Inhibition of Mn2+-Induced Error-Prone DNA Synthesis with Cd2+ and Zn2+
L. V. Gening1*, A. V. Lakhin1, E. V. Stelmashook2, N. K. Isaev2,3, and V. Z. Tarantul1
1Institute of Molecular Genetics, Russian Academy of Sciences, pl. Kurchatova 2, 123182 Moscow, Russia; E-mail: geni@img.ras.ru2Department of Brain Research, Institute of Neurology, Russian Academy of Medical Sciences, Pereulok Obukha 5, 105064 Moscow, Russia
3Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119992 Moscow, Russia
* To whom correspondence should be addressed.
Received May 23, 2013; Revision received June 17, 2013
Bivalent metal cations are key components in the reaction of DNA synthesis. They are necessary for all DNA polymerases, being involved as cofactors in catalytic mechanisms of nucleotide polymerization. It is also known that in the presence of Mn2+ the accuracy of DNA synthesis is considerably decreased. The findings of this work show that Cd2+ and Zn2+ selectively inhibit the Mn2+-induced error-prone DNA polymerase activity in extracts of cells from human and mouse tissues. Moreover, these cations in low concentrations also can efficiently inhibit the activity of homogeneous preparations of DNA polymerase iota (Pol ι), which is mainly responsible for the Mn2+-induced error-prone DNA polymerase activity in cell extracts. Using a primary culture of granular cells from postnatal rat cerebellum, we show that low concentrations of Cd2+ significantly increase cell survival in the presence of toxic Mn2+ doses. Thus, we have shown that in some cases low concentrations of Cd2+ can display a positive influence on cells, whereas it is widely acknowledged that this metal is not a necessary microelement and is toxic for organisms.
KEY WORDS: DNA polymerase iota, manganese, manganism, cadmiumDOI: 10.1134/S0006297913100088
Abbreviations: DMSO, dimethyl sulfoxide; DTT, dithiothreitol; PARP, poly(ADP-ribose) polymerase; PMSF, phenylmethylsulfonyl fluoride; Pol, DNA polymerase.
DNA synthesis in the living cell is a complex process involving dozens
of different enzymes that participate in DNA chain duplication either
directly or through various accessory processes such as unwinding DNA
chains and their stabilization [1, 2]. From about two dozen DNA polymerases in mammals,
only a few enzymes are directly involved in DNA replication in dividing
cells, whereas the majority of DNA polymerases participate in DNA
repair, which occurs in all living cells without exception. Moreover,
numerous regulatory proteins determine in the cell when one or another
DNA polymerase has to work. Taking into consideration this multiformity
of processes during DNA synthesis, it becomes clear that in
vitro studies only on homogenous preparations of DNA polymerases
cannot adequately represent the situation in the living cell. Studies
on DNA synthesis in cell extracts containing the whole set of DNA
polymerases and regulatory factors in the ratios specific for each cell
type can help in resolving this problem.
Bivalent metal cations are key components in the reaction of DNA synthesis. They are necessary for all polymerases as cofactors participating in the catalytic mechanism of nucleotide polymerization. All DNA polymerases are characterized by the presence of a bivalent metal cation-binding site, which includes two or three residues of aspartic or glutamic acid [3, 4]. Because the Mg2+ concentration in cells is much higher than the concentrations of all other bivalent cations, and this ion activates much more efficiently the majority of known DNA polymerases in experiments in vitro, Mg2+ is reasonably believed to be the major activator of DNA synthesis in cells [5, 6]. Nevertheless, many other bivalent cations can act as cofactors, but in this case DNA synthesis is usually associated with a significant decrease in efficiency and accuracy of nucleotide inclusion [7, 8]. Because concentrations of other bivalent metal cations in cells are significantly lower than the Mg2+ concentration, they are supposed to insignificantly influence DNA synthesis in vivo. However, it is difficult to predict the effect on DNA synthesis and genome stability of high doses of any metal if they enter the organism from the environment.
In particular, manganese is a rather widely distributed metal in the environment. Normally, this metal is a necessary microelement, acting as a cofactor of different enzymes [9, 10]. Intoxication with high doses of manganese can lead to many disorders, mainly ones affecting the nervous system of the organism. However, the molecular mechanisms of manganese-induced neurotoxicity are still not known in detail [11, 12]. On one hand, some neurotoxic effects of manganese can be induced by oxidative stress owing to increased production of free radicals in the Fenton reaction [13, 14]. On the other hand, there are some indications that the manganese-induced neurotoxicity can be based on disorders in genome stability because of a decrease in the accuracy of DNA synthesis under the influence of elevated concentrations of this metal ion on some DNA polymerases [15, 16]. Thus, in the works by Loeb it was shown that the presence of Mn2+ in the reaction medium resulted in a significant decrease in the accuracy of DNA synthesis realized by homogenous preparations of human, viral, and bacterial DNA polymerases [17]. Moreover, in our laboratory [18, 19] a sharp decrease in the accuracy of DNA synthesis was found in extracts of mammalian cells in the presence of Mn2+, mainly caused by activation of DNA polymerase iota (Pol ι), which is characterized by a high frequency of errors in DNA synthesis. Therefore, it was supposed that, because of a very pronounced ability for error-prone inclusion of nucleotides in the presence of Mn2+, Pol ι could be somewhat related with the genotoxic effect of this metal. Inhibition of the Pol ι activity was supposed to be able to decrease the pathologic effect of the manganese-induced intoxication.
The purpose of the present work was to determine bivalent metal cations capable of suppressing Pol ι activity and thus abolishing the Mn2+-induced error-prone activity of DNA polymerase and normalizing the accuracy of DNA synthesis.
MATERIALS AND METHODS
Preparation of recombinant proteins. The full-size human Pol ι fused with glutathione-S-transferase on the N-terminus was used. The enzyme was produced in yeast using the expressed yeast vector obtained by us earlier [20]. Purified preparations of human Pol β, Pol κ, and Pol λ isolated from E. coli as described earlier [21-23] were also used. The purity of all enzymes was higher than 95%.
Preparation of cell extracts for performing the DNA polymerase reaction. To prepare cellular extracts, the studied tissue samples were disrupted on ice with a Teflon homogenizer in the presence of glass particles in the following extracting buffer: 10% glycerol, 1% DMSO, 0.5% Tween 20, 2.5 mM DTT, 1 mM PMSF in 1× PBS (pH 7.4). The buffer volume was taken at the ratio of two parts per one part of the homogenized tissue. The resulting homogenate was centrifuged at 4°C for 10 min at 14,000g. The supernatant was used as the enzyme preparation. The protein concentration was determined using Protein Assay reagent (BioRad, USA) and adjusted to 5 mg protein per ml.
Preparation of substrate for the DNA polymerase reaction. Two complementary deoxyribooligonucleotides, a 17-mer primer 5′-GGAAGAAGAAGTATGTT-3′ and a 30-mer template 5′-CCTTCGTCATTCTAACATACTTCTTCTTCC-3′, were used as substrates for determination the activity of DNA polymerase; these deoxyribooligonucleotides form on hybridization a duplex with an overhanging 5′-end. The primer was labeled from the 5′-end using phage T4 polynucleotide kinase (10 units) and 2 MBq [γ-32P]ATP in 70 mM Tris-HCl buffer (pH 7.6) supplemented with 10 mM MgCl2 and 5 mM DTT at 37°C for 30 min. Then the enzyme was inactivated at 70°C for 10 min. The substrate for the enzymatic reaction was obtained upon annealing in 200 µl of 10 pmol labeled primer with 15 pmol template in buffer for polynucleotide kinase supplemented with NaCl to 100 mM at 73°C for 3 min with subsequent cooling to room temperature.
Elongation of the radio-labeled primer. The reaction of DNA synthesis was performed in 20 µl of reaction mixture containing 50 nM substrate with the labeled primer, 50 mM Tris-HCl (pH 8.0), 0.5 mM each dNTP, and 3 µl of the corresponding sample extract. The studied bivalent metals were also added as their chlorides (MCl2, where M is metal) into the reaction mixture as cofactors for DNA synthesis. The basal level of DNA synthesis during the reaction was provided due to close to physiological concentrations of Mg2+ (250 µM) and Mn2+ (50 µM) [6, 24]. The reaction mixture was incubated at 37°C for 5 min in the case of extracts from mouse organ cells and for 20 min in the case of extracts from human uveal melanoma cells. The reaction was stopped, the reaction products were separated by electrophoresis, and a radioautograph was obtained as described earlier [18]. The data were analyzed using the Image Quant program. In all cases, quantitative parameters were determined from results of three to five independent experiments.
Primary cultures of cerebellum. The work was performed using 7-9-day-old cultures of granular cells isolated from the cerebellum of 8-day-old Wistar rats by enzymatic–mechanical dissociation. The cerebellum tissue was dissociated as follows: the isolated cerebellums were placed into a plastic Petri dish filled with phosphate free of calcium and magnesium ions. The tissue fragments were incubated for 15 min at 37°C in phosphate buffer containing 0.05% trypsin and 0.02% EDTA. During the incubation, the tissue was washed twice in phosphate buffer and once in the culture medium and then subjected to mechanical dissociation in the culture medium. The nutrition medium contained 10% fetal calf serum, 2 mM glutamine, and 10 mM HEPES (pH 7.2-7.4). The cell suspension was centrifuged for 1 min at 1000 rpm, the supernatant was removed, and the precipitate was resuspended in the nutrition medium. The culture was maintained in 96-well plastic chambers covered with polylysine. Cell suspension (0.1 ml) was added into every well. The cultures developed in the presence of CO2 at 35.5°C and relative humidity 98%. The culture medium contained 25 mM KCl necessary for survival of the cerebellum granular neurons after the 4th day of in vitro cultivation. The animals were treated in accordance with the European Society Council 86/609/EEC requirement concerning use of animals for experimental studies. Cadmium or manganese chlorides were added into the culture medium on the 2nd day upon plating the cultures.
Evaluation of survival of neurons. After the experiment, the cultures were fixed with a mixture of ethanol–formaldehyde–acetic acid (7 : 2 : 1 v/v) and stained with Trypan Blue. The percent of neuron survival was evaluated by calculating morphologically intact nuclei in the culture of the cerebellum granular cells in five visual fields at objective magnification ×40. Neuron survival in untreated control cultures was taken as 100%, the survival in the experimental cultures being expressed as percent relative to the control.
Statistical analysis of neuron survival. Statistical analysis was performed using the ANOVA test with the Bonferroni or Dunnett post-test, at the significance level of p < 0.05. The results are expressed as mean ± SEM. All data were obtained on at least 9-12 cultures from two-three independent experiments.
RESULTS
Influence of bivalent metal cations on Mn2+-induced error-prone DNA polymerase activity. To study the influence of bivalent metal cations on Mn2+-induced error-prone DNA polymerase activity, the elongation reaction of the radiolabeled primer was performed with cell extracts from C57Bl line mouse brains in the presence of 250 µM Mg2+, 200 µM Mn2+, and 200 µM ion of one of the studied bivalent metals. The elongation reaction resulted in a set of products with different quantities of included nucleotides. These products were separated by electrophoresis into separate bands. The presence of additional bands in the electrophoregrams indicated the generation of products with incorrectly included nucleotides, and analysis with the Image Quant program allowed us to assess the intensity of their generation. Concentrations of bivalent metal cations used in the reaction of DNA synthesis were taken by us on considering that the physiological contents of Mg2+ in the cell as about 250 µM [6, 25] and that this concentration was sufficient for a certain level of DNA polymerase activities in the cell extracts. In preliminary experiments we found that 200 µM Mn2+ provided the generation of the maximal amount of products with noncomplementary included nucleotides. Thus, studies under optimal conditions on changes in the Mn2+-induced error-prone DNA polymerase activity under the influence of different bivalent metal cations allowed us to exclude false-positive results, which could appear because of action of nonspecific factors on the enzymatic activity (e.g. changes in the intensity of DNA synthesis producing incorrect products in response to changes in the solution ionic strength in solution supplemented with an additional amount of the studied metal cation).
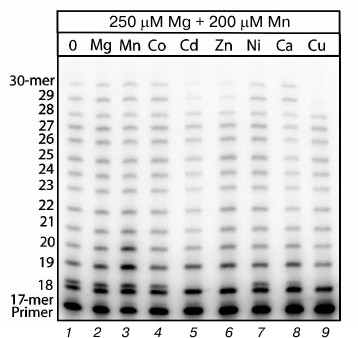
The electrophoregram (Fig. 1) shows that an 18-mer product with an incorrectly included nucleotide opposite to T of the template is the major product of the Mn2+-induced error-prone DNA polymerase activity. Earlier we found [19, 20] that in the reaction of DNA synthesis in the presence of Mn2+ this product was generated in the cell extract owing to Pol ι characterized by preferential inclusion of G opposite to T of the template instead of the complementary A. The major role of Pol ι in the generation of this product with the noncomplementary included nucleotide was confirmed by the absence of generation of this product in cell extracts from the 129 line mice with nonfunctional Pol ι [18]. If in addition to Mg2+ and Mn2+ also Co2+ and Ni2+ were present in the reaction mixture (Fig. 1, lanes 4 and 7), the amount of the generated major incorrect product was unchanged, whereas this product disappeared in the presence of Cd2+, Zn2+, or Cu2+ (Fig. 1, lanes 5, 6, 9). An addition of Ca2+ into the reaction mixture also partially suppressed the generation of the DNA synthesis products with incorrectly included nucleotides. An additional 200 µM Mg2+ and Mn2+ (Fig. 1, lanes 2 and 3) were supplemented as reference ions to show that Cd2+, Zn2+, and Cu2+ acted specifically and not, for instance, through changes in the ionic strength of the reaction mixture.
Fig. 1. Influence of different bivalent metal cations on Mn2+-induced error-prone DNA polymerase activity in extracts of brain cells from C57Bl line mice. The reaction mixtures contained 250 µM Mg2+, 200 µM Mn2+, and 200 µM of the studied ion. The reaction was performed for 5 min, and the concentration of all four dNTPs in the reaction mixture was 500 µM.
Our experiments revealed that the Mn2+-induced error-prone DNA polymerase activity was inhibited specifically in the presence of Cd2+, Zn2+, and Cu2+ because no significant changes occurred in the correct DNA polymerase activity. Because the studied metals at the concentrations used were cytotoxic, it was necessary to elucidate what minimal concentrations of these metals would continue to inhibit the error-prone synthesis.
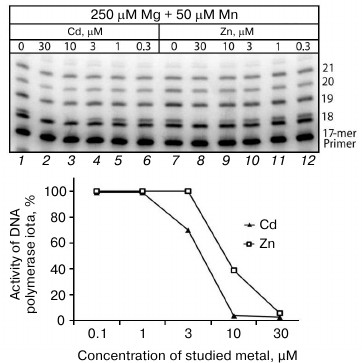
Influence of Cd2+ and Zn2+ on Mn2+-induced error-prone DNA polymerase activity in extracts from mouse brain and liver cells. We performed reactions of DNA synthesis in cell extracts in the presence of different concentrations of Cd2+ and Zn2+ using the misGvA method earlier described by us in medium containing only two nucleotides, dATP and dGTP [18]. This allowed us to specifically isolate products of the Pol ι activity, which was mainly responsible for the Mn2+-induced error-prone DNA polymerase activity. The absence of the two other nucleotides allowed us to unambiguously interpret the findings, because only dGTP could be included opposite to T of the template in the synthesis product with an incorrectly included nucleotide. Studies on the dependence of Pol ι activity on the Cd2+ and Zn2+ concentrations in the extracts from the brain cells of the C57Bl line mice revealed that the Pol ι activity was nearly completely suppressed at Cd2+ concentrations higher than 3 µM (Fig. 2). The inhibitory activity of Zn2+ was lower, and the Pol ι activity was efficiently suppressed only at Zn2+ concentrations higher than 10 µM. Because in both cases the inhibition was observed at rather low concentrations of the metal studied on the background of at least tenfold higher amounts of Mg2+ and Mn2+, it was supposed that Pol ι should be inhibited specifically but not due to a simple competition of ions for the binding site of bivalent metal cations in the enzyme.
Fig. 2. Influence of Cd2+ and Zn2+ on Mn2+-induced error-prone DNA polymerase activity in extracts of brain cells from the C57Bl line mice. The reaction mixtures contained 250 µM Mg2+, 50 µM Mn2+, and some concentrations of the studied ion. The reaction was performed for 5 min, and the concentration of dGTP and dATP in the reaction mixture was 500 µM.
In the extracts from mouse liver cells the Pol ι activity in the presence of Cd2+ and Zn2+ was inhibited similarly, but in this case the error-prone DNA polymerase activity was efficiently suppressed at slightly higher concentrations of these metals (at 10 and 30 µM, respectively).
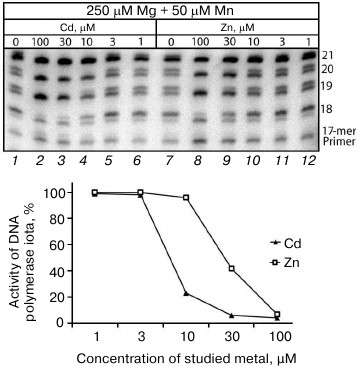
Influence of Cd2+ and Zn2+ on Mn2+-induced error-prone DNA polymerase activity in cell extracts from human uveal melanoma. It is known many enzymes are regulated differently in normal and malignant cells [26, 27]. It has been also shown in our laboratory that DNA synthesis in malignant cells is significantly different from the synthesis in normal cells [18, 19]. In particular, DNA polymerases of at least some malignant tumors can efficiently continue synthesis after inclusion of an incorrect nucleotide under the influence of Pol ι. Many other malignant cells are also characterized by significantly increased activities of error-prone DNA polymerases themselves. Thus, our next goal was to elucidate whether the Mn2+-induced error-prone DNA polymerase activity could change under the influence of Cd2+ and Zn2+ in extracts from malignant cells. We studied DNA synthesis in the presence of Cd2+ and Zn2+ in extracts from human eye uveal melanoma (Fig. 3). The dependence of inhibition of the error-prone Pol ι activity on Cd2+ and Zn2+ concentrations was similar to the activity observed in the extracts from the mouse brain cells. The Pol ι activity in the uveal melanoma cell extracts was efficiently inhibited at Cd2+ concentrations higher than 10 µM and at Zn2+ concentrations higher than 30 µM, which was similar to observations on the mouse liver cell extracts. It should be noted that in the uveal melanoma cell extracts the ions of these metals suppressed both the inclusion of an incorrect nucleotide and post-inclusion synthesis (Fig. 3). The likeness of the inhibition of the Pol ι error-prone activity in extracts from cells of different origin confirmed the universality of mechanisms responsible for regulation of DNA polymerase activity through Cd2+ and Zn2+ in mammalian cells.
Fig. 3. Influence of Cd2+ and Zn2+ on Mn2+-induced error-prone DNA polymerase activity in cell extracts from human uveal melanoma. The reaction mixtures contained 250 µM Mg2+, 50 µM Mn2+, and some concentrations of the studied ion. The reaction was performed for 20 min, and the concentration of dGTP and dATP in the reaction mixture was 500 µM.
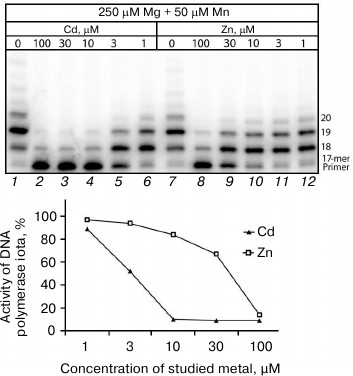
Influence of Cd2+ and Zn2+ on DNA synthesis catalyzed by homogenous preparations of human DNA polymerases. According to the literature data, a homogenous preparation of Pol ι was insignificantly activated in the presence of Cd2+ and Zn2+ [28]. Thus, the present work results seem to contradict these data. However, attention must be given to the DNA synthesis described in the work by Frank and Woodgate [28] that proceeded in the presence only of Cd2+ or Zn2+, whereas in the living cell DNA synthesis is mainly activated by physiological concentrations of Mg2+. The experimental conditions in work [28] in principal did not allow the researchers to detect the suppression of DNA polymerase activity, because due to absence of manganese ions DNA synthesis in the control was absent. To confirm or disprove the literature data on the Pol ι activation, we studied the influence of Cd2+ and Zn2+ on the activity of a homogenous preparation of Pol ι in the presence of close to physiological concentrations of Mg2+ (250 µM) and Mn2+ (50 µM) in both the control and samples. We think that such scheme of experiment can give a more adequate idea concerning all changes in the DNA synthesis induced by a studied metal.
The electrophoregram (Fig. 4) shows that Cd2+ at concentrations higher than 10 µM inhibits nearly totally the DNA polymerase activity of the homogenous preparation of Pol ι, which recovered completely only at Cd2+ concentrations lower than 1 µM. The presence of 100 µM Zn2+ also caused a sharp decrease in the enzyme activity (to 14%) compared to the control, but in this case the Pol ι activity recovered nearly completely on decrease in Zn2+ concentration in the medium to 10-30 µM. Thus, the dependence of inhibition of the Pol ι homogenous preparation activity on concentrations of cadmium and zinc ions was very similar to the dependence observed on all cellular extracts studied. Note that minimal inhibitory concentrations of Cd2+ are rather low and unlikely to be toxic for cells. At least, according to work [29], Cd2+ at concentration 10 µM even stimulated cell growth, whereas the cells died in the presence of 100 µM cadmium ions.
Fig. 4. Influence of Cd2+ and Zn2+ on DNA synthesis catalyzed under the influence of the homogenous Pol ι preparation (15 nM). The reaction mixtures contained 250 µM Mg2+, 50 µM Mn2+, and some concentrations of the studied ion. The reaction was performed for 5 min, and the concentration of all four dNTPs in the reaction mixture was 100 µM.
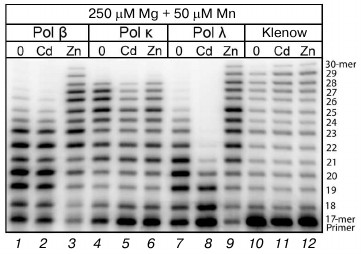
Although the presence of Cd2+ and Zn2+ in the reaction mixture did not lead to decrease in the generation of correct products of DNA synthesis in the cell extracts, it is possible that these metals could inhibit the activity not only of Pol ι but also of some other DNA polymerases. To elucidate this, we studied the influence of Cd2+ and Zn2+ on the activities of homogenous preparations of human DNA polymerase kappa (Pol κ), human DNA polymerase beta (Pol β), human DNA polymerase lambda (Pol λ), and the bacterial Klenow fragment. These three human DNA polymerases are not replicative enzymes, but similarly to Pol ι they are mainly involved in DNA repair. We found (Fig. 5) that the presence of Cd2+ in the reaction mixture in the maximal concentration used by us (100 µM) did not cause significant changes in the DNA synthesis catalyzed by Pol β and the Klenow fragment. However, a sharp decrease in the Pol λ activity and a noticeable decrease in the Pol κ activity were observed under these conditions. On the contrary, the presence of Zn2+ (100 µM) in the reaction medium activated Pol β and Pol λ, whereas the Klenow fragment activity was unchanged. The activity of Pol κ in the presence of both Zn2+ and Cd2+ noticeably decreased. However, significant differences should be noted between the effects of Cd2+ and Zn2+ on homogenous preparations of DNA polymerases and cell extract DNA polymerases.
Fig. 5. Influence of Cd2+ and Zn2+ on DNA synthesis catalyzed under the influence of homogenous preparations of Pol κ (15 nM), Pol β (15 nM), Pol λ (15 nM), and the Klenow fragment (5 nM). The reaction mixtures contained 250 µM Mg2+, 50 µM Mn2+, and 100 µM of the studied ion. The reaction was performed for 5 min, and the concentration of all four dNTPs in the reaction mixture was 100 µM.
Influence of Cd2+ on Mn2+-induced cell death. Based on our studies on the influences of Cd2+ and Zn2+ on DNA synthesis, we supposed that the specific inhibition of Mn2+-induced error-prone DNA polymerase activity could abolish some negative effects caused by intoxication with manganese. To test this hypothesis, we studied the influence of Cd2+ (as the metal most strongly inhibiting the Pol ι activity) on manganese-caused cell death in primary culture of granular cells from the cerebellum of postnatal rats. The neurons in this culture are concurrently glutamatergic and glutamate-sensitive. Such cells are the most frequent type of neurons in the brain, and their population is uniform. These cells were chosen also because the nervous system is mainly affected on intoxication with manganese.
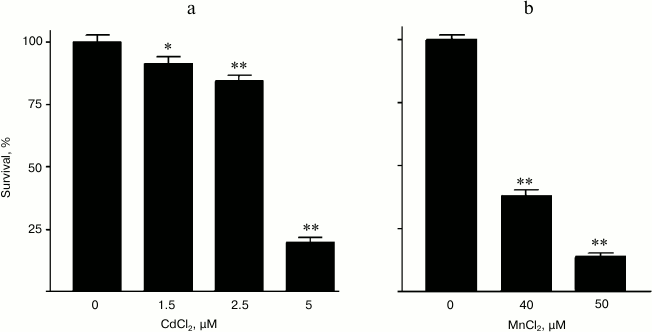
In preliminary experiments, the minimal toxic concentration of Mn2+ for the granular cells was found to be in the range of 40-50 µM, whereas the toxicity of Cd2+ was observed beginning from the concentration of 2.5 µM and higher (Fig. 6). Note that the minimal toxic concentration of Mn2+ for the cells used by us was the same as the minimal toxic concentration of these ions for a similar culture from mouse cerebellum cells [30].
Fig. 6. Survival dependence of rat cerebellum granular cells on concentration of (a) CdCl2 and (b) MnCl2 in the culture medium.
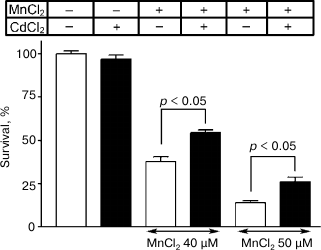
The neurons with the normal morphology were determined after incubation for five days, and neuronal survival in the presence of 40 and 50 µM manganese chloride was found to be 38 ± 3% and 14 ± 2%, respectively. The addition concurrently with manganese of 0.5 µM CdCl2 reliably increased the cell survival to 54 ± 2% (p < 0.05) and 26 ± 3% (p < 0.05), respectively (Fig. 7). The survival of neurons in the presence only of 0.5 µM CdCl2 and in the absence of manganese was 97 ± 2%.
Fig. 7. Influence of 0.5 µM CdCl2 on survival of cultured granular neurons from rat cerebellum on the background of toxic effect of MnCl2.
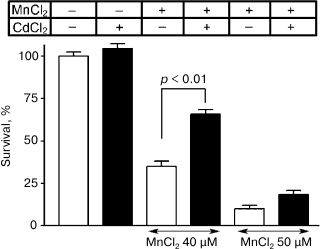
In the other group of experiments, 40 and 50 µM manganese chloride during five days of incubation decreased the survival of neurons in the culture to 35 ± 3% and 10 ± 2%, respectively. The addition concurrently with manganese of 0.25 µM CdCl2 reliably increased the cell survival to 66 ± 3% (p < 0.01) and 19 ± 2%, respectively (Fig. 8). The survival of neurons in the presence only of 0.25 µM CdCl2 was 105 ± 3%.
Fig. 8. Influence of 0.25 µM CdCl2 on survival of cultured granular neurons from rat cerebellum on the background of toxic effect of MnCl2.
DISCUSSION
As we have shown in the present work, Cd2+ and Zn2+ selectively inhibit the Mn2+-induced error-prone DNA polymerase activity in cell extracts. These metal cations also efficiently inhibit the activity of homogenous preparations of Pol ι (the enzyme mainly responsible for Mn2+-induced error-prone DNA polymerase activity in cell extracts) in the presence of rather low concentrations of Cd2+ and Zn2+ in the reaction medium. Moreover, using the procedure of DNA synthesis developed by us, we have shown on both homogenous preparations of DNA polymerases and on cell extracts that for an adequate interpretation of the influence of bivalent cations on DNA synthesis, this reaction must be performed in the presence of basic metal cations in concentrations close to physiological ones. This provides a certain level of DNA polymerase activity and allows researchers to compare with it not only the activating effect of cations under study, but also their probable inhibitory influence.
It is interesting that Cd2+ and Zn2+ can inhibit some DNA polymerases, activate others, and have no influence on the still others (Fig. 5). Such a diverse effect of metal cations seems to be due to cation binding not to DNA polymerases themselves, but due to formation of complexes with DNA as a substrate. This explanation is somewhat confirmed by studies on activities of some other proteins capable of binding to DNA that are involved in DNA repair. Thus, studies on the influence of metal cations on functioning of the Fpg family DNA glycosylases have shown that inhibition of these enzymes is due to Cd2+ and Zn2+ binding just with DNA [31].
Due to phosphodiester groups of the scaffold, nucleic acids carry a large negative charge and therefore are always associated with cations. In addition to oxygen atoms of the phosphate groups, nucleoside residues present numerous possible sites for coordination of metal ions [32, 33]. Hence, inhibition of different DNA polymerases in the presence of Cd2+ and Zn2+ can be explained by different binding sites of these cations with DNA. Thus, according to X-ray crystallography data, Zn2+ binds mainly with peripheral atoms of phosphate groups and only in some cases can interact with the N7 atom of guanine [34, 35]. However, Cd2+ mainly binds not with phosphate groups but with nucleophilic nitrogen atoms of heterocyclic bases [36, 37]. As a consequence, the specific interaction of a metal cation with DNA results in a local change in the charge and electron density in this region of DNA that can prevent functioning of some DNA-binding enzymes because of specific structural features of their DNA-interacting sites.
The data obtained in the present work on the primary culture or granular cells from rat cerebellum have shown that nontoxic concentrations of cadmium ions decrease the toxic effect of manganese ions. Note also that the protective effect of cadmium is displayed at its concentrations in the medium that are significantly lower (at least 10-fold) than the toxic concentrations. We predicted this effect based on the ability of cadmium in relatively low concentrations to suppress the activity of the pure preparation of Pol ι, as well as the activity of this enzyme in cell extracts from mouse brain and liver and from human uveal melanoma.
Hypothetically, the whole mechanism of the positive influence of Cd2+ on cell survival can be as follows. The continuously proceeding reparative synthesis of DNA in the presence of increased Mn2+ concentrations becomes more error-prone, thus decelerating or inhibiting the activity of the protein reparative complex in the site of an incorrect nucleotide inclusion. This can result in accumulation in the genome of a certain critical number of unrepaired regions of DNA that acts as a signal for triggering cell death. Moreover, Mn2+ is shown to rather strongly inhibit the functioning of one of the key enzymes of DNA repair, PARP [38]. This protein is involved in detecting damaged regions of DNA and in the assemblage of a repair complex of enzymes in these regions [39, 40]. Thus, in addition to activation of the error-prone synthesis of DNA, repair processes are inhibited. The above-mentioned factors, as well as some others seem to underlie Mn2+-induced cell death. The Cd2+-mediated elimination of one of the key factors (a decrease in DNA synthesis accuracy!) of the above-described mechanism of cell death is probably responsible for the cell survival in the presence of toxic doses of Mn2+. It also seems likely that in the case of intoxication with high doses of manganese and the associated specific disease, manganism, the suppression of the Mn2+-induced error-prone DNA polymerase activity with Cd2+ or its functional analogs can display a protective effect.
Thus, we have shown for the first time that low concentrations of Cd2+ can positively influence cells, although it is widely acknowledged that this metal is not a necessary microelement and is toxic for the organism, and there are no data on its positive action in any physiological process.
The authors are grateful to O. I. Lavrik for the kindly presented DNA polymerase λ and to A. V. Makarov for the kindly presented polymerase ι.
This work was supported by the Russian Academy of Sciences (program “Molecular and Cell Biology”) and by the Russian Foundation for Basic Research (grant Nos. 11-04-00360a, 13-04-00598, 13-04-00642).
REFERENCES
1.Pavlov, Y. I., Shcherbakova, P. V., and Rogozin, I.
B. (2006) Int. Rev. Cytol., 255, 41-132.
2.Pavlov, Y. I., and Shcherbakova, P. V. (2010)
Mutat. Res., 685, 45-53.
3.Kornberg, A., and Baker, T. (1991) in DNA
Replication, W. H. Freeman and Co., New York.
4.Rothwell, P. J., and Waksman, G. (2005) Adv.
Protein Chem., 71, 401-440.
5.Versieck, J. (1985) Crit. Rev. Clin. Lab.
Sci., 22, 97-184.
6.Gee, J. B. 2nd, Corbett, R. J., Perlman, J. M., and
Laptook, A. R. (2001) Pediatr. Neurol., 25, 304-308.
7.Sirover, M. A., and Loeb, L. A. (1976) Biochem.
Biophys. Res. Commun., 70, 812-817.
8.Zakour, R. A., Kunkel, T. A., and Loeb, L. A.
(1981) Environ. Health Perspect., 40, 197-205.
9.Keen, C. L., Ensunsa, J. L., Watson, M. H., Baly,
D. L., Donovan, S. M., Monaco, M. H., and Clegg, M. S. (1999)
Neurotoxicology, 20, 213-223.
10.Keen, C. L., Ensunsa, J. L., and Clegg, M. S.
(2000) Met. Ions Biol. Syst., 37, 89-121.
11.Pal, P. K., Samii, A., and Calne, D. B. (1999)
Neurotoxicology, 20, 227-238.
12.Lucchini, R. G., Martin, C. J., and Doney, B. C.
(2009) Neuromol. Med., 11, 311-321.
13.Valko, M., Morris, H., and Cronin, M. T. (2005)
Curr. Med. Chem., 12, 1161-1208.
14.Jiao, J., Qi, Y., Fu, J., and Zhou, Z. (2008)
Environ. Toxicol. Pharmacol., 26, 123-127.
15.Beckman, R. A., Mildvan, A. S., and Loeb, L. A.
(1985) Biochemistry, 24, 5810-5817.
16.El-Deiry, W. S., Downey, K. M., and So, A. G.
(1984) Proc. Natl. Acad. Sci. USA, 81, 7378-7382.
17.Zakour, R. A., Kunkel, T. A., and Loeb, L. A.
(1981) Environ. Health Perspect., 40, 197-205.
18.Kazakov, A. A., Grishina, E. E., Tarantul, V. Z.,
and Gening, L. V. (2010) Biochemistry (Moscow), 75,
905-911.
19.Lakhin, A. V., Efremova, A. S., Makarova, I. V.,
Grishina, E. E., Shram, S. I., Tarantul, V. Z., and Gening, L. V.
(2013) Mol. Genet. Mikrobiol. Virusol., 1, 14-20.
20.Makarova, A. V., Grabow, C., Gening, L. V.,
Tarantul, V. Z., Tahirov, T. H., Bessho, T., and Pavlov, Y. I. (2011)
PLoS one, 6, e16612.
21.Conlon, K. A., Miller, H., Rosenquist, T. A.,
Zharkov, D. O., and Berrios, M. (2005) DNA Repair, 4,
419-431.
22.Ohashi, E., Ogi, T., Kusumoto, R., Iwai, S.,
Masutani, C., Hanaoka, F., and Ohmori, H. (2000) Genes Dev.,
14, 1589-1594.
23.Garcia-Diaz, M., Bebenek, K., Kunkel, T. A., and
Blanco, L. (2001) J. Biol. Chem., 276, 34659-34663.
24.Ash, D. E., and Schramm, V. L. (1982) J. Biol.
Chem., 257, 9261-9264.
25.Goldschmidt, V., Didierjean, J., Ehresmann, B.,
Ehresmann, C., Isel, C., and Marquet, R. (2006) Nucleic Acids
Res., 34, 42-52.
26.Herling, A., Konig, M., Bulik, S., and
Holzhutter, H. G. (2011) FEBS J., 278, 2436-2459.
27.Munoz-Pinedo, C., El Mjiyad, N., and Ricci, J. E.
(2012) Cell Death Dis., 3, e248.
28.Frank, E. G., and Woodgate, R. (2007) J. Biol.
Chem., 282, 24689-24696.
29.Gulisano, M., Pacini, S., Punzi, T., Morucci, G.,
Quagliata, S., Delfino, G., Sarchielli, E., Marini, M., and Vannelli,
G. B. (2009) J. Neurosci. Res., 87, 228-237.
30.Hernandez, R. B., Farina, M., Esposito, B. P.,
Souza-Pinto, N. C., Barbosa, F., Jr., and Sunol, C. (2011) Toxicol.
Sci., 124, 414-423.
31.Grin, I. R., Konorovskii, P. G., Nevinskii, G.
A., and Zharkov, D. O. (2009) Biochemistry (Moscow), 74,
1253-1259.
32.Pechlaner, M., and Sigel, R. K. (2012) Met.
Ions Life Sci., 10, 1-42.
33.Muller, J., Sigel, R. K., and Lippert, B. (2000)
J. Inorg. Biochem., 79, 261-265.
34.Soler-Lopez, M., Malinina, L., Tereshko, V.,
Zarytova, V., and Subirana, J. A. (2002) J. Biol. Inorg. Chem.,
7, 533-538.
35.Labiuk, S. L., Delbaere, L. T., and Lee, J. S.
(2003) J. Biol. Inorg. Chem., 8, 715-720.
36.Duguid, J., Bloomfield, V. A., Benevides, J., and
Thomas, G. J., Jr. (1993) Biophys. J., 65, 1916-1928.
37.Eichhorn, G. L., and Shin, Y. A. (1968) J. Am.
Chem. Soc., 90, 7323-7328.
38.Bornhorst, J., Ebert, F., Hartwig, A., Michalke,
B., and Schwerdtle, T. (2010) J. Environ. Monit., 12,
2062-2069.
39.Sousa, F. G., Matuo, R., Soares, D. G.,
Escargueil, A. E., Henriques, J. A., Larsen, A. K., and Saffi, J.
(2012) Carcinogenesis, 33, 1433-1440.
40.Wang, Z., Wang, F., Tang, T., and Guo, C. (2012)
Front. Med., 6, 156-164.