Identification and Differential Expression of Two Dehydrin cDNAs during Maturation of Jatropha curcas Seeds
S. A. Omar1,2, N. I. Elsheery3, H. M. Kalaji4*, M. K. H. Ebrahim5,6, S. Pietkiewicz4, C.-H. Lee7, S. I. Allakhverdiev8,9*, and Zeng-Fu Xu1*
1Key Laboratory of Tropical Plant Resource and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Menglun 666303, Yunnan, China; E-mail: zfxu@xtbg.ac.cn; zengfu.xu@gmail.com2Genetic Department, Faculty of Agriculture, Tanta University, Egypt
3Agricultural Botany Department, Faculty of Agriculture, Tanta University, Tanta, Egypt
4Department of Plant Physiology, Warsaw University of Life Sciences SGGW, Nowoursynowska 159, 02-776 Warsaw, Poland; E-mail: hazem@kalaji.pl
5Biology Department, Faculty of Science, Taibah University, Almadinah Almunawwarah, KSA (Saudi Arabia)
6Botany Department, Faculty of Science, Tanta University, Tanta, Egypt
7Department of Molecular Biology, Pusan National University, Busan 609-735, Korea
8Institute of Plant Physiology, Russian Academy of Sciences, ul. Botanicheskaya 35, 127276 Moscow, Russia; E-mail: suleyman.allakhverdiev@gmail.com
9Institute of Basic Biological Problems, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia
* To whom correspondence should be addressed.
Received January 25, 2013; Revision received February 1, 2013
Plant dehydrin proteins (DHNs) are known to be important for environmental stress tolerance and are involved in various developmental processes. Two full-length cDNAs JcDHN-1 and JcDHN-2 encoding two dehydrins from Jatropha curcas seeds were identified and characterized. JcDHN-1 is 764 bp long and contains an open reading frame of 528 bp. The deduced JcDHN-1 protein has 175 a.a. residues that form a 19.3-kDa polypeptide with a predicted isoelectric point (pI) of 6.41. JcDHN-2 is 855 bp long and contains an open reading frame of 441 bp. The deduced JcDHN-2 protein has 156 a.a. residues that form a 17.1-kDa polypeptide with a predicted pI of 7.09. JcDHN-1 is classified as type Y3SK2 and JcDHN-2 is classified as type Y2SK2 according to the YSK shorthand for structural classification of dehydrins. Homology analysis indicates that both JcDHN-1 and JcDHN-2 share identity with DHNs of other plants. Analysis of the conserved domain revealed that JcDHN-2 has glycoside hydrolase GH20 super-family activity. Quantitative real time PCR analysis for JcDHN-1 and JcDHN-2 expression during seed development showed increasing gene expression of both their transcript levels along with the natural dehydration process during seed development. A sharp increase in JcDHN-2 transcript level occurred in response to water content dropping from 42% in mature seeds to 12% in dry seeds. These results indicate that both JcDHNs have the potential to play a role in cell protection during dehydration occurring naturally during jatropha orthodox seed development.
KEY WORDS: dehydrin, Jatropha curcas, real time PCR, seed developmentDOI: 10.1134/S0006297913050076
Abbreviations: DAF, days after flowering; DHNs, dehydrins; DW, dry weight; FW, fresh weight; LEA, late embryogenesis abundant; qRT-PCR, real time quantitative reverse transcription-polymerase chain reaction; RACE, Rapid Amplification of cDNA Ends; RT-PCR, reverse transcription-polymerase chain reaction; WC, water content.
Dehydrins (DHNs) are a sub-group of the late embryogenesis abundant
(LEA) proteins that have also been called the LEA D-11 family or LEA
type 2 proteins [1]. Expression of the DHNs has
been associated with the protection of various types of plant cells
from osmotic stresses, such as those caused by desiccation, salts, and
low temperatures [1-6].
Over the last few years, direct experimental evidence linking higher expression of DHNs and protection from osmotic stress has begun to appear in the literatures. For example, Arabidopsis plants engineered to overexpress a dehydrin fusion protein were found to have improved survival to low temperature exposures [7]. Similarly, expression of a citrus dehydrin protein in transgenic tobacco has been shown to give increased tolerance to low temperatures [8].
DHN genes are also expressed significantly in seeds towards the end of maturation, a period when the seed undergoes a developmentally programmed reduction in water content. LEA/dehydrin proteins have been estimated to comprise up to 4% of the total seed proteins and are thought to be involved in protecting the embryo and/or other seed tissues from the osmotic stresses associated with the low water content of mature seeds [9, 10].
Although a considerable number of dehydrin proteins have been isolated and studied, the precise physiochemical and/or structural mechanism(s) whereby these proteins function to protect cells from osmotic stress in vivo is(are) unknown [11]. It has been proposed that the short amphipathic K-segments of dehydrin polypeptides interact with solvent-exposed hydrophobic patches on proteins undergoing partial denaturation and thereby interfere with protein aggregate formation [1]. Amphipathic K-helixes could also be involved in binding membrane lipids and thus could play a more specific role in protecting lipoproteins, proteins located in membranes, and/or the membrane structure itself [1, 12]. An alternative proposal for at least part of the protective effect of dehydrins is the ability of these very stable but relatively unstructured proteins to tightly bind and organize water molecules [13]. This latter effect could help in slowing water loss from cells under dehydration conditions and also possibly improve the stability of certain macromolecules by the development of dehydrin-based regions of more tightly bound “ordered” water around these molecules.
Little information is available on these genes in jatropha plants, which are known by their ability to survive under numerous severe environmental conditions [14, 15]. Thus, it was decided to take advantage of the new jatropha expressed sequence tag (EST) analysis [16] to identify cDNAs encoding expressed DHNs and to establish the expression patterns of the DHNs during seed development and natural dehydration. Therefore, we carry out to validate the importance of this kind of protein in acquisition of desiccation tolerance of orthodox jatropha seed as a part of a study project to clarify the biochemical and molecular changes during jatropha seed development.
MATERIALS AND METHODS
Plant materials. Jatropha curcas fruits were collected from plants grown in the Xishuangbanna Tropical Botanical Garden of the Chinese Academy of Sciences, Yunnan, southwest China. The plants were grown under field conditions. When floral buds appeared, flowers were tagged daily throughout the flowering period. Seeds were staged with regard to the time of flowering, fresh weight, and phenotype. Seeds were utilized at different developmental stages, starting from 50 days after flowering (DAF) (28.49% of maximum dry weight (DW)) with water content (WC) 5.23 g H2O/g DW until 100 DAF (100% of maximum DW) with WC 0.12 g H2O/g DW.
Fresh, dry weight, and water content. Seeds at different developmental stages were weighed to determine fresh weight (FW). The DW and WC of whole seeds were determined by reweighing after oven drying at 103°C for 17 h. WC was expressed on the basis of fresh weight (% FW).
Protein analysis. Seeds at different developmental stages were ground to fine powder in liquid nitrogen in a mortar with a pestle. The powder was mixed immediately with extraction buffer (H3PO4, pH 7.0, 0.2% (v/v) Triton X-100, 7 mM β-mercaptoethanol, and 5 mM ascorbic acid). The protein extract was recovered by centrifugation of the homogenate at 4°C for 15 min at 12,000 rpm. After centrifugation, the supernatant was transferred to fresh reaction tubes heated in a water bath at 100ºC for 10 min and then transferred to ice for 5 min. The heat-treated supernatant was then centrifuged at 12,000 rpm for 15 min. The resulting supernatant was transferred to new tubes in 50-100-µl aliquots. The extracts were either directly used or stored at –20°C. Concentration of heat-stable proteins was determined according to the Bradford protocol [17]. SDS-PAGE was carried out with a discontinuous buffer system [18]. Protein samples in 1× SDS gel-loading buffer were denatured by heating at 95°C for 5 min before loading onto the gel. The gels were set up with a Mini Protean 3 Cell (Bio-Rad, USA) apparatus and 10 × 12-cm slab gels unit (Bio-Rad) and run using Laemmli running buffer (pH 8.3). The samples standardized on protein amount were loaded on the gel and separated electrophoretically; low range SDS-PAGE prestained molecular weight standards (Fermentas, Lithuania) were used. The proteins were electrophoretically transferred to polyvinylidene fluoride (PVDF) microporous membrane (Cat. No. IPVH00010; Millipore, USA) from the SDS-PAGE gel using the Trans-BlotP cell (Bio-Rad) following the membrane manufacturer’s instructions. For protein detection, the Western blot membrane was washed three times for 5 min with 20 ml of 1× TBS buffer (20 mM Tris-HCl, pH 8.0, 180 mM NaCl). The unoccupied sites on nitrocellulose were blocked with 1% (w/v) nonfat milk powder in TBS at room temperature. The membranes were washed three times for 5 min with 20 ml of washing buffer (0.1% Tween-20 in 1× TBS) and then incubated for 2 h at room temperature on a rotary shaker in 10 ml of antibody buffer (0.1% Tween-20 and 1% w/v nonfat dry milk in 1× TBS) containing 1 : 1000 dilution of anti-dehydrin antibody raised against the dehydrin consensus K-segment of dehydrins (kindly supplied by Dr. T. J. Close, University of California, Riverside, CA, USA). The membrane was washed three times for 5 min in 20 ml of washing buffer and then incubated for 2 h more in 10 ml of antibody buffer containing 1 : 500 dilution of secondary antibody, alkaline phosphatase anti-rabbit IgG (Vector Laboratories, USA) at room temperature on a rotary shaker. The membrane was then washed two times for 5 min with 20 ml of washing buffer and one time more for 5 min with 20 ml of 1× TBS to remove the Tween 20. Finally, the membrane was stained with freshly prepared developing buffer (100 µl of NBT solution and 100 µl BCIP solution in 10 ml of alkaline phosphatase buffer) [19] until signals were clearly visible. The reaction was stopped by washing the membrane in deionized distilled water (dd-H2O).
Total RNA extraction and DNase treatment. Total RNA from J. curcas seeds at different developmental stages was extracted using a silica particle protocol [20]. Contaminating DNA was removed with 30-min incubation at 37°C by adding 5 µl 10× DNase I buffer (TaKaRa, Japan), 2 µl DNase I (RNase-free, 5 U/µl; TaKaRa), and 0.5 µl RNase inhibitor (RNasin plus RNase inhibitor, 40 U/µl; Promega, USA) to 30 µg RNA in 50 µl of sterile deionized distilled water (sdd-H2O). The DNase treatment was ended by phenol–chloroform extraction.
Isolation and identification of J. curcas dehydrin cDNAs. The EST database of about 4000 J. curcas EST sequences recently generated using RNA isolated from developing seeds [16] were screened for dehydrin sequence search of the unigene annotations with the keyword “dehydrin”. The search yielded five candidates for dehydrin unigenes; DNA sequence analysis of the selected cDNA clones indicated that there were essentially four unique sequences representing two different dehydrin genes. The longest cDNA for both of the two different cDNAs were isolated by reverse transcription-polymerase chain reaction (RT-PCR) on total RNA isolated from dry seed using gene-specific primers (Table 1). The full length of both cDNAs was gained by applying the Rapid Amplification of cDNA 3′ Ends (3′-RACE) method according to the protocol of the Rapid Amplification of cDNA ends (Clontech, USA). Gene-specific primers XT30 and XT66 (Table 1) for both of JcDHN-1 and JcDHN-2 genes, respectively, were used with other primers (provided by the kit) to amplify the 3′ end of cDNA. The 3′-RACE-PCR fragment was purified using a gel extraction kit (Gel Extraction System B; BioDev, Italy) according to the manufacture’s manual and cloned into a cloning vector, pEasy-T3 Cloning Vector (TRANS, China) and sequenced. The full-length cDNAs were obtained by assembling the sequences of 3′-RACE fragment with the EST fragment.
Table 1. Primers used in RT-PCR and
real-time PCR analysis
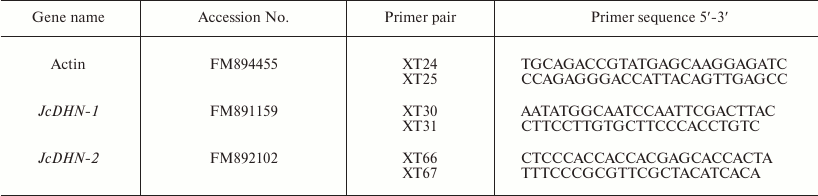
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR). RNA concentration was accurately quantified by spectrophotometric measurements. cDNAs were synthesized from 2 µg DNase-treated total RNA with M-MLV reverse transcriptase (Promega, USA) and oligo d(T)18N (TaKaRa) according to manufacturer’s recommendations. Genes were quantified using real-time PCR, a LightCyclerP instrument 480 (Roche, Germany), and LightCycler 480 Relative Quantification software. Specific primer pairs of the studied genes were designed based on the prepared expressed sequence tags (ESTs) database for developing J. curcas seed [16] using Primer 5 software (Table 1). Each reaction was performed with 2 µl of 1 : 5 (v/v) dilution of the first-strand cDNA in a total reaction volume of 20 µl using the SybrGreen PCR Kit (TRANS) with the following final concentration of constituent components: 1× TransStart Green qPCR SuperMix, primers (0.25 mM each), 1× Passive Reference Dye/PCR Enhancer (50×). Reaction conditions for thermal cycling were the following: starting with a denaturation step of 94ºC for 3 min, followed by 40 cycles of 94ºC for 30 s, 54-59ºC for 50 s, and 72ºC for 50 s, then an elongation step of 1 cycle at 72ºC for 7 min. Amplification specificity was checked with a heat-dissociated protocol (melting curves in the 54-90ºC range) as a final step of the PCR. All primer pairs showed a single peak on the melting curve, and a single band of the expected size was observed using agarose gel electrophoresis. The actin gene (FM894455) of jatropha amplified with the specific primers XT24 and XT25 (Table 1) was used as an internal control for the relative amount of RNA and as a constitutive control. Transcription activity was evaluated as normalized relative expression calculated with qPCR efficiency. The efficiency-corrected quantification performed automatically by the LightCycler 480 Relative Quantification software is based on relative standard curves describing the PCR efficiencies of the target and the reference gene. PCR efficiencies of target and reference genes were determined by providing the running experiment with generated standard curves. The method was based on serial dilutions (1 : 1, 1 : 10, 1 : 100, 1 : 1000, 1 : 10,000) prepared from experimental cDNA sample that contained target gene in preliminary reverse transcription (RT)-PCR analysis. Fifteen samples (three replicates for each dilution) of target standards and another 15 samples of reference standards were used for creation of the relative standard curve. Two replicates were examined for each sample.
Sequencing and bioinformatic analyses. Programs on the website of the National Center for Biotechnology Information (NCBI) were used for finding and analysis of open reading frames (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and conserved domains (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Sequence homology searches for both the nucleotide and protein sequences in GenBank were carried out with the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignment analysis was performed with the ClustalW program (http://www.ch.embnet.org/software/ClustalW.html). Protein calculator v.3.3 online software (http://www.scripps.edu/cgi-bin/cdputnam/protcalc3) was used for determination of proteins molecular weights. Phylogenetic analysis of jatropha DHNs with DHNs from other plants was carried out using the ClustalX program.
RESULTS
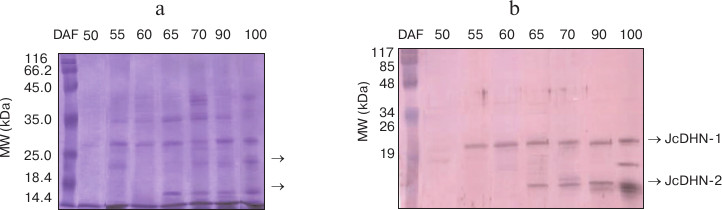
Dehydrins induced by maturation and natural dehydration. The protein profile of heat-stable protein fractions for the seed at different developmental stages showed noticeable changes in number of heat-stable proteins during seed development. This number increased from none in the seed at 50 DAF to about seven polypeptide fractions in the seed at 100 DAF (Fig. 1a). The immunoblot of the corresponding gel probed with anti-dehydrin antibody detected two main dehydrin proteins bands (indicated by arrows) at molecular weight ranging from (20 to 17 kDa) (Fig. 1b). The two detected dehydrin proteins were named JcDHN-1 and JcDHN-2. The JcDHN-1 protein started to be detected at 55 DAF and continued till the end of the seed development, while JcDHN-2 started to be detected at 65 DAF and showed a noticeable increase to reach the maximum expression in the dry seed (100 DAF).
Fig. 1. Western blot analysis of proteins from seeds at different developmental stages. SDS-PAGE of heat-stable protein fractions (100ºC) extracted from the seeds at different developmental stages (a) and Western blot of corresponding gel probed with anti-dehydrin antibody (b). Arrows indicated two main dehydrin proteins at different molecular weight expressed during seed development.
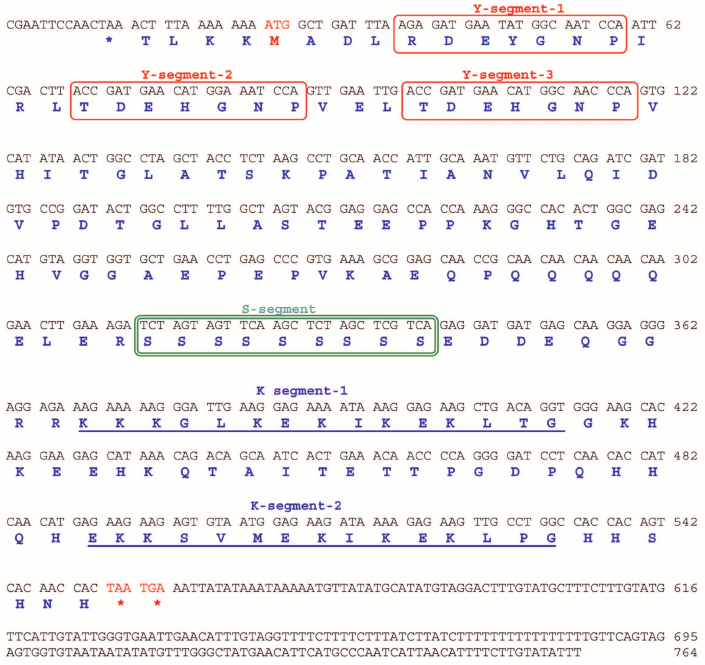
Identification and characterization of J. curcas dehydrin cDNAs. JcDHN-1. The full-length cDNA of JcDHN-1 is 764 bp long and has an open reading frame (ORF) of 528 bp. The ORF of JcDHN-1 starts at nucleotide position 27 and ends at position 544. The JcDHN-1 gene encodes a precursor protein of 175 a.a. residues (Fig. 2). The calculated molecular mass of the protein is approximately 19.3 kDa with a predicated isoelectric point of 6.41. The total number of different amino acids residues in the protein is presented in Table 2. JcDHN-1 does not contain tryptophan or cysteine residues (Table 2). The analysis of conserved domains in the JcDHN-1 precursor protein reveals that it contains three Y-segments, Y1, Y2, and Y3, with amino acid sequences DEYGNP, DEHGNP, and DEHGNP, respectively. It also contains an S-segment consisting of nine serine (S) residues. The conserved K-segment of the DHNs was found in JcDHN-1 in two copies with the sequences KKKGLKEKIKEKLTG and EKKSVMEKIKEKLPG for both K-segment-1 and K-segment-2, respectively (Fig. 2).
Fig. 2. Nucleotide sequence and deduced amino acid sequence of JcDHN-1. The cDNA of JcDHN-1 isolated from jatropha seeds. Numbers on the right indicate nucleotide positions. The deduced amino acid sequence is shown in single-letter code below the nucleotide sequence. The asterisk denotes the translation stop signal. Amino acids in single line boxes indicate the Y-segment and amino acids in double line boxes indicate the S-segment. The two underlined sequences indicate the K-segments.
Table 2. Total number of different amino
acid residues in the JcDHN-1 and JcDHN-2 proteins

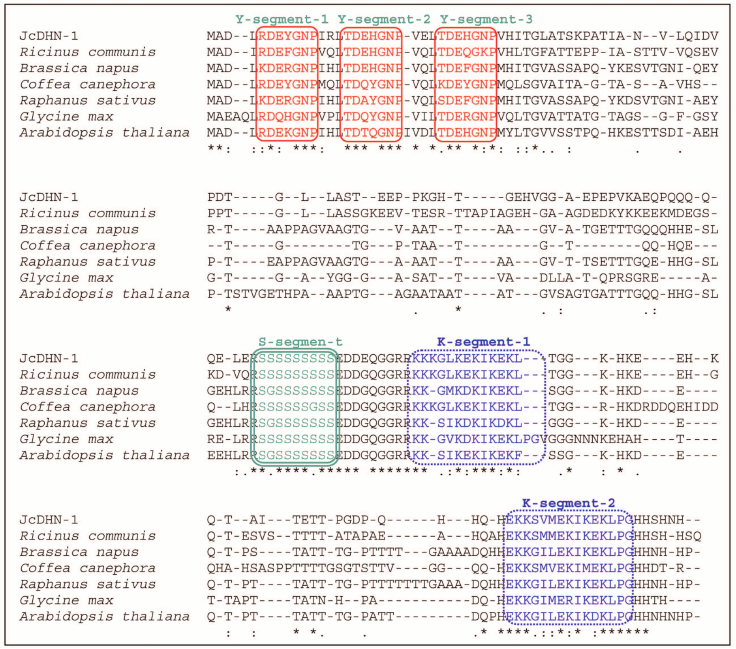
Homology analysis of JcDHN-1 with DHNs from other plants using GenBank Blastp search revealed that JcDHN-1 had the highest sequence homology to the castor bean (Ricinus communis) LEA (EEF49452) with 61% identity and 73% similarity. JcDHN-1 also shared homology with dehydrins from cabbage (Brassica napus DHN, AAQ74768, 50% identity and 61% similarity), coffee (Coffea canephora DH2b, ABC55672, 48% identity and 57% similarity), radish (Raphanus sativus DHLE, P21298, 50% identity and 58% similarity), soybean (Glycine max DHN, AAB71225, 48% identity and 61% similarity) and Arabidopsis (Arabidopsis thaliana DHN, AAM65831, 46% identity and 58% similarity) (Fig. 3).
Fig. 3. Optimized alignment of JcDHN-1 with the most closely related DHNs. Alignment with DHNs from castor bean (Ricinus communis, EEF49452), cabbage (Brassica napus, AAQ74768), coffee (Coffea canephora, ABC55672), radish (Raphanus sativus, P21298), soybean (Glycine max, AAB71225) and Arabidopsis (Arabidopsis thaliana, AAM65831). Gaps are marked with dashes. Y-, S-, and K-segments are shown in frames.
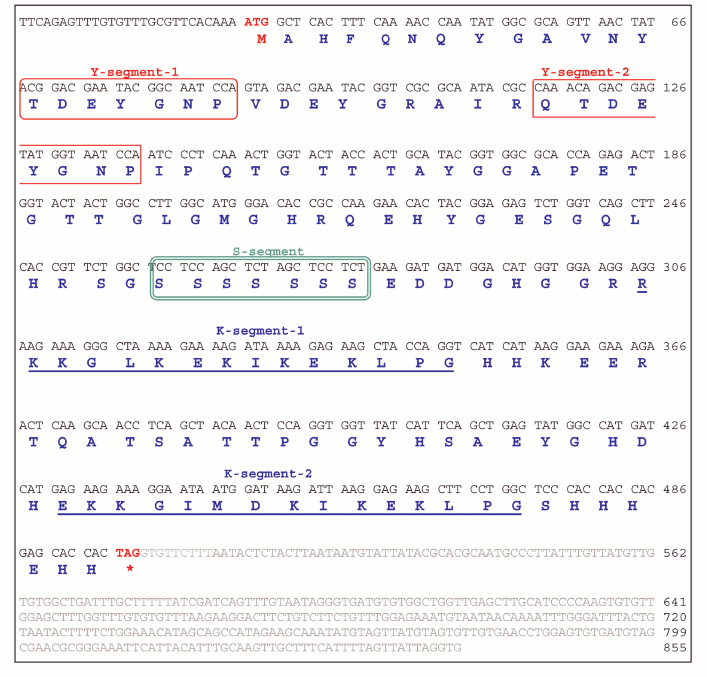
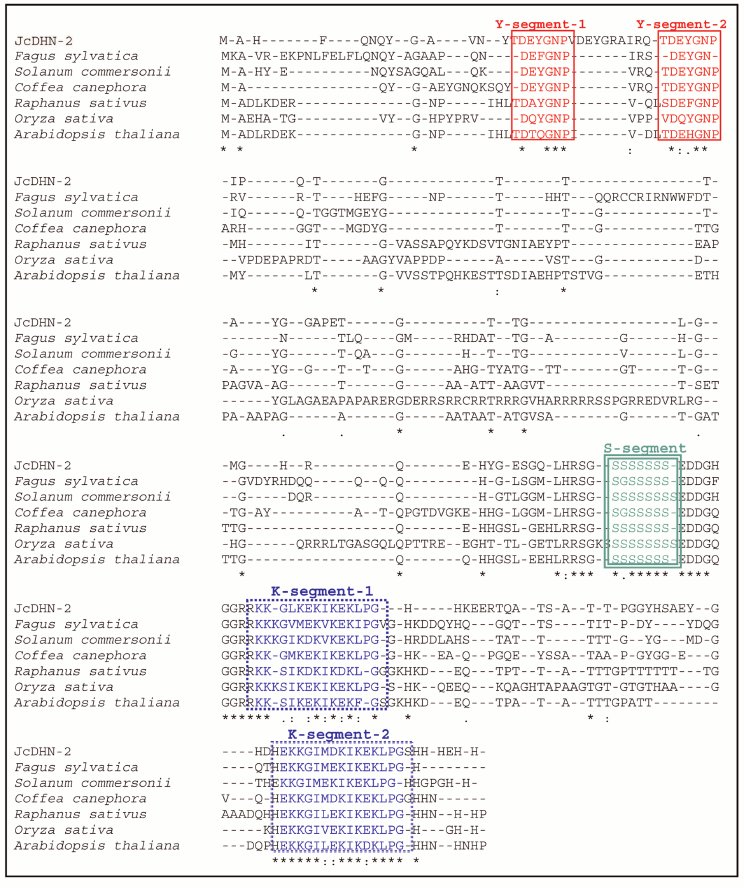
JcDHN-2. The full-length cDNA of JcDHN-2 is 855 bp long and has an ORF of 441 bp. The ORF of JcDHN-1 starts at nucleotide position 29 and ends at position 499. The JcDHN-2 encodes a precursor protein of 156 a.a. (Fig. 4). The calculated molecular mass of the protein is approximately 17.1 kDa with a predicated isoelectric point of 7.09. The total number of different amino acids residues in the protein is presented in Table 2. JcDHN-2 does not contain tryptophan or cysteine residues (Table 2). The precursor protein contains two typical Y-segments, Y1 and Y2, with the same amino acids sequence (TDEYGNP) for both. It also contains an S-segment consisting of seven serine residues. The conserved K-segment of the DHNs was found in JcDHN-2 in two copies with the sequences RKKGLKEKIKEKLPG and EKKGIMDKIKEKLPG for K-segment-1 and K-segment-2, respectively (Fig. 4).
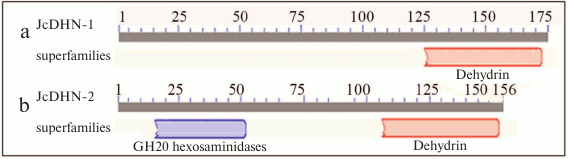
The analysis of conserved domains in the JcDHN-2 precursor protein revealed that it is a member of super-family β-N-acetylhexosaminidases of glycoside hydrolase family 20 (GH20) (Fig. 5).
Fig. 4. Nucleotide sequence and deduced amino acid sequence of JcDHN-2. The cDNA was isolated from jatropha seed. Numbers on the right indicate nucleotide positions. The deduced amino acid sequence is shown in single-letter code below the nucleotide sequence. The asterisk denotes the translation stop signal. Amino acids in single line boxes indicate the Y-segment and amino acids in double line boxes indicate the S-segment. The two underlined sequences indicate the K-segments.
Fig. 5. Analysis of conserved domains for JcDHN-1 (a) and JcDHN-2 (b). Numbers indicate the residue positions.
A GenBank Blastp search revealed that JcDHN-2 has the highest sequence homology to the common beech (Fagus sylvatica) DHN (CAE54590) with 50% identity and 58% similarity. JcDHN-2 also shares homology with wild potato (Solanum commersonii DHn1, CAA75798, 52% identity and 60% similarity), coffee (Coffea canephora DH1b, ABC55671, 66% identity and 73% similarity), radish (Raphanus sativus DHLE, P21298, 40% identity and 53% similarity), rice (Oryza sativa rDRP1, P30287, 56% identity and 61% similarity), and Arabidopsis (Arabidopsis thaliana DHN, CAA45524, 56% identity and 64% similarity) (Fig. 6).
Fig. 6. Optimized alignment of JcDHN-2 with the most closely related DHNs. Alignment with DHNs from common beech (Fagus sylvatica, CAE54590), wild potato (Solanum commersonii, CAA75798), coffee (Coffea canephora, ABC55671), radish (Raphanus sativus, P21298), rice (Oryza sativa, P30287), and Arabidopsis (Arabidopsis thaliana, CAA45524). Gaps are marked with dashes. Y-, S-, and K-segment are shown in frames.
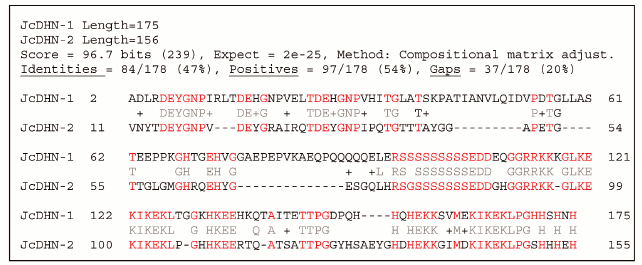
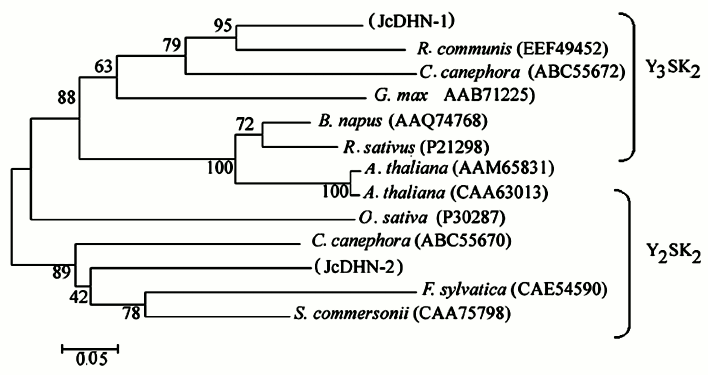
JcDHN-1 and JcDHN-2 alignment. The alignment of the JcDHN-1 and JcDHN-2 amino acid sequences reveals that the two proteins have 47% identity with the conserved sequence of dehydrin protein and 54% similarity with each other (Fig. 7). Phylogenetic relationships among JcDHNs and other DHNs have been established in the form of a phylogenetic tree based on the deduced amino acid sequences (Fig. 8). The result shows that JcDHN-1 and JcDHN-2 probably belong to two types of DHNs. JcDHN-1 is related to Y3SK2 type, while JcDHN-2 is related to Y2SK2 type.
Fig. 7. Optimized alignment of JcDHN-1 and JcDHN-2.
Fig. 8. Phylogenetic tree of jatropha DHNs. A phylogenetic tree based on JcDHN-1, JcDHN-2 and other DHN amino acid sequences shows the phylogenetic relationship between JcDHNs and other plant DHNs. The tree was constructed using the ClustalX method (neighbor-joining method), and the bar indicates 0.05 substitution per amino acid position.
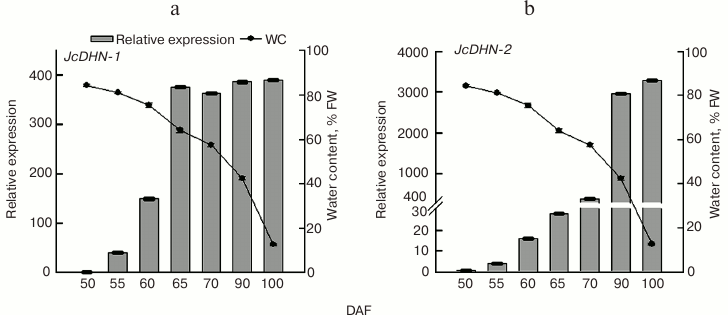
qRT-PCR analysis of JcDHN-1 and JcDHN-2 genes during seed development. To establish the difference in expression patterns of the Jc-DHNs, quantitative analysis was carried out using qRT-PCR. The results (Fig. 9) of analysis by real-time RT-PCR of JcDHN-1 and JcDHN-2 expression presented in Fig. 9 show that there is a difference in expression pattern of the two DHNs. JcDHN-1 transcript increased from 40-fold (comparing to the control gene, actin) to 149-fold from 55 to 60 DAF, then it almost doubled (from 149- to 375-fold) after that to remain without noticeable changes from 65 DAF till the end of the natural dehydration process (100 DAF) (Fig. 9a). On the other hand, the JcDHN-2 transcript analysis shows weak expression during the early stages of about 3-, 14-, and 28-fold (compared to the control gene, actin) for the seeds at 55, 60, and 65 DAF, respectively. The JcDHN-2 transcript level showed a sharp increase from 385-fold at 70 DAF to about 2900-fold for the seeds at 90 DAF and reached the highest level of expression of 3270-fold in the dry seeds (100 DAF) (Fig. 9b).
Fig. 9. qRT-PCR analysis of jatropha dehydrins. Expression analysis of JcDHN-1 (a) and JcDHN-2 (b) detected by quantitative real-time PCR. Real-time PCR was carried out with cDNA obtained from the seeds at different developmental stages. Quantification is based on Cp values that were normalized using the CBtB value corresponding to jatropha actin (housekeeping gene). Two replicates were examined for each sample. Each value is the mean ± SE (n = 2).
DISCUSSION
The presented results show that jatropha seeds synthesize DHNs during maturation and the natural dehydration process (Fig. 1). The analysis of conserved domains in JcDHNs reveals that both JcDHN-1 and JcDHN-2 have a dehydrin super-family sequence near the carboxyl end (Fig. 5). Moreover, JcDHN-2 showed the presence of superfamily β-N-acetylhexosaminidases of glycoside hydrolase family 20 (GH20) (Fig. 5). Enzymes of GH20 are broadly distributed in microorganisms, plants, and animals, and they play roles in various key physiological and pathological processes. These processes include cell structural integrity, energy storage, cellular signaling, fertilization, pathogen defense, and viral penetration [21]. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by the solvent or the enzyme, but by the substrate itself [21]. To our knowledge, this is the first record for GH20 activity in DHNs, which can add to the protection roles and functions of DHNs.
There was a great difference between the transcript level of JcDHN-1 and JcDHN-2 in the mature and dry seed, where the transcript level of JcDHN-1 was at its maximum level about 400-fold of control gene actin while the maximum value for JcDHN-2 was 3200-fold of the same control gene. Also the presence of GH20 activity in JcDHN-2 (Fig. 5) illustrates the specification and importance of JcDHN-2 expression during the actual dehydration process where water content dropped from about 40 to 12% FW (Fig. 9b).
Expression analysis of the two DHNs (JcDHN-1 and JcDHN-2) during jatropha seed development supports the idea about the types of DHN participation in seed development. DHNs participate in two important and rather specific aspects of seed development. First, they are widely perceived to participate, with other LEA proteins, in the dehydration process that occurs during the late stages of seed maturation by assisting the acclimatization of seed tissues to the lower water content found in mature seeds [1, 22]. This kind of participation is consistent with the expression pattern of JcDHN-1, which is expressed early in seed development and continues during the dehydration process at a semi-stable level (Figs. 1b and 9a). Second, it is presumed that the DHNs are synthesized in seeds during maturation, but this does not mean that they are active, and it is activated with increasing stress [23] to stabilize the associated cellular structures during further dehydration or seed quiescence. In this latter context, it should also be noted that it has recently been proposed that DHNs may also possess a radical-scavenging capability [8] and have metal-binding properties [24], both characteristics that are likely to be useful in avoiding accumulation of reactive oxygen species during further dehydration or during long periods of seed storage. This kind of participation could harmonize with the expression pattern of JcDHN-2, which showed a sharp increase in expression during the dehydration process occurring from 70 to 100 DAF (Fig. 9b). This result also supports the idea that the protection proteins produced at the late stages and during the dehydration of jatropha seeds play an important role in cell protection against the deleterious effects of dehydration. In view of the results about antioxidant enzymes [15], which reveal the poor participation of antioxidant enzymes during the late stages of jatropha seed development, and the expression patterns of DHN genes, this probably suggests the importance of the protection function of DHNs during jatropha seed development.
This work was supported by the Knowledge Innovation Programs of the Chinese Academy of Sciences (grant No. KSCX2-YW-Z-0723) and the Top Science and Technology Talents Scheme of Yunnan Province (grant No. 2009CI123) to Z.-F.Xu. C-H. Lee is grateful for support by the National Research Foundation (NRF) of Korea grant funded by MEST (No. 2012-0004968). This work was also supported by grants from the Russian Foundation for Basic Research, by “Molecular and Cell Biology” Programs of the Russian Academy of Sciences, by BMBF (No. 8125) Bilateral Cooperation between Germany and Russia, and by the “Brain Pool” Program of the Ministry of Education, Science and Technology (MEST) and the Korean Federation of Science and Technology Societies (KOFST) to SIA.
REFERENCES
1.Close, T. J. (1996) Physiol. Plant.,
97, 795-803.
2.Allagulova, C. R., Gimalov, F. R., Shakirova, F.
M., and Vakhitov, V. A. (2003) Biochemistry (Moscow), 68,
945-951.
3.Close, T. J. (1997) Physiol. Plant.,
100, 291-296.
4.Ha, Y-L., Lim, J-M., Ko, S. M., Ryol Liu, J. R.,
and Choi, D-W. (2006) J. Plant Biol., 49, 205-211.
5.Hartwigsen, J. A., and Goggi, S. A. (2002) J.
Plant Biol., 45, 225-229.
6.Ingram, J., and Bartels, D. (1996) Ann. Rev.
Plant Biol., 47, 377-403.
7.Puhakainen, T., Hess, M. W., Makela, P., Svensson,
J., Heino, P., and Palva, E. T. (2004) Plant Mol. Biol.,
54, 743-753.
8.Hara, M., Terashima, S., Fukaya, T., and Kuboi, T.
(2003) Planta, 217, 290-298.
9.Roberts, J. K., DeSimone, N. A., Lingle, W. L., and
Dure, L., III (1993) Plant Cell, 5, 769-780.
10.Wise, M. J., and Tunnacliffe, A. (2004) Trends
Plant Sci., 9, 13-17.
11.Hinniger, C., Caillet, V., Michoux, F., Ben Amor,
M., Tanksley, S., Lin, C., and McCarthy, J. (2006) Ann. Bot.,
97, 755-763.
12.Koag, M. C., Fenton, R. D., Wilkens, S., and
Close, T. J. (2003) Plant Physiol., 131, 309-316.
13.Soulages, J. L., Kim, K., Arrese, E. L., Walters,
C., and Cushman, J. C. (2003) Plant Physiol., 131,
963-975.
14.Hammer, K., and Heller, J. (1997) Promoting
the Conservation and Use of Underutilized and Neglected Crops in Physic
Nut Jatropha curcas, The International Plant Genetic Resources
Institute (IPGRI), Gatersleben/International Plant Genetic Resources
Institute, Rome, pp. 14-123.
15.Omar, S., Elsheery, N., Kalaji, M. H., Xu, Z-F.,
Song, S-Q., Carpentier, R., Lee, C-H., and Allakhverdiev, S. I. (2012)
J. Plant Biol., 55, 469-480.
16.Mao-Sheng Chen, Gui-Juan Wang, Ru-Ling Wang, Jun
Wang, Song-Quan Song, and Zeng-Fu Xu (2011) Plant Sci.,
181, 696-700.
17.Bradford, M. M. (1976) Anal. Biochem.,
72, 248-254.
18.Laemmli, U. K. (1970) Nature, 227,
680-685.
19.Ahumada, A. A. P. (2006) Antioxidant Defense
and the Role of the Peroxiredoxins during Silique Development and
Seedling Establishment in Oil Seed Plants, Bielefeld.
20.Ding, L. W., Sun, Q. Y., Wang, Z. Y., Sun, Y. B.,
and Xu, Z. F. (2007) Anal. Biochem., 374, 426-428.
21.Marchler-Bauer, A., Anderson, J. B., Derbyshire,
M. K., DeWeese-Scott, C., Gonzales, N. R., Gwadz, M., Hao, L., He, S.,
Hurwitz, D. I., and Jackson, J. D. (2007) Nucleic Acids Res.,
35, D237-243.
22.Nylander, M., Svensson, J., Palva, E. T., and
Welin, B. V. (2001) Plant Mol. Biol., 45, 263-279.
23.Xiao, H., and Nassuth, A. (2006) Plant Cell
Rep., 25, 968-977.
24.Alsheikh, M. K., Heyen, B. J., and Randall, S. K.
(2003) J. Biol. Chem., 278, 40882-40889.