance) for 1-4-day-old seedlings of A. thaliana [20].
We assume that synthesis of nitric oxide in the control (water) occurs due to use of endogenous L-arginine as substrate in the reaction catalyzed by an enzyme similar to animal NO-synthase. On the other hand, trace amounts of nitrate and nitrite in roots of pea seedlings in the variant with water suggest insufficient amount of a substrate (nitrate) for nitrate reductase (NR) activity. These assumptions proved to be true according to results of our experiments with inhibitors of animal NO-synthase and the inhibitor of NR – sodium tungstate.
Further analysis of the data (Fig. 1) showed elevation of the level of NO in all the variants with respect to the control: with SNP (4 mM) after 24 h – by 1.9-fold; with KNO3 (20 mM) after 30 min – by 1.7-fold; with NaNO2 (2 mM) after 24 h – by 2.3-fold; with L-arginine (2 mM) after 30 min – by 2.1-fold. Therefore, the substances given to the seedlings exogenously increase the fluorescence intensity for roots of the seedlings. To prove that NO is formed in cells of the root, we applied PTIO as a scavenger for NO. This substance is not a biological compound, and its chemical effect in the cells of an organism is based on oxidation of NO to NO2− [21]. The goal of application of PTIO in our investigations in vitro and in vivo was to prove that NO is synthesized in cells and that it depresses biological activity of NO.
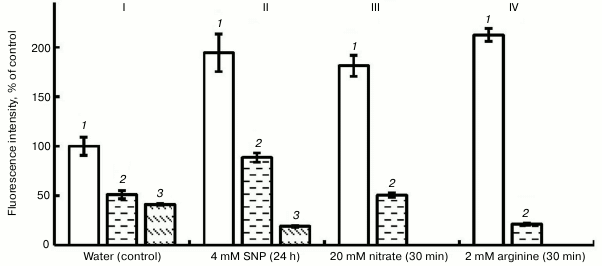
Figure 2 shows that exposure of roots to PTIO (100 µM) on the background of water, SNP, nitrate, and L-arginine lowered the fluorescence intensity by 65 to 90%. This shows the presence of NO in pea root cells and the fact of its binding with PTIO. Hemoglobin, under the influence of which (at concentration 4 µM) the intensity of fluorescence in variants with H2O and SNP decreased by approximately 2-fold (Fig. 2), also has a similar scavenging effect.
Fig. 2. Influence of 100 µM PTIO and 4 µM hemoglobin (Hb) on the level of NO in roots of etiolated pea seedlings. I (control): 1) water; 2) water + Hb; 3) water + PTIO; II: 1) SNP; 2) SNP + Hb; 3) SNP + PTIO; III: 1) nitrate; 2) nitrite + PTIO; IV: 1) arginine; 2) arginine + PTIO.
The mechanism of binding of NO with hemoglobin involves its interaction with heme of hemoglobin, which presumably forms nitrosyl hemoglobin [22]. We can assume that in plants nonsymbiotic (not involving symbiotic biological nitrogen fixation in nodules of bean plants) forms of hemoglobin can play a protective role against nitrosative stress and modulate the signaling function for NO [22]. Transgenic tobacco and alfalfa plants (characterized by increased synthesis of nonsymbiotic hemoglobin of class 1) had lower sensitivity to nitrosative stress caused in cells by the effect of exogenous donors of NO as compared with normal (wild) forms of the plants [23, 24]. Possibly, nonsymbiotic forms of hemoglobin play a role in the regulation of NO concentration in cells at earlier stages of legume-rhizobial symbiosis [25]. Although the physiological role of plant hemoglobins in NO metabolism is obvious, it necessitates serious investigations, say, in the aspect of possible regulators of NO concentration in plant cells and from the viewpoint of protecting plants from nitrosative stress [26].
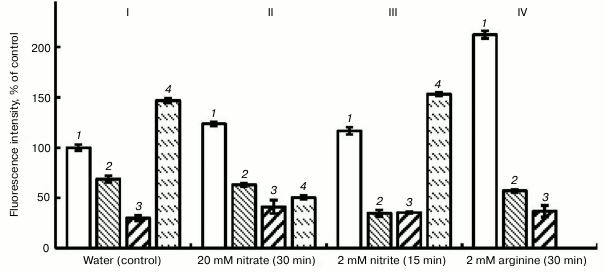
The data of the literature indicate the inhibition of NO synthesis in plants by compounds that are inhibitors of animal NOS [26]. In our investigations, we used two inhibitors of animal NOS, i.e. aminoguanidine hydrochloride [27] and L-NAME, and also an inhibitor of plant nitrate reductase (sodium tungstate) [28]. Figure 3 shows that in the variant with water (control) aminoguanidine (1 mM) lowered the intensity of fluorescence by 37%, and L-NAME (1 mM) lowered it by 68%. Substantial lowering the intensity of fluorescence was observed also in the variant with nitrite, and especially on the background of arginine (Fig. 3).
Fig. 3. Influence of inhibitors of animal NO-synthase (1 mM L-NAME and 1 mM aminoguanidine (AG)) and nitrate reductase (150 µM sodium tungstate) on the NO level in roots of etiolated pea seedlings. I (control): 1) water; 2) water + AG; 3) water + L-NAME; 4) water + sodium tungstate; II: 1) nitrate; 2) nitrate + AG; 3) nitrate + L-NAME; 4) nitrate + sodium tungstate; III: 1) nitrite; 2) nitrite + AG; 3) nitrite + L-NAME; 4) nitrite + sodium tungstate; IV: 1) arginine; 2) arginine + AG; 3) arginine + L-NAME.
In the variant with KNO3, the intensity of fluorescence increased by 20% with respect to the control (Fig. 3, II). This probably shows the inclusion of an additional pathway of NO synthesis, i.e. the reduction of nitrate to NO. But in this case the inhibitors of animal NOS, likewise in the variant with the water, lowered the intensity of fluorescence: aminoguanidine – by 49%, L-NAME – by 66%. So, we conclude that after 30-min exposure the principal pathway of NO generation in pea roots involves arginine-dependent formation of NO. However, reduction of nitrate reductase activity by 60% under the effect of sodium tungstate (150 µM) confirms the assumption that this enzyme is also involved in the process of NO generation (Fig. 3, II).
Participation of cytosolic NR in generation of NO is presently considered to be certain [26]. Still, there remains the problem of determining the quantities in which NO may form with the participation of this enzyme. According to Rockel et al. [29], NO production in cases when concentrations of the substrate (nitrate) are close to saturation is only 1% of the nitrate-reducing capability of NR. But this quantity of NO is more than that sufficient for its functioning as a signaling molecule [30]. Although, in cases when plants are effected by extreme factors and the process of NO generation in cells increases by many times, it is problematic to explain NO synthesis only by NR activity. It can be assumed that there are other forms of nitrate reducing enzymes that participate in the formation of NO in plant cells. In this connection, it is expedient to consider the results of investigations substantiating the localization of nitrate reductase (PM-NR) and nitrite-NO-reductase (NI-NOR) in tobacco root cells, these substances being root specific and associated with plasmalemma [11, 12]. The operation of these enzymes is coordinated, and the rate of enzymatic production of NO in this case depends on the power of the stress effect.
In our experiments, as far as we can see the absence of negative influence of the inhibitor of nitrate reductase activity, sodium tungstate, on the concentration of NO in pea seedling roots in the variant with nitrite (Fig. 3, III) gives evidence that the reduction of nitrite (NO2−) is catalyzed by another enzyme (or enzymes) on whose activity the inhibitor of nitrate reductase has no influence. However, the plausible elevation of NO concentration in roots under the effect of sodium tungstate during 15-min exposures in the variants with water and with nitrite (Fig. 3, I and III) cannot yet be explained. Probably this is due to a nonspecific effect of sodium tungstate upon NO synthesis. Viktorova et al. [31] found that the largest generation of NO in the apoplastic space of the leaves of autumn wheat seedlings was observed on infiltration of nitrite salt solution into the tissues, to compare with that in the case of combination of nitrate salt and SNP. This fact suggests different mechanisms of reduction of nitrites.
NO synthesis by an arginine-dependent mechanism confirm our data on substantial (3.7-fold) reduction in the fluorescence intensity under the effect of 1 mM aminoguanidine and 1 mM L-NAME after the exposure of pea seedlings to a solution of 2 mM L-arginine for 30 min (Fig. 3, IV). The same figure suggests the data of for inhibiting influence of aminoguanidine and L-NAME upon the concentration of NO in roots for the variants with water, nitrate, and nitrite. Potassium ferrocyanide as an inactive analog of SNP reduced the intensity of fluorescence in comparison with SNP (Fig. 1).
Therefore, the present paper has discussed the possibilities of regulation of NO synthesis in roots of etiolated pea seedlings by exogenous nitrogen-containing compounds – nitrate, nitrite, and L-arginine. Partial blockage of NO synthesis in roots of seedlings by inhibitors of animal NO-synthase as well as inhibition of the activity of plant nitrate reductase with sodium tungstate have been demonstrated. However, the concentration of NO in these variants tends to decrease also under the influence of inhibitors of animal NOS. The results of the experiments suggest that there are at least two mechanisms of NO generation functioning in the seedlings simultaneously: a nitrate/nitrite-generating mechanism and an L-arginine-dependent mechanism. It is known that, grounded on the experimentally confirmed fact that A. thaliana mutants, which are deficient with respect to NR and nitrate, contain 10 times less L-arginine than the wild species, Modolo et al. [32] hypothesized that NO synthesis via the nitrate/nitrite-reduction mechanism is related to the metabolism of arginine. This hypothesis has been confirmed to some extent by the results published by Zonia et al. [20]. The experiments show 3- to 5-fold increase in the content of free arginine in seedlings of A. thaliana, together with their growth in a solution of NH4NO3, to compare with seedlings growing in water. On the whole, the issue related to the mechanisms of NO synthesis in plants remains open for further investigations.
The author would like to thank Dr. N. B. Mitanova and Dr. A. V. Stepanov for their assistance in the experiments.
REFERENCES
1.Proskuryakov, S. Ya., Konoplyanikov, A. G.,
Ivannikov, A. I., and Skvortsov, V. G. (1999) Uspekhi Sovrem.
Biol., 119, 380-395.
2.Neill, S., Bright, J., Desikan, R., Hancock, J.,
Harrison, J., and Wilson, I. (2008) J. Exp. Bot.,
59, 25-35.
3.Kolupaev, Yu. E., and Karpets, Yu. V. (2010)
Formation of Plants Adaptive Reactions to Abiotic Stressors
Influence [in Ukrainian], Osnova Publishing, Kiiv.
4.Zumft, W. G. (1997) Microbiol. Mol. Biol.
Rev., 61, 533-616.
5.Dubovskaya, L. V., Kolesneva, E. V., Knyazev, D.
M., and Volotovskii, I. D. (2007) Russ. J. Plant Physiol.,
54, 755-762.
6.Van der Vliet, A., Eiserich, J. P., Kaur, H.,
Cross, C. E., and Halliwell, B. (1996) Methods
Enzymol., 269, 175-184.
7.Grun, S., Lindermayr, S., and Durner, J. (2006)
J. Exp. Bot., 57, 507-516.
8.Tarchevsky I. A. (2002) Signal Systems of Plant
Cells [in Russian], Nauka, Moscow.
9.Crawford, N. M. (2005) J. Exp. Bot.,
57, 471-478.
10.Shi, F-M., and Li, Y-Z. (2008) BMB Rep.,
41, 79-85.
11.Stohr, C., Strube, F., Marx, G., Ullrich, W. R.,
and Rockel, P. (2001) Planta, 212, 835-841.
12.Stohr, C., and Stremlau, S. (2006) J. Exp.
Bot., 57, 463-470.
13.Moreau, M., Lee, G. L., Wang, Y., Crane, B. R.,
and Klessig, D. F. (2008) J. Biol. Chem., 283,
32957-32967.
14.Bethke, P. C., Badger, M. P., and Jones, R. L.
(2004) Plant Cell., 16, 332-341.
15.Tun, N. N., Santa-Catarina, C., Begum, T.,
Silveira, V., Handro, W., Floh, E. I. S., and Scherer, G. F. F. (2006)
Plant Cell Physiol., 47, 346-354.
16.Flores, T., Todd, C. D., Tovar-Mendez, A.,
Dhanoa, P. K., Corra-Aragunde, N., Hoyos, M. E., Brownfield, D. M.,
Mullen, R. T., Lamattina, L., and Polacco, J. C. (2008) Plant
Physiol., 147, 1936-1946.
17.Nakatsuboa, N., Kojimaa, H., Kikuchia, K.,
Nagoshib, H., Maedaa, D., Imaia, Y., Irimuraa, T., and Naganoa, T.
(1998) FEBS Lett., 427, 263-266.
18.Cataldo, D. A., Haroom, M., Schrader, L. E., and
Young, V. L. (1975) Commun. Soil Sci. Plant Anal.,
6, 71-80.
19.Tewari, R. K., Kim, S., Hahn, E-J., and Paek,
K-Y. (2008) Plant Biotech. Rep., 2, 113-122.
20.Zonia, L. E., Stebbins, N. E., and Polacco, J. C.
(1995) Plant Physiol., 107, 1097-1103.
21.Maeda, H., Akaike, T., Yoshida, M., and Suga, M.
(1994) J. Leukocyte Biol., 56, 588-592.
22.Perazzolli, M., Romero-Puertas, M. C., and
Delledonne, M. (2006) J. Exp. Bot., 57, 479-488.
23.Dordas, C., Rivoal, J., and Hill, R. D. (2003)
Ann. Bot., 91, 173-178.
24.Seregelyes, C., Igamberdiev, A. U., Maassen, A.,
Hennig, J., Dudits, D., and Hill, R. D. (2004) FEBS Lett.,
571, 61-66.
25.Glyan’ko, A. K., and Vasil’eva, G. G.
(2010) Appl. Biochem. Microbiol., 46, 15-22.
26.Besson-Bard, A., Pugin, A., and Wendehenne, D.
(2008) Annu. Rev. Plant Biol., 59, 21-39.
27.Corpas, F. J., Barroso, J. B., Carreras, A.,
Valderrama, R., Palma, J. M., Leon, A. M., and del Rio, L. (2006)
Planta, 224, 246-254.
28.Assai, S., Ohta, T., and Yoshioka, H. (2008)
Plant Cell, 20, 1390-1406.
29.Rockel, P., Strube, F., Rockel, A., Wild, J., and
Kaizer, W. M. (2002) J. Exp. Bot., 53,
103-110.
30.Meyer, C., Lea, U. S., Provan, F., Kaiser, W. M.,
and Lillo, C. (2005) Photosynth. Res., 83, 181-189.
31.Viktorova, L. V., Maksyutova, N. N., Trifonova,
T. V., and Andrianov, V. V. (2010) Biochemistry (Moscow),
75, 95-100.
32.Modolo, L. V., Augusto, O., Almeida, I. M. G.,
Pinto-Maglio, C. A. F., Oliveira, H. C., Seligman, K., and Salgado, I.
(2006) Plant Sci., 171, 34-40.