Expression of Chicken CTCF Gene in COS-1 Cells and Partial Purification of CTCF Protein
E. S. Kotova, I. V. Sorokina, S. B. Akopov*, L. G. Nikolaev, and E. D. Sverdlov
Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, ul. Miklukho-Maklaya 16/10, 117997 Moscow, Russia; fax: (495) 330-6538; E-mail: akser@ibch.ru* To whom correspondence should be addressed.
Received April 12, 2013
The chicken gene for transcription factor CTCF was expressed in COS-1 mammalian cells. The CTCF protein containing polyhistidine tag was partially purified using metallo-affinity and ion-exchange chromatography. The expressed protein localized in the cell nucleus and was shown to be functionally active in the electrophoretic mobility shift assay and specifically interacted with anti-CTCF antibodies.
KEY WORDS: transcription factor CTCF, expression in mammalian cells, COS-1DOI: 10.1134/S0006297913080038
The binding of transcription factor CTCF to DNA plays a major role in gene regulation in vertebrates [1]. CTCF binding sites can be found within promoters, silencers, insulators, and other genome regulatory elements. The CTCF protein interacts with a variety of cellular proteins and with nuclear matrix; it ensures the functioning of insulator elements and other cellular systems (reviewed in [2, 3]).
The human CTCF polypeptide is 727 a.a. in length. The central DNA-binding domain contains one Cys2-His-Cys and 10 Cys2-His2 zinc fingers and flanked by lysine- and arginine-rich sequences. The glycine-rich sequence characteristic for ATP- and GTP-binding proteins and nuclear localization signal are located near the C-terminus of CTCF [1, 4]. Chicken CTCF contains 728 a.a., and its structure and properties are very similar to that of human CTCF [5].
The 82-kDa CTCF protein has an abnormal electrophoretic mobility in SDS-PAGE due to peculiarities in its amino acid composition and, possibly, to posttranslational modifications; it migrates as a 130-155 kDa protein [6].
Previously, human CTCF was expressed in bacterial cells [7] and in baculoviruses [8, 9]. However, the expression of vertebrate genes in prokaryotic and invertebrate cells can lead to incorrect posttranslational modifications and/or erroneous folding. We developed a simple and fast method of expression and partial purification of chicken CTCF protein from vertebrate cells. To this end, we prepared a construct expressing CTCF with N-terminal histidine tag in COS-1 cells and purified the protein using metallo-affinity and Q-Sepharose columns. Nuclear localization and functional activity of the expressed CTCF were demonstrated.
MATERIALS AND METHODS
CTCF expression. The pHHc plasmid [6] containing a full-length chicken CTCF cDNA insert in expression vector pSG5 (Stratagene, USA) was modified by insertion of a polyhistidine coding sequence. To this end, a double-stranded adapter AATTCGAAATGGGACACCACCACCACCACCAC and AATTGTGGTGGTGGTGGTGGTGTCCCATTTCG was cloned into the EcoRI site located between the SV40 promoter and the CTCF initiation codon. The resulting plasmid was named pHHc-his, and its structure verified by sequencing.
COS-1 cells [10] were grown in DMEM-F12 HAM (Gibco, USA) with 10% fetal calf serum (Gibco) at 37°C and 5% CO2. The cells at 80% confluency were used for transfection.
The COS-1 cells were transfected with 1-2 µg of the pHHc-his plasmid per 106 cells using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s protocol. At 72 h after transfections, the cells were collected using Versene solution, washed with PBS, and used for analysis and CTCF purification.
Preparation of nuclear and cytoplasmic extracts and CTCF-enriched fractions. The cells were collected by centrifugation and resuspended at ~107 cells/ml in 2 ml of 10 mM HEPES-KOH, pH 7.9, 10% glycerol, 25 mM KCl, 1 mM EDTA, 0.15 mM spermine, 0.5 mM spermidine, 0.5 mM AEBSF (Sigma, USA), 0.5 mM DTT (buffer A) with the addition of protease inhibitor cocktail (1 µl/ml; Sigma; P8340). To prepare nuclear and cytoplasmic extracts, the cells were disrupted with a Dounce homogenizer, and nuclei were pelleted by centrifugation for 5 min at 3000 rpm and 4°C. Supernatant (cytoplasmic extract) was collected and stored at –70°C. The nuclear pellet was resuspended in 500 µl of buffer A, and KCl was added to final concentration of 0.3 M. The nuclei were incubated under constant stirring for 30 min at 4°C and centrifuged at 12,000 rpm for 10 min and 4°C in a microcentrifuge. The supernatant was collected, dialyzed for 2 h against 100 volumes of 12 mM HEPES-KOH, pH 7.9, 12% glycerol, 60 mM KCl, 0.3 mM EDTA, 0.6 mM DTT, 0.5 mM AEBSF and protease inhibitor cocktail, centrifuged at 12,000 rpm and 4°C for 10 min, and the supernatant (nuclear extract) was stored at –70°C.
Cellular lysate for CTCF purification was prepared by sonication of cellular suspension in 50 mM NaHPO4, pH 8.0, 300 mM NaCl (buffer 1×LEW; Macherey-Nagel, USA) with the addition of 2 mM AEBSF, 8 µl/ml protease inhibitor cocktail, 2 µg/ml leupeptin, and 8 µg/ml aprotinin (all from Sigma) in ultrasonic processor model CV303 (Cole Parmer, USA). Sonication was done on ice with 3 s pulse and 9.9 s intervals for 2 min total. The lysate was applied on Protino Ni-IDA 150 Packed Column (Macherey-Nagel), washed, and protein eluted according to the manufacturer’s protocol. The eluate was further diluted 10-fold with 20 mM HEPES-KOH, pH 7.9, 10% glycerol, 0.2 mM EDTA, 0.5 mM DTT, 2 mM AEBSF with the addition of 8 µl/ml protease inhibitor cocktail, 2 µg/ml leupeptin, and 8 µg/ml aprotinin (buffer B) and loaded on an 80-µl Q-Sepharose (Q-1126; Sigma) column equilibrated with the same buffer. The column was washed with 1 ml of buffer B with the addition of NaCl up to 40 mM and eluted with the same buffer stepwise supplied with 200-450 mM NaCl.
Protein gel electrophoresis and Western blotting. Proteins were resolved by 10% SDS-PAGE [11], and the gels were either stained with Brilliant Blue R or silver stain [12] or electroblotted to PVDF membrane (Immobilon P; Millipore, USA) using a Trans-Blot SD cell (Bio-Rad, USA) and the manufacturer’s protocol.
The membrane was blocked with 5% nonfat dry milk in PBS with the addition of 0.1% Tween 20 (PBS-T) for 1 h at room temperature, incubated in the same solution with primary antibodies overnight at 4°C, and washed three times with PBS-T. The anti-GAPDH (Santa Cruz, USA; sc-47724, dilution 1 : 1000), anti-human CTCF N-terminal peptide (Santa-Cruz; sc-5916, dilution 1 : 500), and rabbit polyclonal antibodies against the N-terminal part of chicken CTCF (dilution 1 : 2000, provided by O. V. Iarovaia) were used as primary antibodies. The membrane was then incubated for 1 h at room temperature in PBS-T/5% dry milk solution containing anti-rabbit IgG peroxidase conjugated antibodies (Cell Signaling 7074, 7076, dilution 1 : 1000 or Santa Cruz sc-2304, dilution 1 : 500). Anti-goat, mouse, or rabbit antibodies were used depending on the primary antibodies source. The membrane was washed with PBS-T and visualized using the Immun-Star HRP Chemiluminescent detection kit (Bio-Rad) and Bio-Rad VersaDoc MP4000 imager.
Electrophoretic mobility shift assay (EMSA). The following DNA fragments were used as CTCF-binding EMSA probes: the fragment of 5′-cHS4 insulator from chicken β-globin locus [13, 14], F1 element of the chicken lysozyme gene silencers [15, 16], and the promoter region of the human MYC gene [4]. The fragment of enhancer from chicken β-globin locus, which does not bind CTCF [17], was used as a negative control. The DNA fragments were PCR amplified using 20 ng human or chicken genome DNA as a template and the following primers: AAGCCCCCAGGGATGTAAT and AGGTGTCTGCAGGCTCAAAG (5′-cHS4 insulator); CAGCACAGTTCTGGCTATGAAA and CCTCAGCTG GGGTCAATAAGT (lysozyme gene silencer); GGGATCGCGCTGAGTATAAA and GGATCTCCCTTCCCAGGAC (MYC gene promoter); GGGCAGGTTGCAGATAAACA and TAACCCCCTCTCTTCCCTCA (β-globin enhancer). The DNA fragments were amplified for 26-29 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 90 s, purified by electrophoresis in 1.2% agarose gel, and used as a templates for PCR labeling as described previously [18].
For EMSA, ~5 ng (30,000-50,000 cpm) of the labeled DNA fragment, 1 µg of poly(dI-dC), and 1-2 µg of nuclear or cytoplasmic extract protein or 2 µl of purified CTCF protein fraction were mixed in a 20-µl final volume of 12 mM HEPES-KOH, pH 7.9, 12% glycerol, 60 mM KCl, 0.3 mM EDTA, 0.6 mM DTT. For supershift, 4.5 µg of anti-chicken CTCF antibodies (provided by O. V. Iarovaia) or 3 µl of monoclonal anti-polyhistidine antibodies (Sigma; H1029) were added, and the mixture was incubated for 20 min at room temperature. The mixture was then resolved in 5 or 7.5% polyacrylamide gel prepared in 50 mM Tris-borate buffer, 0.5 mM EDTA, pH 8.3, and autoradiographed for 16-40 h.
RESULTS AND DISCUSSION
To express chicken CTCF protein with an N-terminal polyhistidine tag (see “Materials and Methods”), we modified the pHHc plasmid [6] bearing the chicken CTCF cDNA sequence. The pHHc plasmid was constructed based on the pSG5 vector (Stratagene, USA) that contains SV40 origin of replication for efficient expression in COS-1 cells. Next, the double-stranded oligonucleotide adapter encoding six histidine residues was inserted into the unique EcoRI site of pHHc located between the SV40 promoter and the CTCF coding sequence. The adapter also contained the ATG codon within the Kozak consensus sequence [19].
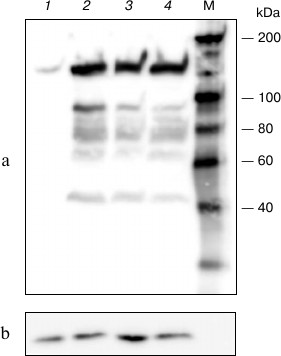
The resulting plasmid (pHHc-his) was used to transfect COS-1 cells at different conditions, and the lysate from transfected cells was analyzed by Western blotting (Fig. 1). As seen in the figure, lysate from transfected cells contains significantly more CTCF than that from non-transfected cells. Also, the content of the expressed CTCF only weakly depended on the conditions of transfection, like the plasmid and Lipofectamine concentrations.
Fig. 1. Western blot analysis of lysates prepared from COS-1 cells transfected with pHHc-his plasmid. Lanes: 1) non-transfected cells; 2) 6 µg of the plasmid, 15 µl of Lipofectamine; 3) 3 µg of the plasmid, 20 µl of Lipofectamine; 4) 3 µg of the plasmid, 10 µl of Lipofectamine. a) Stained using antibodies to chicken CTCF; b) stained using antibodies to glyceraldehyde 3-phosphate dehydrogenase. M, molecular mass marker.
The ability of the expressed CTCF to bind specifically its cognate DNA sequences was verified by EMSA. Well-characterized CTCF-binding sequences, namely 5′-cHS4 insulator from chicken β-globin locus [13, 14], F1 element of the chicken lysozyme gene silencers [15, 16], and promoter region of the human MYC gene [4] were used as probes, and the fragment of enhancer from chicken β-globin locus that does not bind CTCF [17] was used as a negative control.
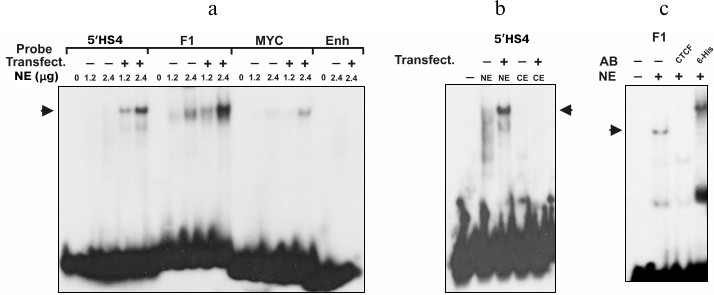
As seen in Fig. 2a, after incubation of nuclear extract of the transfected COS-1 cells with the DNA fragments containing 5′-cHS4 insulator, F1 element, and MYC promoter, DNA–protein complexes were formed. When the same fragments were incubated with the nuclear extract from non-transfected COS-1, the bands corresponding to the DNA–protein complexes were slightly visible, which confirms elevated expression of CTCF in the transfected cells. No DNA–protein complexes were formed after incubation of nuclear extract with negative control – the fragment of enhancer from chicken β-globin locus (Fig. 2a).
Fig. 2. EMSA analysis of interaction between expressed CTCF and different DNA fragments. Probes: 5′HS4, 5′-cHS4 insulator from chicken β-globin locus; F1, F1 element of the chicken lysozyme gene silencer; MYC, promoter region of the human MYC gene; Enh, fragment of enhancer from chicken β-globin locus. NE, nuclear extract; CE, cytoplasmic extract from COS-1 cells. Transfect. +/– indicates cells transfected/non-transfected with pHHc-his plasmid. Arrows indicate the positions of the CTCF–DNA complexes. a) Binding of CTCF from transfected and non-transfected COS-1 cells with different DNA sequences. The figures above the lanes indicate the amount (in µg) of nuclear extract protein added. b) Binding of 5′-cHS4 insulator from chicken β-globin locus with nuclear and cytoplasmic extracts from COS-1 cells. c) Binding of CTCF-F1 complexes with anti-chicken CTCF and anti-polyhistidine antibodies (AB).
It is known that CTCF protein contains nuclear localization signal and is transported to the nucleus after synthesis [4, 5]. We examined the binding of the labeled DNA fragments with the cytoplasmic extract from the transfected and non-transfected COS-1 cells (Fig. 2b). As seen from the figure, the band corresponding to the DNA–protein complex is absent in both cytoplasmic extracts, indicating that both endogenous and overexpressed CTCF was localized mainly in the nucleus. The expressed CTCF is also able to bind both anti-CTCF and anti-polyhistidine antibodies (Fig. 2c). Hence, the transfection of COS-1 cells with pHHc-his leads to overexpression of the CTCF protein containing the N-terminal polyhistidine tag.
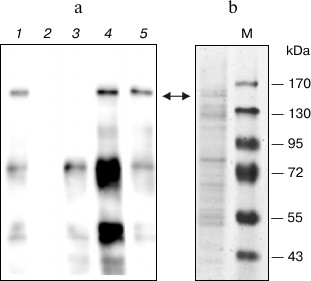
As a first step of purification of the CTCF from COS-1 cells lysate, we used the Protino Ni-IDA 150 Packed Column (Macherey-Nagel) according to the manufacturer’s recommendations. The protein was eluted with buffer containing 250 mM imidazole. As seen in the Western blot (Fig. 3a, lane 1), the eluted fraction contained two main bands interacting with the anti-CTCF antibodies. The first band with a molecular mass of more than 130 kDa corresponded to the full-length CTCF with histidine tag. The second interacting with the antibodies band with molecular mass of about 70 kDa did not change its intensity upon addition of different proteolysis inhibitors and can be a result of either premature termination of the polypeptide chain or a product of specific proteolysis. The presence of CTCF-specific protease in mammalian cells was suggested earlier [20].
Fig. 3. Purification of expressed CTCF protein. a) Western blot analysis of eluates from different purification steps. Lanes: 1) protein purified by Ni affinity chromatography; 2) Q-Sepharose flow-through; 3) elution from Q-Sepharose with 200 mM NaCl buffer; 4) the same as (3) with 300 mM NaCl; 5) the same as (3) with 450 mM NaCl. b) SDS-PAGE of fraction eluted from Q-Sepharose with 450 mM NaCl buffer. Arrows indicate the position in gel of the full-length CTCF protein.
The analysis of the resulting fraction by SDS-PAGE with silver staining revealed insufficient purity of the protein (not shown). For additional purification, we employed Q-Sepharose chromatography similar to that described previously [21]. The eluate from the Ni-IDA column was diluted 10-fold and applied to the Q-Sepharose column, and the protein was eluted stepwise with buffer containing 200, 300, and 450 mM NaCl (Fig. 3a). As seen, the major contaminating 70-kDa protein and other contaminants are eluted mostly at 200 mM NaCl, whereas CTCF is eluted at 450 mM. The additionally purified protein was then analyzed by SDS-PAGE with Coomassie staining (Fig. 3b). As seen in the figure, the two-stage purification allowed us to obtain protein fraction containing about 10% of CTCF. The protein obtained is able to specifically interact with several known CTCF binding sites.
The authors are grateful to E. M. Klenova for gift of pHHc plasmid and to O. V. Iarovaia and S. V. Razin for anti-CTCF antibodies.
This work was supported by the Scientific School program (project NSh. 5638.2010.4), the Program of the Russian Academy of Sciences on Molecular and Cellular Biology, and by the Russian Foundation for Basic Research (project 10-04-01365). DNA was sequenced at the Genome Center (http://www.genome-centre.ru/) supported by the Russian Foundation for Basic Research.
REFERENCES
1.Ohlsson, R., Renkawitz, R., and Lobanenkov, V.
(2001) Trends Genet., 17, 520-527.
2.Filippova, G. N. (2008) Curr. Top. Dev.
Biol., 80, 337-360.
3.Nikolaev, L. G., Akopov, S. B., Didych, D. A., and
Sverdlov, E. D. (2009) Curr. Genom., 10, 294-302.
4.Filippova, G. N., Fagerlie, S., Klenova, E. M.,
Myers, C., Dehner, Y., Goodwin, G., Neiman, P. E., Collins, S. J., and
Lobanenkov, V. V. (1996) Mol. Cell Biol., 16,
2802-2813.
5.Klenova, E. M., Nicolas, R. H., Paterson, H. F.,
Carne, A. F., Heath, C. M., Goodwin, G. H., Neiman, P. E., and
Lobanenkov, V. V. (1993) Mol. Cell Biol., 13,
7612-7624.
6.Klenova, E. M., Nicolas, R. H. U. S., Carne, A. F.,
Lee, R. E., Lobanenkov, V. V., and Goodwin, G. H. (1997) Nucleic
Acids Res., 25, 466-474.
7.Jiang, L., Qin, Y., Sun, Z. L., Wu, J., Yang, W.
L., and Zhang, J. Z. (2009) Sichuan Da Xue Xue Bao Yi Xue Ban,
40, 1-5.
8.Chernukhin, I. V., Shamsuddin, S., Robinson, A. F.,
Carne, A. F., Paul, A., El-Kady, A. I., Lobanenkov, V. V., and Klenova,
E. M. (2000) J. Biol. Chem., 275, 29915-29921.
9.Chau, C. M., Zhang, X. Y., McMahon, S. B., and
Lieberman, P. M. (2006) J. Virol., 80, 5723-5732.
10.Gluzman, Y. (1981) Cell, 23,
175-182.
11.Laemmli, U. K. (1970) Nature, 227,
680-685.
12.Merril, C. R., Dunau, M. L., and Goldman, D.
(1981) Anal. Biochem., 110, 201-207.
13.Bell, A. C., West, A. G., and Felsenfeld, G.
(1999) Cell, 98, 387-396.
14.Chung, J. H., Bell, A. C., and Felsenfeld, G.
(1997) Proc. Natl. Acad. Sci. USA, 94, 575-580.
15.Baniahmad, A., Steiner, C., Kohne, A. C., and
Renkawitz, R. (1990) Cell, 61, 505-514.
16.Kohne, A. C., Baniahmad, A., and Renkawitz, R.
(1993) J. Mol. Biol., 232, 747-755.
17.Klochkov, D., Rincon-Arano, H., Ioudinkova, E.
S., Valadez-Graham, V., Gavrilov, A., Recillas-Targa, F., and Razin, S.
V. (2006) Mol. Cell Biol., 26, 1589-1597.
18.Vetchinova, A. S., Akopov, S. B., Chernov, I. P.,
Nikolaev, L. G., and Sverdlov, E. D. (2006) Anal. Biochem.,
354, 85-93.
19.Kozak, M. (1987) Nucleic Acids Res.,
15, 8125-8148.
20.Dunn, K. L., Zhao, H., and Davie, J. R. (2003)
Exp. Cell Res., 288, 218-223.
21.Arnold, R., Burcin, M., Kaiser, B., Muller, M.,
and Renkawitz, R. (1996) Nucleic Acids Res., 24,
2640-2647.