REVIEW: Human Cardiac Troponin Complex. Structure and Functions
I. A. Katrukha
Department of Biochemistry, Biological Faculty, Lomonosov Moscow State University, 119991 Moscow, Russia; E-mail: katrukhai@gmail.com
Received July 15, 2013
Troponin complex is a component of skeletal and cardiac muscle thin filaments. It consists of three subunits – troponin I, T, and C, and it plays a crucial role in muscle activity, connecting changes in intracellular Ca2+ concentration with generation of contraction. In spite of more than 40 years of studies, many aspects of troponin functioning are still not completely understood, and several models describing the mechanism of muscle contraction exist. Being a key factor in the regulation of cardiac muscle contraction, troponin complex is utilized in medicine as a target for some cardiotonic drugs used in the treatment of heart failure. A number of mutations in troponin subunits are associated with development of different types of cardiomyopathy. Moreover, for the last 25 years cardiac isoforms of troponin I and T have been widely used for immunochemical diagnostics of pathologies associated with cardiomyocyte death (myocardial infarction, myocardial trauma, and others). This review summarizes the existing evidence on the structure and function of troponin complex subunits, their role in the regulation of cardiac muscle contraction, and their clinical applications.
KEY WORDS: troponin complex, muscle contraction, phosphorylation, troponin I, troponin T, troponin CDOI: 10.1134/S0006297913130063
Abbreviations: a.a., amino acid residue; cMyBP-C, cardiac myosin binding protein-C; c/ssTnC, cardiac/slow skeletal isoform of human troponin C; DCM, dilated cardiomyopathy; FRET, Forster resonance energy transfer; HCM, hypertrophic cardiomyopathy; hcTnI, human cardiac troponin I; hcTnT, human cardiac troponin T; ID, troponin I inhibitory domain; MD, troponin I mobile domain; NMR, nuclear magnetic resonance; PKA, protein kinase A; PKC, protein kinase C; PP1 and PP2, protein phosphatases 1 and 2; RCM, restrictive cardiomyopathy; RD, troponin I regulatory domain; SDSL-EPR, site-directed spin labeling electron paramagnetic resonance; TnC, troponin C; TnI, troponin I; TnT, troponin T.
Cardiac muscle contraction is a complex process in which cardiomyocyte
stimulation, occurring as a result of cell membrane depolarization and
subsequent increase in intracellular Ca2+ level, is coupled
with the generation of mechanical force. The key role in the regulation
of muscle contraction belongs to one of the components of thin
filament – troponin complex. This protein consists of three
subunits – troponin C (TnC), which binds Ca2+,
troponin I (TnI), which inhibits the ATPase activity of actomyosin
complex, and troponin T (TnT), which interacts with tropomyosin.
Complex changes in troponin structure that occur after binding of
Ca2+ enable the ATP-dependent interaction of myosin with
actin and development of muscle contraction. Developmental isoform
change, alternative splicing, and various posttranslational
modifications of troponin complex proteins allow the fine adjustment of
cardiac contraction.
Troponin complex also has important medical implications. This protein is a target for some cardiotonics used in therapy of heart failure. A number of mutations in troponin genes lead to the development of different types of cardiomyopathies. In addition, for the last 25 years isoforms of TnI and TnT specific for cardiac muscle tissue have been used for diagnostics of pathologies associated with cardiomyocyte necrosis (myocardial infarction, myocardial trauma, and others).
This review summarizes the existing data on structure and functioning of troponin complex components and their role in the regulation of cardiac muscle activity.
EXPRESSION OF TROPONIN COMPLEX SUBUNITS
Troponin I
In humans, troponin I is expressed in three isoforms: fast and slow skeletal isoforms and specific cardiac isoform [1-3]. Genes of all three TnI isoforms are arranged in tandems with TnT genes and are paralogs formed by the triplication of the ancestor TnI/TnT pair of genes [4, 5]. During embryonic development the fast and slow skeletal isoforms of TnI are expressed in all types of skeletal musculature, but in the adult state these isoforms are present only in the fast and slow skeletal muscles, respectively [6]. The slow skeletal TnI isoform is also expressed in cardiac muscle during embryonic development [7-9], and it is completely substituted by cardiac TnI shortly after birth. The cardiac isoform of TnI is expressed exclusively in heart [10-12]. The gene of cardiac TnI (TNNI3) is situated on the 19th chromosome (19q13.4) and consists of eight exons and seven introns [3]. Exon 3 is absent in genes of the skeletal isoforms, and it codes the main part of the unique N-terminal domain of cardiac TnI [3].
Developmental changes of TnI isoforms in heart. During ontogenesis of many animals and humans, changes in TnI isoforms in heart occur [8, 13-16]. In the embryonic stage of heart development the slow skeletal TnI isoform is mainly expressed. During the last weeks of human fetal development the rate of hcTnI expression increases [7, 9], and by the ninth month after birth skeletal TnI is completely substituted by the cardiac isoform [9]. The molecular mechanism that regulates this substitution is not fully understood. It seems that this process is controlled on both transcriptional and translational levels. In experiments with mouse myoblasts, it was shown that transcriptional factor Yin Yang 1 may participate in the inhibition of the slow skeletal TnI gene promoter [17]. In another series of experiments with embryonic rat myocardium, increased level of antisense RNA impeding the translation of hcTnI was observed. The level of this antisense RNA decreased with maturation of the organism [18].
Experiments on cardiac myocytes of mice [19, 20], rats [21, 22], and rabbits [23, 24] show that slow skeletal TnI incorporated in cardiac troponin complex provides higher affinity for Ca2+ and is less sensitive to acidosis than the cardiac TnI isoform. So expression of slow skeletal TnI in embryonic myocytes makes cardiac muscle more resistant to hypoxia and acidosis that may occur during fetal development and labor.
Troponin T
Troponin T is expressed in humans by three genes coding slow and fast skeletal and cardiac isoforms of the protein. The cardiac troponin T gene (TNNT2) is located on chromosome 1 (1q32.3) [25, 26], is 17-kb long, and consists of 17 exons and 16 introns [27]. It was shown that apart from heart tissue, cardiac TnT is transiently expressed in skeletal muscles during fetal development [28-31].
Alternative splicing and isoform changing of cTnT during ontogenesis. In a few studies dedicated to the analysis of expression of TnT isoforms during development of mouse [32, 33] and rat [34] cardiac muscle, a transient increase in mRNA coding the slow skeletal isoform of TnT was observed. An increased level of this mRNA was also detected in fetal human heart muscle and in cardiomyocytes of patients with end-stage heart failure [34]. However, it is worth mentioning that in these works no evidence of the translation of these mRNAs was presented. The absence of skeletal TnT isoforms in heart during different stages of development was confirmed by many works [30, 35-37], and so it is possible to say that the cardiac isoform of TnT is the only one that is expressed in cardiomyocytes.
In avian [38] and mammal [39-43] heart muscle, several forms of cardiac troponin T are expressed. These isoforms are produced as a result of alternative splicing of the cardiac troponin T gene. In humans, several forms of TnT are formed by alternative splicing of exons 4, 5, and 13 [25, 26, 30, 38, 42, 44, 45] (see further Fig. 2c). The amino acid sequence that is coded by exon 5 is expressed only in the embryonic form of cardiac TnT and is absent in the structure of TnT in adult heart [13, 30, 38, 43, 45]. Exons 4 and 13 can be alternatively spliced during any developmental stage [28, 42, 45, 46]. In mice and rats the expression of the embryonic form of cardiac troponin T ceases by, respectively, the 5th and 14th day of postnatal development [28]. The exact time of cessation of human embryonic cardiac TnT expression is not known, but by the ninth month of postnatal development embryonic TnT is completely substituted by the adult form in which the amino acid sequence coded by exon 5 is absent [26]. In the heart of healthy adults, troponin T is mainly present as isoform TnT3, which is 287-a.a. long and incorporates the amino acid sequences coded by exons 4 and 13 [30, 46].
The amino acid sequence coded by exon 5 (EEEDWREDED) is highly negatively charged. So alternative splicing of this exon leads, first, to the shortening of the protein and, second, to the substitution of a more acidic isoform by a less acidic one. These changes can affect the functioning of the troponin complex. It was shown that the expression of the adult isoform of troponin T enhances the maximal myofibril force and sensitivity of myocytes to Ca2+ [47-49]. Splicing of exon 4 may also be functionally important. Expression of TnT lacking the sequence coded by exon 4 was observed in hearts of patients with end-stage heart failure [30, 46].
The molecular mechanism that regulates the alternative splicing of TnT is not fully understood. So far only factors influencing the splicing of TnT exon 5 have been investigated. It was shown that binding of transcriptional factors CUG-BP and ETR-3 to the UG-rich part of the intronic sequence downstream of exon 5 leads to the introduction of this exon into the mRNA [50]. Expression of CUG-BP and ETR-3 is downregulated during development of the organism [51]. Two other proteins – PTB and MBNL – are antagonists of CUG-BP and ETR-3. Binding of PTB and MBNL to the intronic sequences both upstream and downstream from exon 5 prevents its incorporation into the mRNA [52].
Troponin C
Troponin C is expressed in humans by two genes: gene TNNC1 that is located on chromosome 3 (3p21.1) and codes the cardiac/slow skeletal isoform of TnC (c/ssTnC) [53] and gene TNNC2 that is located on chromosome 20 (20q12) and codes the fast skeletal isoform of troponin C [54, 55]. TNNC1 consists of six exons and five introns [56] and is expressed in cardiac and slow skeletal muscles [57]. In experiments with chickens, it was shown that TNNC1 is also transiently expressed during fetal development of fast skeletal musculature [58, 59]. Gene TNNC2 is expressed exclusively in fast skeletal muscles independently of the stage of development [57].
STRUCTURE OF HUMAN CARDIAC TROPONIN COMPLEX COMPONENTS
Troponin I
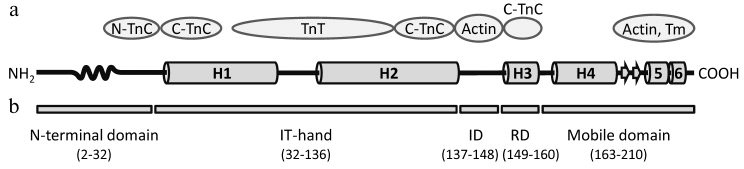
Translation of hcTnI mRNA leads to the development of a 210-a.a. protein. During synthesis, the first methionine is removed and the subsequent alanine residue is acetylated [60, 61]. Thus, the mature molecule of TnI is 209 a.a. long and has a molecular mass of 24 kDa. The cardiac isoform of troponin I consists of five domains: N-terminal domain, IT-arm, inhibitory domain, regulatory domain, and C-terminal mobile domain (Fig. 1b).
Fig. 1. Domain structure of human cardiac troponin I. a) Scheme of the hcTnI secondary structure. The structure of the N-terminal domain of TnI is represented according to [62], arrangement of α-helices H1-H4 in the TnI molecule according to [67], and structure of the mobile domain according to [86]. The wavy curve represents the Xaa-Pro region of TnI (residues 12-18) forming a proline helix. Short β-strands 1 and 2 are marked by arrows. Proteins of the thin filament that interact with the relevant regions of the TnI molecule are indicated in ovals [62, 63, 65, 67, 88]. N-TnC and C-TnC, N- and C-terminal domains of TnC, respectively; Tm, tropomyosin. b) Domain organization of hcTnI [67]. ID, inhibitory domain; RD, regulatory domain.
The N-terminal domain (residues 2-32)1 is present exclusively in the cardiac isoform of troponin I and plays a crucial role in the interaction of TnI with TnC and in the regulation of muscle contraction. This domain consists of an acidic part (residues 2-11), Xaa-Pro motif (residues 12-18), and a part that carries two serine residues (S23 and S24) that can be phosphorylated (residues 19-32) [60, 62]. By means of dot blotting [63] and NMR [62, 64-66], it was shown that residues 19-33 of the N-terminal domain of dephosphorylated TnI are able to interact with the N-terminal globule of TnC (Fig. 1a) [63, 65]. Phosphorylation of S23 and S24 leads to the formation of a new α-helix motif formed by residues 21-30 of TnI [62]. This structural change disrupts the interaction of TnC with the N-terminal domain of TnI and dislocates the latter. It is proposed that the acidic N-terminal domain of biphosphorylated TnI interacts with the positively charged inhibitory domain of TnI, and this interaction has an influence on the Ca2+-dependent regulation of muscle contraction [62].
1 Numbering of amino acid residues in TnI and TnT corresponds to the products of human cardiac TnI and TnT gene expression and includes the first methionines if not stated otherwise. The IT-arm (residues 42-136), which follows the N-terminal domain, is composed of two α-helices (H1 (residues 43-79) and H2 (residues 90-135)) connected with a short linker [67]. The IT-arm is the least flexible part of the TnI molecule, and it plays a structural role, providing contact with troponins C and T and orientating TnI in the troponin complex. An amphiphilic part of the H1 α-helix (residues 43-65) contacts the C-terminal domain of TnC [67, 68]. The C-terminal part of the H1 α-helix (residues 66-79) interacts with the H2 α-helix of TnT [67, 69]. Residues 80-89 of TnI make up a flexible linker between helices H1 and H2. The H2 α-helix of TnI forms a coiled-coil structure with the H2 α-helix of TnT (residues 226-271 of hcTnT) [67, 70, 71].
The inhibitory domain of hcTnI spans residues 137-148 [67, 72] (somewhat different borders of this region (residues 129-148) are indicated in papers of Lindhout and Sykes [73] and Brown et al. [74]). In the absence of Ca2+, residues 138-148 of the inhibitory domain interact with actin [72, 75] and shift the tropomyosin molecule so as to impede the actomyosin complex formation [76, 77]. A few variants of the inhibitory domain structure are presented in the literature. In the work of Brown et al. [74] on artificial troponin complex composed of different animal troponins, by means of site-directed spin labeling (SDSL-EPR) it was shown that the region limited by residues 138-145 does not possess a stable secondary structure. In the work by Takeda et al. [67], they were not able to crystallize the inhibitory region, which also suggests a high flexibility of this part of the molecule. The data of Lindhout and Sykes [73] indicate that in the presence of Ca2+, residues 132-135 of hcTnI interact with a hydrophobic region on the surface of the C-terminal domain of TnC, the following residues 135-140 form an α-helix that interacts with E- and H-helices of TnC, while residues 141-148 do not possess secondary structure [73]. But a few other studies argue that the inhibitory domain of TnI may have an arranged structure. Data on chicken troponin I structure obtained by FRET suggest that in the absence of Ca2+ the region limited by residues 138-149 (residues 137-148 of hcTnI) interacts with actin and accepts a compact conformation [75]. Increase in Ca2+ concentration leads to a conformational change in the inhibitory region of chicken TnI, which forms a quasi-α-helix that interacts with the linker region of TnC [75, 78]. Data obtained by H/D mass spectrometry also suggest that the inhibitory domain of TnI may obtain secondary structure [79].
The regulatory domain of hcTnI (residues 149-163) includes the short H3 α-helix (residues 150-159) [67]. At high Ca2+ concentrations, this domain interacts with a hydrophobic cleft on the surface of the TnC N-terminal domain, forming a third region of contact of hcTnI with TnC [80]. Interaction of the regulatory domain with TnC leads to the dissociation of the TnI inhibitory domain from actin and to the shift of tropomyosin that enables the formation of the actomyosin complex [69, 80, 81].
The mobile domain of hcTnI (residues 163-210) contains the H4 α-helix (residues 164-188) and the C-terminal part of the molecule (residues 190-210). The structure of the latter is not fully understood. In the work of Takeda et al. [67], in which the troponin complex was crystallized in a Ca2+-saturated state, they could not define the structure of residues 192-210 that, according to the authors, indicates high mobility of this region in the presence of Ca2+ [67]. Interestingly, the C-terminal part of skeletal troponin I has a high degree of homology with the same region of hcTnI, and in studies of skeletal troponin I a few variants of structure of this part of the molecule have been proposed. NMR studies of the troponin complex reconstructed in vitro from recombinant TnI, TnC, and a fragment of TnT [82], as well as studies of the chimeric proteins [83], suggest that the C-terminal part of TnI remains in the unordered state. However, the data of King et al. [84], obtained by means of small-angle neutron scattering, indicate that in the presence of Ca2+ this region of chicken skeletal TnI may adopt a compact conformation that is presumably formed by α-helices. At low Ca2+ concentrations, it changes to elongated coiled-coil. In the work of Murakami et al. [85] using NMR, it was shown that the C-terminal part of chicken skeletal troponin I adopts secondary structure formed by consecutive α-helix, two short anti-parallel β-strands, and two following α-helices. An analogical structure was proposed in a recent paper of Wang et al. [86] performed using FRET. Finally, in a work of Zhou et al. [87] performed on rat cardiac TnI, it was shown that in the presence of Ca2+ the C-terminal part of TnI adopts an arranged secondary structure, while at low Ca2+ concentration it stays in the unordered state.
In a study of chicken recombinant skeletal troponin [85], rat recombinant cardiac troponin I [87], or different recombinant fragments of human cardiac TnI (residues 131-210 [88] or 1-192 [89]), it was demonstrated that at low Ca2+ concentrations the mobile domain of TnI interacts with tropomyosin and the C-terminal part of actin. It is believed that these interactions play a crucial role in the regulation of Ca2+-dependent contraction and stabilization of the troponin complex on the surface of the thin filament [88, 89].
Troponin T
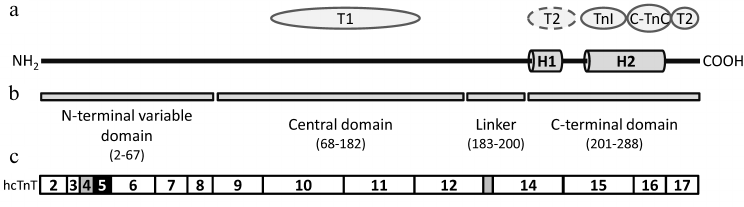
Troponin T plays the main role in the fixation of the troponin complex on the actin filament, organizes the subunits in the complex, and also participates in the regulation of muscle contraction [90, 91]. As stated earlier, in adult human heart muscle the 35.9 kDa isoform TnT3 is predominantly expressed. The N-terminal methionine is cleaved during the processing of the protein and the consecutive serine is acetylated, so the mature human cardiac troponin T molecule is 287-a.a.-long. So far only a small C-terminal portion of the molecule has been crystallized [67], and most of the information about the structure of the protein has been gathered by analysis of its fragments.
The hcTnT molecule is stretched along the thin filament [88, 92] and consists of the N-terminal variable domain (residues 2-68) and conservative central (residues 69-200) and C-terminal domains (residues 201-288) (Fig. 2b). The structure of the N-terminal domain varies among different species of TnT [93], and, like in hcTnI, has a unique sequence of ~32 a.a. that is not present in the skeletal isoforms of TnT [94]. This region is highly polar and negatively charged because by more than half it consists of aspartate or glutamate residues. Some of the early studies performed on the isolated N-terminal domain of TnT indicate that this region does not interact with other proteins of the thin filament [95, 96], although others presume that the first 40 amino acid residues of cardiac TnT may contact with other components of the troponin complex and tropomyosin [97]. According to studies performed on different chimeric proteins and deletion mutants of TnT, the N-terminal part of the protein influences the conformation of the complex [98-100] and the interaction of troponin complex with actin and tropomyosin [100, 101], and effects the Ca2+ sensitivity of the muscle [94, 102, 103] and the development of the maximum force of contraction [103, 104].
Fig. 2. Domain structure of human cardiac troponin T. a) Scheme of the hcTnT secondary structure. Arrangement of α-helices H1 and H2 in the hcTnT molecule is represented according to [67]. Proteins of the thin filament that interact with the relevant regions of the TnT molecule are indicated in ovals [67, 105, 113]. T1 and T2, regions of interaction with tropomyosin. b) Domain organization of hcTnT molecule. c) Arrangement of the exons coding the relevant parts of the hcTnT molecule [91, 93]. Gray boxes, exons 4 and 13 that are alternatively spliced; black box, exon 5 that is expressed only in the embryonic form of the protein.
The central domain of hcTnT embodies the first site (T1) of interaction with tropomyosin (residues 98-136 of hcTnT) [105]. T1 contacts tropomyosin in the place of binding of two tropomyosin dimers [92, 106], and the force of this contact does not depend on the concentration of Ca2+ [107]. The exact structure of T1 is not fully clarified, but it is supposed that this region is mainly α-helical [108, 109]. Residues 183-200, following T1, form a flexible linker that connects the central part of the molecule with its C-terminal domain [67, 109].
The C-terminal domain of hcTnT contains α-helices H1 (residues 204-220) and H2 (residues 226-271) [67]. The H2 α-helix participates in the interaction with TnI, forming coiled-coil with the H2 α-helix of the latter [67, 70]. With its C-terminus (residues 256-270), the H2 α-helix of hcTnT interacts with the Ca2+-binding loops of the TnC C-terminal domain [67]. The structure of the last 16 residues of hcTnT was not resolved in the crystal, which indicates the structural flexibility of this region [67]. Apart from contacting with TnI and TnC, the C-terminal domain of hcTnT contains a second site of interaction with tropomyosin (T2). T2 connects to the region around the C190 of the tropomyosin molecule [110, 111], and this interaction becomes stronger in the absence of Ca2+ [107]. Data on the exact localization of the T2 region is controversial. In the paper of Jin et al. [105] using immunoanalysis of mouse slow skeletal TnT fragments, it was shown that T2 spans residues 180-204 (homologous to residues 197-239 of hcTnT). But other works suggest that the T2 region is formed by the last 16 residues of TnT [111-113]. Interacting with tropomyosin and, probably, actin, this C-terminal part of the molecule stabilizes the troponin complex in the non-active state, because the deletion of the last 14 amino acid residues of hcTnT resulted in the enhancement of actin-activated ATPase activity in vitro [113].
Troponin C
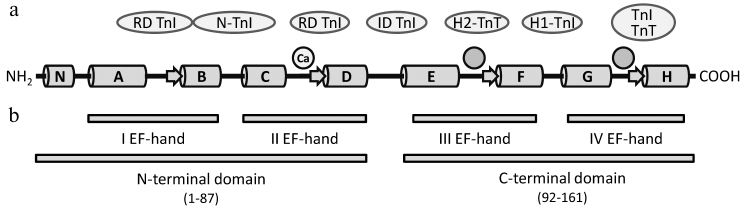
The isoform of TnC expressed in human heart has molecular mass of 18.4 kDa and consists of 161 a.a. [114]. As opposed to hcTnI and hcTnT, the N-terminal methionine of the mature c/ssTnC is not removed, and it is acetylated [114, 115]. TnC consists of a short N-terminal domain (residues 1-13) that is formed by the first α-helix (N-α-helix) and four Ca2+-binding EF-hands that are combined pairwise into the N-terminal (residues 14-87) and C-terminal (residues 92-161) globular domains (Fig. 3). Each EF-hand consists of two α-helices (α-helices A-H), between which is located the Ca2+-binding loop. Each loop incorporates a short β-strand that forms an antiparallel β-sheet with the β-strand of the neighboring Ca2+-binding loop [67, 116]. The N- and C-terminal globular domains of TnC are connected by a short linker situated between α-helices D and E (D–E linker) [116].
Fig. 3. Domain structure of human cardiac/slow skeletal troponin C. a) Scheme of c/ssTnC secondary structure according to [67, 116]. α-Helices N and A-H are represented by cylinders, β-strands by arrows. Two circles without designation mark the low-specificity regions that bind both Ca2+ and Mg2+. The circle designated with “Ca” represents the high-specificity region that binds Ca2+. Proteins of the thin filament that interact with the relevant regions of the TnT molecule are indicated in ovals [63, 65, 67, 80]. N-TnI, N-terminal domain of TnI; RD, regulatory domain; ID, inhibitory domain. b) Domain structure of c/ssTnC [67].
In the fast skeletal isoform of TnC, all four EF-hands are able to bind Ca2+ or Mg2+ [117, 118]. A few nonconservative amino acid substitutions in the first EF-hand of the c/ssTnC isoform impede the ion binding [116, 119], and c/ssTnC has only three Ca2+-binding sites: two C-terminal sites with high affinity for Ca2+ (Kd ~ 107 M–1) and one site with low affinity (Kd ~ 105 M–1) [120-122].
The high affinity sites of the c/ssTnC C-terminal domain (EF-hands III and IV) have relatively low specificity for Ca2+ and under physiological conditions are constantly filled with Mg2+ or Ca2+ [118] (Fig. 3a). The C-terminal domain provides the interaction of TnC with other proteins of the troponin complex and does not directly participate in the Ca2+-dependent regulation of muscle contraction [122, 123]. The N-terminal domain EF-hand II has a low affinity but high selectivity for Ca2+ and plays a crucial role in the regulation of contraction [122, 124].
ROLE OF TROPONIN COMPLEX IN REGULATION OF MUSCLE
CONTRACTION
Sarcomere Structure and the Mechanism of Muscle Contraction
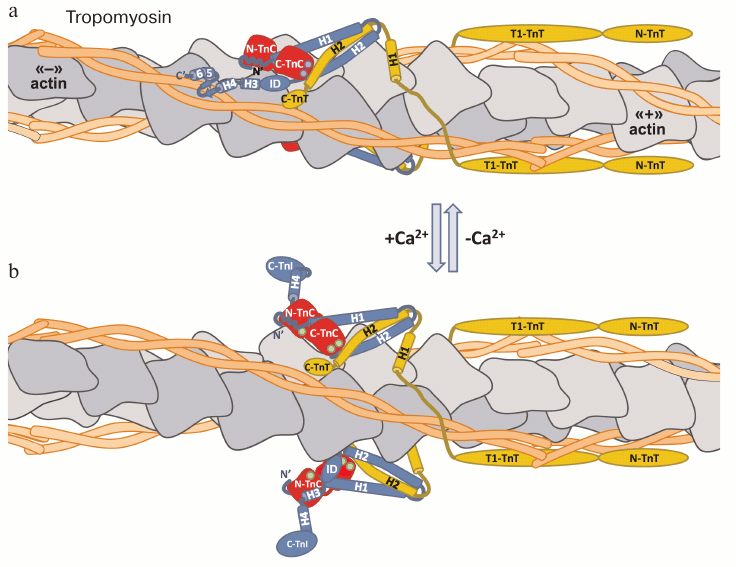
The functional unit providing contraction of the muscle is the sarcomere. It consists of thick filaments formed by myosin molecules, each surrounded by six thin filaments anchored by their “+”-ends in Z-disks. The thin filament is composed of molecules of fibrous actin (F-actin) that are connected with two fibers of tropomyosin and molecules of troponin complex. The F-actin fibril is formed by polymerization of molecules of globular actin, and it appears in electron microscopy as a two-stranded right-handed helix [125, 126] with a groove on each side (Fig. 4; see color insert). The tropomyosin strand is assembled by polymerization of the α-helical dimers that form a coiled-coil structure [127]. During polymerization, the ends of each tropomyosin dimer partially overlap with the neighboring dimers in a “head-to-tail” fashion, forming two continuous tropomyosin threads that lie in each actin groove [128]. Interactions of tropomyosin with actin are noncovalent and rather flexible [129], and each tropomyosin dimer contacts seven actin monomers [130].
Fig. 4. Troponin complex structure at low (a) and high (b) Ca2+ concentrations. TnI is blue, TnC red, and TnT yellow. Arrangement of the proteins and their interactions are shown according to the models, represented in [62, 67, 86, 87, 109]. At low Ca2+ concentrations, EF-hand II in the N-terminal domain of TnC (N-TnC) does not contain Ca2+, while EF-hands III and IV of the C-terminal domain TnC (C-TnC) bind Ca2+ or Mg2+ (violet circles) [118]. The inhibitory domain (ID) of TnI interacts with actin (gray). It is presumed that the mobile domain of TnI possesses secondary structure [85] and interacts with actin and tropomyosin (orange) [85, 87-89]. The C-terminal part of TnT (C-TnT) also interacts with actin and tropomyosin, additionally stabilizing the troponin complex on the thin filament [111-113]. Tropomyosin sterically blocks the site of interaction of actin with myosin. Increase in intracellular concentration of Ca2+ and binding of Ca2+ to the N-terminal domain of TnC (green circles) changes the conformation of TnC and leads to the dissociation of the inhibitory and mobile domains of TnI from actin. Detached from actin, the mobile domain of TnI loses its secondary structure and becomes disordered [86, 87, 109]. These structural changes enable the tropomyosin molecule to move into the groove of the F-actin helix, which opens the site of interaction of myosin with actin and leads to the development of muscle contraction.
The troponin complex is located on every seventh monomer of actin, so the stoichiometric ration of actin/tropomyosin/troponin in the thin filament is 7 : 1 : 1 [131] (Fig. 4). The troponin complex is connected to the thin filament mainly by the interactions of TnT with tropomyosin. The inhibitory and mobile domains of TnI also participate in the binding, interacting with actin at low Ca2+ concentrations [132, 133]. It is also assumed that the mobile domain of TnI can interact with tropomyosin [134, 135].
Cardiac muscle contraction is developed by the sliding of actin filaments along myosin. This process is energized by the cyclic ATP-dependent interaction of myosin and actin. In the first step of the cycle, myosin binds an ATP molecule and hydrolyses it to ADP and phosphate, but at this stage it is not able to get rid of the products of the reaction. The head of the myosin molecule (S1-myosin subfragment), containing the ADP and phosphate, is capable of weak interactions with actin. The latter stimulate the dissociation of phosphate from the S1-myosin active center. Escape of phosphate and following dissociation of ADP from the active center leads to conformational change in the “neck” of the myosin molecule and drawing of the actin filament along the myosin. Binding of a new ATP molecule leads to the dissociation of S1-myosin from actin. The subsequent hydrolysis of ATP enables a new “step” of myosin on actin [136, 137].
Role of Troponin Complex in Regulation of Cardiac Muscle Contraction
Muscle contraction is regulated by the changes of the Ca2+ concentration in the cytoplasm of the myocyte. But as opposed to smooth muscle, where the formation of actomyosin complex is mostly regulated by Ca2+-dependent phosphorylation of myosin light chain (MLC20) [138], Ca2+-dependent regulation of cardiac and skeletal muscle contraction is based on alteration in thin filament structure [139, 140].
At present, a few models of regulation of cardiac muscle contraction are discussed: a three-state model [140-142], a “fly-casting” model [82, 143], and a four-state model [87, 144].
According to the three-state model, thin filament proteins can accept three structural states: a blocked B-state, a closed C-state, and an open myosin-bound M-state [141]. At low (~100 nM [145]) cytoplasmic Ca2+ concentrations during diastole, the thin filament remains in the B-state. The inhibitory domain of TnI binds to F-actin and shifts the tropomyosin molecule towards the periphery of the actin groove, which leads to steric blocking of the myosin-binding site on the surface of the thin filament [146]. The displacement of tropomyosin is also supported by the binding of the T2 region of TnT to tropomyosin, which is enhanced in the absence of Ca2+ [107].
Spreading of cardiac action potential leads to a release of Ca2+ from the sarcoplasmic reticulum of the myocyte and an increase in free Ca2+ concentration in the cytoplasm (up to ~1 μM [145]). Binding of Ca2+ to the EF-hand II of c/ssTnC leads to changes in TnC conformation. The α-helices B and C move away from α-helices A and D [147-149], which leads to the formation a hydrophobic region on the surface of the N-terminal domain of TnC and subsequent binding of the regulatory domain of TnI to it [80, 150, 151]. As opposed to the fast skeletal isoform of TnC, Ca2+ binding to the EF hand II does not result in a complete change in the TnC conformation from “closed” (with hidden hydrophobic sites and almost antiparallel orientation of α-helices flanking the Ca2+-binding loop) to the completely “open” (with accessible hydrophobic regions and α-helices orientated at almost 90° angle to each other) [116, 150]. After the binding of Ca2+, the N-terminal domain of TnC turns to the “half-open” conformation [116, 152], and it is considered that the binding of the hcTnI regulatory domain is necessary for stabilization of this conformation [80, 153]. Interaction of the TnI regulatory domain with TnC leads to the dissociation of the TnI inhibitory domain from actin, a certain shift of the tropomyosin molecule to the center of the F-actin groove, and consecutive formation of weak contact between S1-myosin and actin (C-state). This contact does not fully activate the actomyosin complex, but it promotes the further movement of tropomyosin into the actin groove [154]. In the M-state, tropomyosin is completely displaced into the groove of F-actin, which opens the myosin binding site on the actin surface [155-157] and leads to the formation of strong actomyosin complex and the development of muscle contraction [140]. Decreasing intracellular Ca2+ concentration leads to the dissociation of Ca2+ from the N-terminal domain of TnC and change in conformation of the protein back to the “closed” state. This promotes the dissociation of the TnI regulatory domain from TnC and movement of the TnI inhibitory domain towards actin. Resumed interaction of the inhibitory domain with actin moves the tropomyosin molecule back to the edge of the groove, which blocks the myosin-binding site, prevents the formation of actomyosin complex, and leads to the relaxation of the muscle. The three-state model was further developed in the work of Robinson et al. [158], who by means of FRET experiments showed that in the absence of S1-myosin, only 50% of Ca2+-saturated actin–troponin–tropomyosin complex was in the activated state. These results demonstrate that increase in Ca2+ level alone is not sufficient for the complete activation of contraction. Addition of S1-myosin and formation of strong actomyosin complexes stabilized the active state of the thin filament. Robinson et al. proposed that myosin also influences the affinity of TnC to Ca2+, thus regulating the dissociation rate of the TnI regulatory domain from the N-terminal domain of TnC [158]. Similar results were obtained by the group of Houmeida [159], who using stop-flow spectroscopy of native porcine cardiac troponin complex showed that the addition of both Ca2+ and S1-myosin is essential for the full activation of the thin filament, while the addition of just one of these components leads to only ~70% activation. Studies of the structure and function of actomyosin complex components led to the development of new models of the regulation of muscle contraction.
Structural studies of skeletal troponin I resulted in the proposal of the “fly-casting” model of muscle contraction. According to this model, the critical regulatory role is assigned to the C-terminal mobile domain of skeletal TnI [143]. It is presumed that in the absence of Ca2+ this part of the molecule adopts a stable secondary structure composed of the H4 α-helix that is followed by two short antiparallel β-strands arranged in a β-sheet and two consecutive short α-helices. In this conformation, the mobile domain binds to actin, stabilizing the non-active state of the thin filament. The regulatory domain of TnI does not have any stabilized conformation, and it fluctuates freely between actin and the N-terminal domain of TnC. Increasing Ca2+ concentration provokes the adoption of α-helical structure by the regulatory domain of TnI and its interaction with TnC [143]. Next, the TnI mobile domain dissociates from actin, which is accompanied by the turning of the mobile domain into the flexible unarranged state. These conformational changes stimulate the dissociation of the TnI inhibitory domain from actin and its association with the D–E linker of TnC. Decreasing of the Ca2+ concentration leads to the association of the mobile domain with actin, which promotes the dissociation of, first, the TnI regulatory and, then, the inhibitory domains from TnC, interaction of the inhibitory domain with actin, and transition of the muscle into the relaxed state [143].
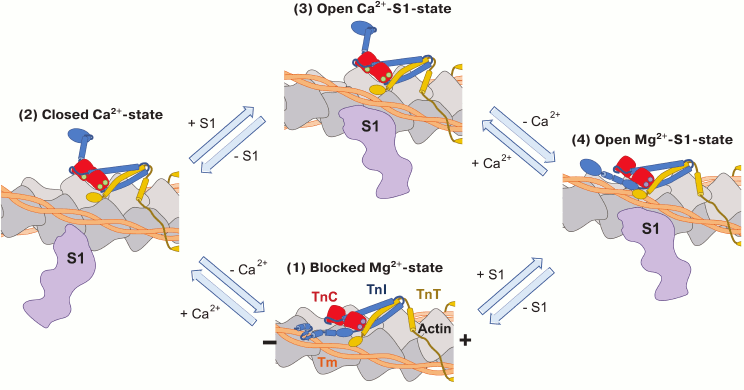
The four-state model of muscle contraction was first proposed by Lehrer [144] and developed in the recent work of Zhou et al. [87]. This model, on the whole, unites and supplements the theories that were proposed before. According to this model, the thin filament can remain in four structural states: blocked Mg2+-state, closed Ca2+-state and, open Mg2+-S1- and Ca2+-S1-states [87, 144] (Fig. 5; see color insert). In the blocked Mg2+-state, which corresponds to the B-state of the three-state model, the interaction of the TnI inhibitory domain with actin shifts the tropomyosin and impedes the formation of cross-bridges. The mobile domain of TnI additionally stabilizes the troponin complex on the surface of the thin filament, contacting with actin and, presumably, tropomyosin. Increase in Ca2+ concentration induces relatively fast dissociation of the mobile domain from the thin filament and its shift to the disordered state. The subsequent interaction of the TnI regulatory domain with the N-terminal part of TnC provokes the dissociation of the TnI inhibitory domain from actin (closed Ca2+-state that is similar to the C-state of the three-state model). The tropomyosin molecule shifts into the actin groove, opening the site of contact of S1-myosin with actin, which leads to the formation of strong cross-bridges (open Ca2+-S1-state corresponding to the M-state of the three-state model). In the Mg2+-S1 state, which presents an alternative way of thin filament activation, S1-myosin interacts with actin in the absence of Ca2+. It is assumed that this interaction shifts the tropomyosin molecule and leads to the dissociation of the TnI mobile domain from actin and association of the regulatory domain with TnC, increasing the sensitivity of TnC to Ca2+ [144]. The transition to the Mg2+-S1-state is not sufficient for the development of muscle contraction, which takes place only after binding of Ca2+ to the TnC N-terminal domain, but this interaction accelerates the shift of the thin filament into the active state and facilitates the formation of strong cross-bridges [87].
Fig. 5. Schematic representation of the four-state model of muscle contraction. TnI is blue, TnC red, and TnT yellow. In the blocked Mg2+-state (1), the inhibitory and mobile domains of TnI interact with actin (gray) and tropomyosin (orange) and inhibit the formation of actomyosin complex. The N-terminal EF-hand of TnC is not saturated with Ca2+, while both the C-terminal EF-hands bind Ca2+ or Mg2+ (violet circles). Depolarization of the cardiomyocyte membrane leads to increase in intracellular concentration of Ca2+ and binding of Ca2+ (green circles) to the EF-hand in the N-terminal domain of TnC. This interaction leads to conformational change in TnC, dissociation of TnI from actin, and subsequent association of actin with TnC. The tropomyosin molecule moves towards the actin groove, which enables the formation of weak interactions between S1-myosin (violet) and actin (closed Ca2+-state (2)). Interactions of myosin with actin provoke the further movement of tropomyosin into the actin groove and formation of the strong actomyosin complex (open Ca2+-S1-state (3)). Formation of the open Mg2+-S1-state (4) represents an alternative way of myofilament activation. This state is stimulated by the strong binding of S1-myosin to actin in the absence of Ca2+. It is presumed that the formation of the Mg2+-S1-state can stimulate the binding of Ca2+ to the N-terminal domain of TnC and, thus, the formation of the open Ca2+-S1-state [87, 160].
According to the four-state model, the relaxation of muscle occurs in two steps distinguished by their rate. During the first (fast) step, Ca2+ dissociates from the N-terminal domain of TnC, which leads to the interaction of the TnI mobile domain with actin and dissociation of the regulatory domain from TnC. These structural alterations stimulate the slower removal of the inhibitory domain from TnC and association of the inhibitory domain with actin (second step). The presence of S1-myosin impedes the interaction of the mobile domain with actin, thus stabilizing the open conformation of TnC and hindering muscle relaxation. As a result, the rate of TnI inhibitory domain switching becomes comparable with the speed of cross-bridge formation, and it is presumed that this step can play a significant role in the regulation of the rate of muscle contraction [87, 160].
Interactions of S1-myosin with actin in the absence of Ca2+ and the resulting formation of the open state of actin filament may also explain such features of cardiac muscle contraction as length-dependent activation of cardiomyocytes [161], slow rate of myocyte relaxation after the decrease in intracellular Ca2+ concentration, and increased Ca2+-sensitivity of the thin filament after the formation of cross-bridges [144].
According to the existing models, troponin complex plays an essential role in the regulation of cardiac muscle contraction. Conformational changes in troponin complex subunits provide the development of muscle contraction during the increase in cytoplasmic Ca2+ concentration and muscle relaxation after its decrease. Moreover, structural features of the troponin complex enable the fine regulation of these processes, giving cardiac muscle an opportunity to adapt to various physiological and pathological conditions. Some of these regulatory mechanisms and their role in the adaptation of cardiac muscle to different conditions are discussed in the next chapters.
Troponin Complex and Length-Dependent Regulation of Muscle Contraction. Frank–Starling Law
According to the Frank–Starling law, the force of muscle contraction increases proportionally to the degree of muscle stretching. This relation is particularly pronounced in cardiac muscle, where the strength of myocardial contraction increases after the greater filling of ventricles with blood [162]. This mechanism synchronizes the cardiac output volume with the volume of blood coming into the heart from veins. The molecular mechanisms of this action are not yet fully understood. It is presumed that the increase in cardiac output can be provided by such features as the increase in cardiac myocyte Ca2+-sensitivity, reduction of the distance between thin and thick filaments during stretching, and increase in cardiac muscle cooperativity of contraction [163, 164].
The data of a few investigations show that the troponin complex may participate in the regulation of length-dependence of cardiac muscle contraction [165, 166]. In experiments with transgenic mice, it was shown that the substitution of cardiac TnI with the skeletal isoform led to a decrease in cardiac muscle length-dependence and increase in its Ca2+-sensitivity [165, 167]. On the contrary, the substitution of skeletal TnI with the cardiac isoform in skinned rabbit skeletal muscles increased its length-dependence and decreased the affinity of TnC to Ca2+ [166]. It was also demonstrated that phosphorylation of TnI S23 and 24 by protein kinase A (PKA) has a pronounced influence on the Frank–Starling relation in myocytes, leading to decrease in muscle Ca2+-sensitivity [167, 168].
The reasons by which cardiac isoform of TnI has a bigger influence on the length-dependence than skeletal isoforms are not fully understood. In a paper of Tachampa et al. [169], it was presumed that residue T144 can play a key role in the regulation of this process. This residue is located in the inhibitory domain of cardiac TnI, and in skeletal isoforms it is substituted by proline. Mutation of this residue in skeletal TnI to threonine led to increase in length-dependence of skeletal muscle almost to the level characteristic of the cardiac muscle [169].
Influence of Acidosis on Cardiac Muscle Contraction
Heart muscle is very sensitive to decrease in intracellular pH (acidosis). Decline in pH beyond the physiological values decreases the efficiency of cardiac contraction, violates the Ca2+-dependent regulation, and can lead to arrhythmia [170-172]. Physiological fluctuations of pH appear during change in cardiac loading and increased cardiac rate [173]. Dangerous decline in intracellular pH (down to values 6.5 and lower [174, 175]) stipulated by accumulation of metabolic products including lactate [176] is observed during prolonged ischemia.
The troponin complex plays a significant role in the sensitivity of cardiac muscle to the decline in intracellular pH [21, 177, 178]. It was noted that embryonic cardiomyocytes expressing the slow skeletal isoform of TnI are less susceptible to the influence of acidosis than adult myocytes [14, 179]. Substitution of cardiac TnI with the slow skeletal isoform in primary culture of rat cardiomyocytes led to decrease in the threshold for Ca2+-activated tension, molecular cooperativity and reduced the acidic pH-induced (pH 6.2) desensitization of Ca2+-activated tension [21]. Similar results were obtained after the substitution of cardiac TnI with the slow skeletal isoform in hearts of transgenic mice [19].
In experiments on mice, it was shown that the difference in the sensitivity of TnI isoforms to decrease in pH can be caused by the substitution of one amino acid residue located in the regulatory domain of cardiac TnI isoform – A164 (homologous to A163 in hcTnI). In skeletal troponins this residue is a histidine. Perfusion of isolated hearts of wild-type mice with low pH buffer (pH 6.8) led to the fast violation of left ventricle functioning and decrease in the developed pressure. However, hearts of mutant mice carrying the cardiac troponin in which H164 was substituted with alanine were much more resistant to the decline in pH [180]. This mutation decreased the negative effect of ischemia–reperfusion and the duration of arrhythmia [180]. In confirmation of this effect, experiments in vitro with hcTnI showed that the A163H mutation leads to the recovery of Ca2+-sensitivity of mutated troponin I compared with the wild-type protein [178]. A recent investigation conducted on the hcTnI inhibitory domain peptide with introduced A163H mutation showed that H163 interacts with residues E15 and E19 of c/ssTnC [181]. The authors suggest that an increase in the positive charge of the histidine residue at low pH values leads to the enhancement of interaction of the TnI inhibitory domain with TnC. This contact stabilizes the open conformation of TnC and increases the Ca2+-sensitivity of the complex [181].
Troponin T can also influence the functioning of cardiac muscle during acidosis, but, as opposed to TnI, cardiac TnT is more resistant to the decline in intracellular pH than the skeletal isoforms of the protein. Substitution of cardiac TnT with the fast skeletal isoform led to increase in the influence of low pH values (pH 6.5) on the muscle contraction in vitro. Hearts of transgenic mice carrying skeletal TnT showed more pronounced decrease in maximal tension and pCa50 than hearts of wild-type animals [182].
POSTTRANSLATIONAL MODIFICATIONS OF TROPONIN COMPLEX SUBUNITS AND
REGULATION OF MUSCLE CONTRACTION
One of the most important mechanisms of regulation of protein functioning is phosphorylation. Many studies have shown that TnT and TnI can be phosphorylated by various protein kinases, and these posttranslational modifications have a considerable effect on the structure and properties of troponin complex. Disruption of troponin phosphorylation mechanisms is associated with different pathologies of cardiac muscle, which also confirms the important role of these modifications in troponin complex functioning.
Phosphorylation of Troponin I
By means of computer analysis in silico [183] and in experiments in vitro and in vivo [183-194], it was shown that at least 17 amino acid residues of cardiac TnI can be phosphorylated by different protein kinases (Fig. 6; see color insert). The functional significance of some of those modifications is not yet well studied in vivo, but the importance of other phosphorylation sites is well documented, and there is no doubt that this type of posttranslational modifications plays and important role in the regulation of cardiac muscle contraction.
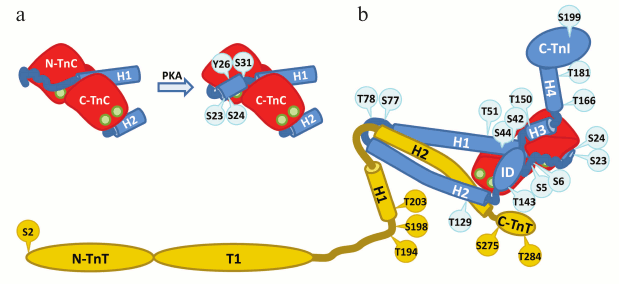
Fig. 6. Troponin I and T phosphorylation sites. TnI is blue, TnC red, and TnT yellow. Phosphorylation sites of TnI are shown in blue circles, TnT sites in yellow circles. Ca2+ ions are shown as green circles. Troponin complex is present in an open Ca2+-bound state. a) Schematic representation of TnI N-terminal domain conformational change after the phosphorylation of residues S23 and S24 according [62]. In the dephosphorylated state, TnI residues 1-31 interact with the N-terminal domain of TnC. It is supposed that phosphorylation of residues S23 and S24 leads to the formation of α-helical region by residues 21-30 and shifting of the acidic N-terminal part of TnI (residues 2-11) towards the inhibitory domain (ID) of TnI (Fig. 6b). In addition, two putative phosphorylation sites (Y26 and S31) that are hidden behind the TnC N-terminal domain on Fig. 6b are shown. b) Schematic representation of the described TnI and TnT phosphorylation sites [183-194, 242-247].
Protein kinase A. During increased load (for example, during intensive physical exercises) an organism needs more intensive supply of certain muscles and organs with oxygen-rich blood. This demand is ensured by augmentation of heart output and by increase in heart rate. These processes are controlled by the sympathetic nervous system via cardiomyocyte β-adrenergic receptors [195]. β-Adrenergic stimulation results in an increase in cAMP and subsequent activation of PKA. This enzyme phosphorylates many proteins participating in muscle contraction including a number of myofilament proteins – cardiac myosin binding protein C (cMyBP-C) [196], titin [197, 198], TnI [199-201], and proteins participating in the binding and transportation of Ca2+ (phospholamban [202], dihydropyridine, and ryanodine receptors [203, 204]). PKA-dependent phosphorylation of myocyte proteins increases cardiac output. This can be obtained both by increase in strength of contraction of heart muscle (inotropic effect) and by increase in is relaxation rate (lusitropic effect), which is essential for the proper filling of the chambers with blood during diastole. The positive inotropic effect of PKA is achieved mainly by phosphorylation of Ca2+-channels (dihydropyridine and ryanodine receptors), which leads to increased flow of Ca2+ from the endoplasmic reticulum and outside of the cell into the cytoplasm [203]. The lusitropic effect of PKA is achieved by the phosphorylation of cMyBP-C, phospholamban, and TnI, which increases the rate of cardiac relaxation. This in turn can be realized through decrease in Ca2+-sensitivity of the muscle and by augmentation of the cross-bridge formation rate. It is supposed that phosphorylation of TnI by PKA can affect both ways of cardiac muscle relaxation [205-208].
Phosphorylation of hcTnI by PKA is of particular interest because this enzyme phosphorylates amino acid residues S23 and S24 [60, 184, 185, 209] situated in the N-terminal domain of hcTnI, which is lacking in skeletal isoforms of the protein. Thus, phosphorylation by PKA is a unique regulatory mechanism expressed only in cardiac muscle. According to mass-spectrometric study of troponin that was affinity purified from cardiac muscle of humans and different animals, under normal conditions almost 50% of TnI molecules are in the dephosphorylated state, while the rest is present in monophosphorylated or biphosphorylated (on S23 and S24) molecules. In the work of Sancho Solis et al., it was shown that 41 ± 3% of troponin I purified from rat hearts was dephosphorylated, 46 ± 1% was monophosphorylated, and 13 ± 3% was biphosphorylated [210]. Similar results were obtained in studies of human cardiac troponin I by Van der Velden et al. (48.9 ± 10.0% dephosphorylated, 21.6 ± 9.8% monophosphorylated, and 29.5 ± 11.3% biphosphorylated) [184] and Zhang et al. (50.5 ± 5.9% dephosphorylated, 38.6 ± 3.8% monophosphorylated, and 10.9 ± 2.2% biphosphorylated) [185]. In studies [185, 210] it was also noted that the monophosphorylated form of cardiac TnI was present only with phosphorylation on residue S23.
In the work of Howarth et al. [62] using NMR, it was shown that biphosphorylation on S23 and S24 of the peptide containing residues 1-32 of cardiac TnI results in significant conformational changes in the peptide. It is presumed that in the absence of phosphorylation the N-terminal part of hcTnI remains in the unstructured state and contacts the N-terminal domain of TnC. Phosphorylation on S23 and S24 leads to the formation of α-helical structure by residues 21-30, dissociation of the N-terminal domain of TnI from TnC, and shift of the N-terminal domain towards the inhibitory domain of TnI (Fig. 6a). The authors suggested that it is the interactions of the N-terminal TnI domain with the inhibitory domain of TnI that mediate the effects of cardiac TnI phosphorylation by PKA [62].
The role of S23 and S24 phosphorylation by PKA in the decrease in cardiac muscle sensitivity to Ca2+ and in increase in the relaxation rate of cardiac myocytes has been confirmed by many studies both in vitro and in vivo. In in vitro experiments on fragments and whole molecules of mouse cardiac TnI and TnC, phosphorylation of S23 and S24 led to decrease in TnI–TnC complex sensitivity to Ca2+ and decrease in TnI affinity to TnC [211, 212]. The substitution of the cardiac TnI by the slow skeletal isoform led to decrease in the cardiac muscle relaxation rate in vivo. Myocytes of mutant mice also demonstrated higher Ca2+-sensitivity compared to wild-type myocytes [20, 213]. Similar effects were demonstrated on mutant mice carrying TnI in which residues S23 and S24 were substituted by unphosphorylatable alanines [214, 215]. And, vice versa, substitution of S23 and S24 by aspartates, which imitates the phosphorylation, led to enhanced relaxation of the left ventricular myocytes of the mutant mice in vivo [216]. Similar results were obtained in vitro on preparations of skinned human cardiomyocytes in which the native troponin complex was substituted by reconstructed complex including the mutant on both serine residues of hcTnI [208]. In these experiments, it was shown that the decrease in Ca2+-sensitivity was observed only when both serine residues were mutated, and it was not significant when mutants mutated in one of the residues were used [208, 217].
However, the influence of cardiac TnI phosphorylation on the rate of actomyosin complex formation and the force of cardiomyocyte contraction is controversial. Studies conducted on skinned mouse or porcine myocytes showed that the PKA-dependent phosphorylation of TnI increased the rate of actomyosin complex formation as well as the rate and force of myocyte contraction [206, 207]. Similar results were obtained during the study of skinned ventricular rat myocytes [201]. But in another investigation conducted on skinned rat trabecules and myocytes, no changes in PKA-dependent Ca2+-activated isometric actomyosin formation rate was observed [217]. Also, no significant changes in the rate of actomyosin complex formation and in the force of myocyte contraction were found in transgenic mice carrying the S23A and S24A mutated cardiac TnI [215, 216]. In works of Stelzer et al. [218] and Chen et al. [219], it was shown that PKA-dependent increase in actomyosin complex formation rate occurs mainly because of the phosphorylation of cMyBP-C, but not TnI.
PKA-dependent phosphorylation of TnI can play a significant role in the development of cardiac pathologies. It is known that hearts of patients with end-stage cardiomyopathies have higher sensitivity to Ca2+ than normal hearts [184]. At the same time, the rate of phosphorylation of troponin I S23 and S24 was decreased in hearts of patients with heart failure and cardiomyopathy [183, 185, 220]. But at present it is not known whether these changes in TnI phosphorylation induce or appear as the result of these pathologies.
Protein kinase C. The term “protein kinase C” (PKC) describes a vast family of serine/threonine protein kinases. This family consists of three subgroups: so-called conventional kinases (isoforms PKCα, β1, β2, and γ), novel kinases (isoforms PKCδ, ε, η, and θ), and atypical kinases (PKCζ and ιλ), which differ in the structure of their intracellular membrane localization domains [221]. In normal human adult cardiomyocytes, five isoforms of PKC (PKCα, β1, β2, δ, and ε) are expressed [221, 222]. It was shown that these isoforms interact with many proteins that regulate cardiac contraction, including cardiac TnI [223], TnT [224], cMyBP-C [225], and titin [226].
Different in vitro studies have shown that TnI is phosphorylated by PKC mainly on residues S23, S24 [186-188], S42, S44, and T143 [189] (numbered according to the hcTnI sequence). However, PKC isoforms differ in their specificity. Experiments utilizing bovine cardiac TnI showed that PKCα phosphorylated mainly amino acid residues S42 and S44, while PKCδ phosphorylated S23 and S24 [186]. Phosphorylation of mouse TnI by PKCβ and PKCε in vitro showed that these protein kinases modify residues S23, S24, and T144 (T143 in the hcTnI sequence), but not S42 and S44 [188]. But in the another in vitro study it was observed that PKCβ phosphorylates mainly residue S44 [227]. In vitro experiments with an “atypical” PKCζ that was expressed in rat cardiomyocytes [228] showed that this enzyme modifies exclusively the T143 residue of rat TnI [229].
Data of the influence of TnI phosphorylation by PKC on the functioning of troponin complex are controversial. Generally, in vitro studies show that the physiological effect of PKC phosphorylation is opposite to that of PKA [230]. Substitution of S23 and S24 (residues modified by PKA) in murine TnI with aspartates led to increase in the relaxation rate and muscle contraction during overload. At the same time, simultaneous mutation S23D, S24D and S43D, S45D, T144D (residues phosphorylated by PKC, homologous, respectively, to S42, S44, and T143 of hcTnI) leveled this effect [231]. Myocytes of transgenic mice carrying troponin I in which S43 and S45 were substituted by alanines were more prone to ischemia-induced contracture [232] than the wild-type cells. Transgenic mice expressing S43E, S45E, and T144E TnI mutant in their hearts showed decrease in Ca2+-sensitivity, maximum tension, and maximum sliding speed [233]. On the contrary, substitution of native TnI by the same mutants in skinned murine myocytes in vitro led to decrease in both muscle contraction rate and Ca2+-sensitivity [234]. Similar results (decrease in maximal Ca2+-activated force of muscle contraction, ATPase activity, and Ca2+-sensitivity) were observed in vitro in reconstructed thin filament containing mutated hcTnI [235].
The functioning of PKC can be important in the development of different cardiac pathologies. Expression of PKCα and PKCδ was increased in rats with spontaneous hypertension, while the level of PKA and PKCβ remained unchanged. Animals of this model exhibited higher level of TnI phosphorylation on residues S23, S24, S42, and S44 (rat TnI sequence) [236]. In another study [225], it was observed that PKCα- and PKCε-dependent phosphorylation of TnI in cardiomyocytes of patients with dilated cardiomyopathy led to decrease in Ca2+-sensitivity with no effect on the maximal developed tension. It is presumed that PKC-dependent decrease in contraction activity can have a compensatory effect, preventing energy depletion of cardiomyocytes under conditions of Ca2+-overload [237].
Other protein kinases. In in vitro experiments, it was demonstrated that the cardiac isoform of TnI can be phosphorylated by protein kinase D [238, 239] and cGMP-dependent protein kinase (PKG) [240] on residues S23 and S24, by p21-activated kinase 3 (PAK3) (on residue S150) [190, 191], by protein kinase Mst-1 (predominately on residue T31, but also on residues T51, T129, and T143) [192], and by AMP-dependent protein kinase (AMPK) (on residue S150) [193, 194] (Fig. 6b).
Phosphorylation of troponin I was also studied in vivo by mass-spectrometry of affinity-purified troponin from heart muscle. In some of these studies eight more putative phosphorylation sites (S4, S5, Y25, S76, T77, S166, T180, and S198 (numeration without the first methionine)) were determined [61, 183]. In contrast, in other studies of troponin purified from mouse [241] and rat [210] myocardium, it was shown that in vivo TnI is phosphorylated only on residues S23 and S24. The same results (phosphorylation on only two residues) were observed in both healthy [184, 185] and diseased [185] human hearts.
Phosphorylation by protein kinase A has an important physiological effect, decreasing the sensitivity of troponin complex to Ca2+ and increasing the rate of cardiac muscle relaxation. Data concerning the physiological effects of TnI phosphorylation by other protein kinases has been obtained mostly from in vitro experiments or with the use of different animal models. Results described in studies of phosphorylation of human TnI by PKC and other protein kinases are controversial, suggesting that more research on phosphorylation of hcTnI in vivo is required.
Posttranslational Modification of Troponin T
Phosphorylation of hcTnT. Many in vitro experiments have demonstrated that, like hcTnI, hcTnT can also be phosphorylated by different protein kinases. In vitro experiments have shown that hcTnT can be phosphorylated by troponin T kinase on residue S2 [242-244], by apoptosis signal-regulating kinase 1 (ASK1) on residues T194 and S198 [245], by Rho-A-associated protein kinase-II (ROCK-II) on residues S275 and T284 [246], and by Raf-1 kinase on residue T203 [247] (Fig. 6b). Experiments conducted on bovine heart showed that unlike TnI, TnT is not phosphorylated by PKA [248, 249]. The best-studied protein kinases that can phosphorylate hcTnT are enzymes of the PKC family [248-250]. In vitro studies conducted on rat cardiomyocytes have shown that the main isoforms that can phosphorylate hcTnT are PKC α, β, and ζ [229]. Using the bovine cardiac TnT isoform, it was shown that PKC phosphorylates residues T190, S194, T199, and T280 (homologous, respectively, to T194, S198, T203, and T284 of hcTnT) [189, 251, 252]. Phosphorylation by PKC inhibited the Ca2+-activation of thin filament and decreased the maximum force generated by cardiomyocytes [251, 253, 254].
According to the in vitro data, hcTnT has many putative phosphorylation sites. Very unexpectedly, recent mass-spectroscopic analyses of affinity-purified TnT from human [255], rat [210], or mouse [255] hearts have shown that in vivo the protein is phosphorylated only on one residue, namely S2. These data confirm the first results of bovine TnT phosphorylation [256] in which S2 was also the only phosphorylated residue. This amino acid is phosphorylated by troponin T kinase [242], and the degree of its phosphorylation is very high [210, 255, 256]. The physiological role of this modification is not fully understood, for it was shown that phosphorylation on S2 does not affect the Ca2+-sensitivity and the formation of the actomyosin complex [41].
Dephosphorylation of TnI and TnT. In spite of the fact that dephosphorylation of proteins is of the same importance for the regulation of cardiac muscle contraction as their phosphorylation [257-259], the role of phosphatases in the modulation of troponin complex activity has been studied rather superficially. TnI and TnT are dephosphorylated by protein phosphatases 1 and 2A (PP1 and PP2A) [260-263]. These widespread phosphatases [264] participate in many intracellular processes including cell division, protein synthesis, carbohydrate metabolism, and muscle contraction [265, 266]. One can hope that future studies will explain the role of phosphatases in the regulation of troponin complex activity.
Restricted proteolysis of TnT. One of the recently discovered mechanisms of cardiac muscle adaptation is the restricted proteolysis of TnT. Proteolysis of the N-terminal part of cardiac TnT (residues 1-71) was detected in mouse and rat hearts that were subjected to experimental ischemia–reperfusion and ex vivo left ventricular pressure overload [267, 268]. A proteolytic fragment of TnT was also detected in model experiments of Ca2+-overload [269]. It was shown that cleavage of residues 1-71 is performed by the protease calpain-1 (μ-calpain) [267].
Proteolysis of the N-terminal part of cardiac TnT changes the conformation of the central and C-terminal parts of the protein and reduces the affinity of TnT to TnI, TnC, and tropomyosin [100, 267]. In ex vivo experiments on the hearts of transgenic mice that expressed the cleaved part of TnT (residues 72-291 of mouse TnT), it was shown that this substitution is not lethal and does not result either in hypertrophy of the heart muscle or in decrease in contractile activity [100]. At the same time, hearts expressing the mutated protein were more resistant to overloading under pressure. The authors suggest that proteolysis of the N-terminal part of TnT might be a mechanism of cardiac muscle adaptation to ischemic stress [268].
TROPONIN COMPLEX AND HEART DISEASES
Troponin complex and therapy of heart failure. Being one of the key regulators of cardiac contraction, troponin complex plays an important part in therapy of heart failure – a pathological state in which the heart is incapable of pumping the required volume of blood. One of the reasons for the onset of this pathology is the decrease in cardiac muscle sensitivity to Ca2+, which leads to reduction in the force of contraction and systolic heart function. As stated above, the affinity of TnC to Ca2+ and interactions between TnI and TnC play a key role in the Ca2+-sensitivity of the thin filament. Currently, a number of substances that interact with troponin complex subunits and enhance the sensitivity of thin filament to calcium are known. Among these are bepridil, levosimendan, EMD 57033, and some others [270, 271]. Studies have demonstrated that these substances enhance Ca2+-sensitivity in different ways. Using NMR, it was shown that the therapeutic effect of bepridil and levosimendan is realized through the interaction with the hydrophobic region on the surface of the N-terminal domain of troponin C [272-274]. This interaction stabilizes an open conformation of TnC similar to the effect of the inhibitory domain of TnI [274]. The presence of the drug leads to decrease in the rate of dissociation of Ca2+ from TnC and increase in thin filament Ca2+-sensitivity. EMD 57033 interacts with the C-terminal domain of TnC [275, 276], disrupting its contact with residues 34-71 of hcTnI [277]. It is supposed that this action increases the interaction between the TnI inhibitory domain and TnC [270].
One of the advantages of agents that modulate the Ca2+-sensitivity of troponin complex is the absence of increase in intracellular Ca2+ concentration [278] that is observed during therapy with such drugs as cardiotonic steroids, catecholamines, and type III cAMP phosphodiesterase inhibitor [279]. This property of agents interacting with TnC decreases the risk of side effects related to Ca2+ overload [270, 279]. Thus, development of pharmaceuticals based on an agent that modulates the sensitivity of thin filament to Ca2+ is a promising direction for the therapy of heart failure.
Troponin complex and cardiomyopathy. Cardiomyopathy is a pathology induced by the alteration of myocardial structure and/or development of systolic or diastolic heart dysfunction in the absence of inherited valvular defect, arterial hypertension, or pathologies of coronary arteries [280]. According to the modern classification of the European Cardiac Society, five types of cardiomyopathies are distinguished: hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), restrictive cardiomyopathy (RCM), arrhythmogenic right ventricular cardiomyopathy, and unclassified cardiomyopathies [280]. These pathologies are associated with increased risk of development of heart failure, myocardial infarction, and sudden death [281-283].
Currently, more than 90 identified mutations in troponin subunits are associated with HCM, DCM, and RCM [284-287]. Among these are point mutations of TnI, TnT, or TnC associated with deletion or substitution of one amino acid residue [288-290], mutations induced by the shift of the reading frame in TnC [287], and by the deletion of the C-terminal part of TnT and its substitution with seven nonsense amino acid residues [291]. Mutations in troponin genes associated with HCM and RCM generally lead to increase in the thin filament Ca2+-sensitivity, while the mutations associated with DCM mainly decrease the Ca2+-sensitivity of the filament [284].
Troponin complex and diagnostics of myocardial infarction. As mentioned above, in the heart of healthy adults, troponins I and T are presented exclusively in the specific cardiac isoforms that are not expressed in other tissues. This makes it possible to use these proteins as highly specific markers of cardiomyocyte damage. One of the most common reasons for necrosis of heart muscle cells is myocardial infarction induced by prolonged ischemia of the myocardium. Intracellular proteins of dead cardiomyocytes move to the blood flow, where they can be detected by different immunochemical methods.
The utilization of TnI and TnT as protein markers of myocardial infarction was first proposed more than 20 years ago [292, 293]. Since then diagnostic systems based on immunochemical measurement of hcTnI or hcTnT concentrations in the patient’s blood sidelined the traditional methods of diagnostics of myocardial infarction. High specificity and selectivity of methods based on the detection of troponins led to the inclusion of troponin concentration measurement into the criteria of definition of myocardial infarction [294-296]. Modern diagnostic systems can reliably diagnose infarction already 3-6 h after the onset of heart attack [296]. Hopefully, the expected introduction into practice of more sensitive next-generation systems will lead to the shortening of this time to 2-3 h [297-299]. The limited length of this paper does not permit more detailed review of the peculiarities of troponins utilization in diagnostics, but the interested reader can get more information in some recent reviews [300-303].
The troponin complex is a unique protein complex that by different biochemical mechanisms regulates the contractile activity of the heart. A prominent role in this regulation is played by the posttranslational modifications of troponin complex subunits. In spite of more than 40 years of investigation, many properties of troponin complex are still not fully understood. We can hope that further studies will elucidate yet unknown features of troponin functioning and its role in cardiac muscle contraction, thus enabling the development of more thorough approaches for diagnostics and therapy of cardiac disorders.
The author is grateful to A. E. Kogan and A. V. Vylegzhanina for their help in preparing this review.
REFERENCES
1.Wade, R., Eddy, R., Shows, T. B., and Kedes, L.
(1990) Genomics, 7, 346-357.
2.Tiso, N., Rampoldi, L., Pallavicini, A., Zimbello,
R., Pandolfo, D., Valle, G., Lanfranchi, G., and Danieli, G. A. (1997)
Biochem. Biophys. Res. Commun., 230, 347-350.
3.Bhavsar, P. K., Brand, N. J., Yacoub, M. H., and
Barton, P. J. (1996) Genomics, 35, 11-23.
4.Lundin, L. G. (1993) Genomics, 16,
1-19.
5.Cullen, M. E., Dellow, K. A., and Barton, P. J.
(2004) Mol. Cell Biochem., 263, 81-90.
6.Dhoot, G. K., and Perry, S. V. (1979)
Nature, 278, 714-718.
7.Bhavsar, P. K., Dhoot, G. K., Cumming, D. V.,
Butler-Browne, G. S., Yacoub, M. H., and Barton, P. J. (1991) FEBS
Lett., 292, 5-8.
8.Hunkeler, N. M., Kullman, J., and Murphy, A. M.
(1991) Circ. Res., 69, 1409-1414.
9.Sasse, S., Brand, N. J., Kyprianou, P., Dhoot, G.
K., Wade, R., Arai, M., Periasamy, M., Yacoub, M. H., and Barton, P. J.
(1993) Circ. Res., 72, 932-938.
10.Bhavsar, P. K., Dellow, K. A., Yacoub, M. H.,
Brand, N. J., and Barton, P. J. (2000) J. Mol. Cell Cardiol.,
32, 95-108.
11.Dellow, K. A., Bhavsar, P. K., Brand, N. J., and
Barton, P. J. (2001) Cardiovasc. Res., 50, 24-33.
12.Bodor, G. S., Porterfield, D., Voss, E. M.,
Smith, S., and Apple, F. S. (1995) Clin. Chem., 41,
1710-1715.
13.Sabry, M. A., and Dhoot, G. K. (1989) J.
Muscle. Res. Cell Motil., 10, 85-91.
14.Saggin, L., Gorza, L., Ausoni, S., and
Schiaffino, S. (1989) J. Biol. Chem., 264,
16299-16302.
15.Ausoni, S., De Nardi, C., Moretti, P., Gorza, L.,
and Schiaffino, S. (1991) Development, 112,
1041-1051.
16.Kruger, M., Kohl, T., and Linke, W. A. (2006)
Am. J. Physiol. Heart Circ. Physiol., 291, H496-506.
17.Nan, C., and Huang, X. (2009) Biochem.
Biophys. Res. Commun., 378, 62-67.
18.Podlowski, S., Bramlage, P., Baumann, G., Morano,
I., and Luther, H. P. (2002) J. Cell Biochem., 85,
198-207.
19.Wolska, B. M., Vijayan, K., Arteaga, G. M.,
Konhilas, J. P., Phillips, R. M., Kim, R., Naya, T., Leiden, J. M.,
Martin, A. F., De Tombe, P. P., and Solaro, R. J. (2001) J.
Physiol., 536, 863-870.
20.Fentzke, R. C., Buck, S. H., Patel, J. R., Lin,
H., Wolska, B. M., Stojanovic, M. O., Martin, A. F., Solaro, R. J.,
Moss, R. L., and Leiden, J. M. (1999) J. Physiol., 517,
143-157.
21.Westfall, M. V., Rust, E. M., and Metzger, J. M.
(1997) Proc. Natl. Acad. Sci. USA, 94, 5444-5449.
22.Metzger, J. M., Michele, D. E., Rust, E. M.,
Borton, A. R., and Westfall, M. V. (2003) J. Biol. Chem.,
278, 13118-13123.
23.Morimoto, S., Harada, K., and Ohtsuki, I. (1999)
J. Biochem., 126, 121-129.
24.Morimoto, S., and Goto, T. (2000) Biochem.
Biophys. Res. Commun., 267, 912-917.
25.Mesnard, L., Logeart, D., Taviaux, S., Diriong,
S., Mercadier, J. J., and Samson, F. (1995) Circ. Res.,
76, 687-692.
26.Townsend, P. J., Farza, H., Macgeoch, C., Spurr,
N. K., Wade, R., Gahlmann, R., Yacoub, M. H., and Barton, P. J. (1994)
Genomics, 21, 311-316.
27.Farza, H., Townsend, P. J., Carrier, L., Barton,
P. J., Mesnard, L., Bahrend, E., Forissier, J. F., Fiszman, M., Yacoub,
M. H., and Schwartz, K. (1998) J. Mol. Cell Cardiol., 30,
1247-1253.
28.Jin, J. P. (1996) Biochem. Biophys. Res.
Commun., 225, 883-889.
29.Saggin, L., Gorza, L., Ausoni, S., and
Schiaffino, S. (1990) Development, 110, 547-554.
30.Anderson, P. A., Malouf, N. N., Oakeley, A. E.,
Pagani, E. D., and Allen, P. D. (1991) Circ. Res., 69,
1226-1233.
31.Yonemura, I., Mitani, Y., Nakada, K., Akutsu, S.,
and Miyazaki, J. (2002) Zool. Sci., 19, 215-223.
32.Krishan, K., Morgan, M. J., Zhao, W., and Dhoot,
G. K. (2000) J. Muscle Res. Cell Motil., 21, 527-536.
33.Wang, Q., Reiter, R. S., Huang, Q. Q., Jin, J.
P., and Lin, J. J. (2001) Anat. Rec., 263, 72-84.
34.Barton, P. J., Felkin, L. E., Koban, M. U.,
Cullen, M. E., Brand, N. J., and Dhoot, G. K. (2004) Mol. Cell
Biochem., 263, 91-97.
35.Anderson, P. A., and Oakeley, A. E. (1989)
Circ. Res., 65, 1087-1093.
36.Samson, F., Mesnard, L., Mihovilovic, M., Potter,
T. G., Mercadier, J. J., Roses, A. D., and Gilbert, J. R. (1994)
Biochem. Biophys. Res. Commun., 199, 841-847.
37.Jin, J. P., Chen, A., and Huang, Q. Q. (1998)
Gene, 214, 121-129.
38.Cooper, T. A., and Ordahl, C. P. (1985) J.
Biol. Chem., 260, 11140-11148.
39.Risnik, V. V., Verin, A. D., and Gusev, N. B.
(1985) Biochem. J., 225, 549-552.
40.Leszyk, J., Dumaswala, R., Potter, J. D., Gusev,
N. B., Verin, A. D., Tobacman, L. S., and Collins, J. H. (1987)
Biochemistry, 26, 7035-7042.
41.Tobacman, L. S., and Lee, R. (1987) J. Biol.
Chem., 262, 4059-4064.
42.Jin, J. P., Wang, J., and Zhang, J. (1996)
Gene, 168, 217-221.
43.Jin, J. P., and Lin, J. J. (1989) J. Biol.
Chem., 264, 14471-12277.
44.Jin, J. P., Huang, Q. Q., Yeh, H. I., and Lin, J.
J. (1992) J. Mol. Biol., 227, 1269-1276.
45.Townsend, P. J., Barton, P. J., Yacoub, M. H.,
and Farza, H. (1995) J. Mol. Cell Cardiol., 27,
2223-2236.
46.Anderson, P. A., Greig, A., Mark, T. M., Malouf,
N. N., Oakeley, A. E., Ungerleider, R. M., Allen, P. D., and Kay, B. K.
(1995) Circ. Res., 76, 681-686.
47.Gomes, A. V., Venkatraman, G., Davis, J. P.,
Tikunova, S. B., Engel, P., Solaro, R. J., and Potter, J. D. (2004)
J. Biol. Chem., 279, 49579-49587.
48.Mcauliffe, J. J., Gao, L. Z., and Solaro, R. J.
(1990) Circ. Res., 66, 1204-1216.
49.Nassar, R., Malouf, N. N., Kelly, M. B., Oakeley,
A. E., and Anderson, P. A. (1991) Circ. Res., 69,
1470-1475.
50.Ladd, A. N., Charlet, N., and Cooper, T. A.
(2001) Mol. Cell Biol., 21, 1285-1296.
51.Ladd, A. N., Stenberg, M. G., Swanson, M. S., and
Cooper, T. A. (2005) Dev. Dyn., 233, 783-793.
52.Charlet, B. N., Logan, P., Singh, G., and Cooper,
T. A. (2002) Mol. Cell, 9, 649-658.
53.Song, W. J., Van Keuren, M. L., Drabkin, H. A.,
Cypser, J. R., Gemmill, R. M., and Kurnit, D. M. (1996) Cytogenet.
Cell Genet., 75, 36-37.
54.Townsend, P. J., Yacoub, M. H., and Barton, P. J.
(1997) Ann. Hum. Genet., 61, 457-459.
55.Gahlmann, R., and Kedes, L. (1990) J. Biol.
Chem., 265, 12520-12528.
56.Schreier, T., Kedes, L., and Gahlmann, R. (1990)
J. Biol. Chem., 265, 21247-21253.
57.Gahlmann, R., Wade, R., Gunning, P., and Kedes,
L. (1988) J. Mol. Biol., 201, 379-391.
58.Toyota, N., and Shimada, Y. (1981) J. Cell
Biol., 91, 497-504.
59.Toyota, N. (1993) Int. J. Dev. Biol.,
37, 531-537.
60.Mittmann, K., Jaquet, K., and Heilmeyer, L. M.,
Jr. (1990) FEBS Lett., 273, 41-45.
61.Zabrouskov, V., Ge, Y., Schwartz, J., and Walker,
J. W. (2008) Mol. Cell Proteom., 7, 1838-1849.
62.Howarth, J. W., Meller, J., Solaro, R. J.,
Trewhella, J., and Rosevear, P. R. (2007) J. Mol. Biol.,
373, 706-722.
63.Ferrieres, G., Pugniere, M., Mani, J. C.,
Villard, S., Laprade, M., Doutre, P., Pau, B., and Granier, C. (2000)
FEBS Lett., 479, 99-105.
64.Ward, D. G., Brewer, S. M., Gallon, C. E., Gao,
Y., Levine, B. A., and Trayer, I. P. (2004) Biochemistry,
43, 5772-5781.
65.Ward, D. G., Brewer, S. M., Calvert, M. J.,
Gallon, C. E., Gao, Y., and Trayer, I. P. (2004) Biochemistry,
43, 4020-4027.
66.Finley, N., Abbott, M. B., Abusamhadneh, E.,
Gaponenko, V., Dong, W., Gasmi-Seabrook, G., Howarth, J. W., Rance, M.,
Solaro, R. J., Cheung, H. C., and Rosevear, P. R. (1999) FEBS
Lett., 453, 107-112.
67.Takeda, S., Yamashita, A., Maeda, K., and Maeda,
Y. (2003) Nature, 424, 35-41.
68.Takeda, S. (2005) J. Electron Microsc.
(Tokyo), 54, Suppl. 1, i35-41.
69.Vassylyev, D. G., Takeda, S., Wakatsuki, S.,
Maeda, K., and Maeda, Y. (1998) Proc. Natl. Acad. Sci. USA,
95, 4847-4852.
70.Stefancsik, R., Jha, P. K., and Sarkar, S. (1998)
Proc. Natl. Acad. Sci. USA, 95, 957-962.
71.Takeda, S., and Maeda, Y. (2003)
Seikagaku, 75, 1540-1545.
72.Kobayashi, T., Patrick, S. E., and Kobayashi, M.
(2009) J. Biol. Chem., 284, 20052-20060.
73.Lindhout, D. A., and Sykes, B. D. (2003) J.
Biol. Chem., 278, 27024-27034.
74.Brown, L. J., Sale, K. L., Hills, R., Rouviere,
C., Song, L., Zhang, X., and Fajer, P. G. (2002) Proc. Natl. Acad.
Sci. USA, 99, 12765-12770.
75.Dong, W. J., Robinson, J. M., Stagg, S., Xing,
J., and Cheung, H. C. (2003) J. Biol. Chem., 278,
8686-8692.
76.Talbot, J. A., and Hodges, R. S. (1981) J.
Biol. Chem., 256, 2798-2802.
77.Farah, C. S., Miyamoto, C. A., Ramos, C. H., Da
Silva, A. C., Quaggio, R. B., Fujimori, K., Smillie, L. B., and
Reinach, F. C. (1994) J. Biol. Chem., 269, 5230-5240.
78.Dong, W. J., Xing, J., Robinson, J. M., and
Cheung, H. C. (2001) J. Mol. Biol., 314, 51-61.
79.Kowlessur, D., and Tobacman, L. S. (2010) J.
Biol. Chem., 285, 2686-2694.
80.Li, M. X., Spyracopoulos, L., and Sykes, B. D.
(1999) Biochemistry, 38, 8289-8298.
81.Wang, X., Li, M. X., and Sykes, B. D. (2002)
J. Biol. Chem., 277, 31124-31133.
82.Blumenschein, T. M., Stone, D. B., Fletterick, R.
J., Mendelson, R. A., and Sykes, B. D. (2006) Biophys. J.,
90, 2436-2444.
83.Julien, O., Mercier, P., Allen, C. N., Fisette,
O., Ramos, C. H., Lague, P., Blumenschein, T. M., and Sykes, B. D.
(2011) Proteins, 79, 1240-1250.
84.King, W. A., Stone, D. B., Timmins, P. A.,
Narayanan, T., Von Brasch, A. A., Mendelson, R. A., and Curmi, P. M.
(2005) J. Mol. Biol., 345, 797-815.
85.Murakami, K., Yumoto, F., Ohki, S. Y., Yasunaga,
T., Tanokura, M., and Wakabayashi, T. (2005) J. Mol. Biol.,
352, 178-201.
86.Wang, H., Chalovich, J. M., and Marriott, G.
(2012) PLoS One, 7, e50420.
87.Zhou, Z., Li, K. L., Rieck, D., Ouyang, Y.,
Chandra, M., and Dong, W. J. (2012) J. Biol. Chem., 287,
7661-7674.
88.Galinska-Rakoczy, A., Engel, P., Xu, C., Jung,
H., Craig, R., Tobacman, L. S., and Lehman, W. (2008) J. Mol.
Biol., 379, 929-935.
89.Galinska, A., Hatch, V., Craig, R., Murphy, A.
M., Van Eyk, J. E., Wang, C. L., Lehman, W., and Foster, D. B. (2009)
Circ. Res., 106, 705-711.
90.Tobacman, L. S. (1996) Annu. Rev.
Physiol., 58, 447-481.
91.Perry, S. V. (1998) J. Muscle Res. Cell
Motil., 19, 575-602.
92.White, S. P., Cohen, C., and Phillips, G. N., Jr.
(1987) Nature, 325, 826-828.
93.Wei, B., and Jin, J. P. (2011) Arch. Biochem.
Biophys., 505, 144-154.
94.Gollapudi, S. K., Mamidi, R., Mallampalli, S. L.,
and Chandra, M. (2012) Biophys. J., 103, 940-948.
95.Pearlstone, J. R., and Smillie, L. B. (1982)
J. Biol. Chem., 257, 10587-10592.
96.Pearlstone, J. R., and Smillie, L. B. (1985)
Can. J. Biochem. Cell Biol., 63, 212-218.
97.Verin, A. D., and Gusev, N. B. (1988)
Biokhimiya, 53, 1235-1246.
98.Wang, J., and Jin, J. P. (1998)
Biochemistry, 37, 14519-14528.
99.Jin, J. P., Chen, A., Ogut, O., and Huang, Q. Q.
(2000) Am. J. Physiol. Cell Physiol., 279,
C1067-C1077.
100.Biesiadecki, B. J., Chong, S. M., Nosek, T. M.,
and Jin, J. P. (2007) Biochemistry, 46, 1368-1379.
101.Chandra, M., Montgomery, D. E., Kim, J. J., and
Solaro, R. J. (1999) J. Mol. Cell Cardiol., 31,
867-880.
102.Sumandea, M. P., Vahebi, S., Sumandea, C. A.,
Garcia-Cazarin, M. L., Staidle, J., and Homsher, E. (2009)
Biochemistry, 48, 7722-7731.
103.Mamidi, R., Michael, J. J., Muthuchamy, M., and
Chandra, M. (2013) FASEB J., 27; doi:
10.1096/fj.13-232363.
104.Gollapudi, S. K., Gallon, C. E., and Chandra,
M. (2013) J. Mol. Biol., 425, 1565-1581.
105.Jin, J. P., and Chong, S. M. (2010) Arch.
Biochem. Biophys., 500, 144-150.
106.Mak, A. S., and Smillie, L. B. (1981) J.
Mol. Biol., 149, 541-550.
107.Pearlstone, J. R., and Smillie, L. B. (1983)
J. Biol. Chem., 258, 2534-2542.
108.Murakami, K., Stewart, M., Nozawa, K., Tomii,
K., Kudou, N., Igarashi, N., Shirakihara, Y., Wakatsuki, S., Yasunaga,
T., and Wakabayashi, T. (2008) Proc. Natl. Acad. Sci. USA,
105, 7200-7205.
109.Manning, E. P., Tardiff, J. C., and Schwartz,
S. D. (2011) Biochemistry, 50, 7405-7413.
110.Chong, P. C., and Hodges, R. S. (1982) J.
Biol. Chem., 257, 9152-9160.
111.Morris, E. P., and Lehrer, S. S. (1984)
Biochemistry, 23, 2214-2220.
112.Tanokura, M., Tawada, Y., and Ohtsuki, I.
(1982) J. Biochem., 91, 1257-1265.
113.Franklin, A. J., Baxley, T., Kobayashi, T., and
Chalovich, J. M. (2012) Biophys. J., 102, 2536-2544.
114.Roher, A., Lieska, N., and Spitz, W. (1986)
Muscle Nerve, 9, 73-77.
115.Van Eerd, J. P., and Takahshi, K. (1976)
Biochemistry, 15, 1171-1180.
116.Sia, S. K., Li, M. X., Spyracopoulos, L.,
Gagne, S. M., Liu, W., Putkey, J. A., and Sykes, B. D. (1997) J.
Biol. Chem., 272, 18216-18221.
117.Kretsinger, R. H. (1980) CRC Crit. Rev.
Biochem., 8, 119-174.
118.Potter, J. D., and Gergely, J. (1975) J.
Biol. Chem., 250, 4628-4633.
119.Van Eerd, J. P., and Takahashi, K. (1975)
Biochem. Biophys. Res. Commun., 64, 122-127.
120.Barskaya, N. V., and Gusev, N. B. (1982)
Biochem. J., 207, 185-192.
121.Holroyde, M. J., Robertson, S. P., Johnson, J.
D., Solaro, R. J., and Potter, J. D. (1980) J. Biol. Chem.,
255, 11688-11693.
122.Pan, B. S., and Solaro, R. J. (1987) J.
Biol. Chem., 262, 7839-7849.
123.Zot, H. G., and Potter, J. D. (1982) J.
Biol. Chem., 257, 7678-7683.
124.Putkey, J. A., Sweeney, H. L., and Campbell, S.
T. (1989) J. Biol. Chem., 264, 12370-12378.
125.Holmes, K. C., Popp, D., Gebhard, W., and
Kabsch, W. (1990) Nature, 347, 44-49.
126.Oda, T., Iwasa, M., Aihara, T., Maeda, Y., and
Narita, A. (2009) Nature, 457, 441-445.
127.Nevzorov, I. A., and Levitsky, D. I. (2011)
Biochemistry (Moscow), 76, 1507-1527.
128.Spudich, J. A., Huxley, H. E., and Finch, J. T.
(1972) J. Mol. Biol., 72, 619-632.
129.Lorenz, M., Poole, K. J., Popp, D., Rosenbaum,
G., and Holmes, K. C. (1995) J. Mol. Biol., 246,
108-119.
130.Phillips, G. N., Jr., Fillers, J. P., and
Cohen, C. (1986) J. Mol. Biol., 192, 111-131.
131.Ebashi, S., Endo, M., and Otsuki, I. (1969)
Q. Rev. Biophys., 2, 351-384.
132.Hitchcock, S. E., Huxley, H. E., and
Szent-Gyorgyi, A. G. (1973) J. Mol. Biol., 80,
825-836.
133.Potter, J. D., and Gergely, J. (1974)
Biochemistry, 13, 2697-2703.
134.Tripet, B., Van Eyk, J. E., and Hodges, R. S.
(1997) J. Mol. Biol., 271, 728-750.
135.Geeves, M. A., Chai, M., and Lehrer, S. S.
(2000) Biochemistry, 39, 9345-9350.
136.Koubassova, N. A., and Tsaturyan, A. K. (2011)
Biochemistry (Moscow), 76, 1484-1506.
137.Geeves, M. A., and Holmes, K. C. (2005) Adv.
Protein Chem., 71, 161-193.
138.Ikebe, M., and Hartshorne, D. J. (1985) J.
Biol. Chem., 260, 10027-10031.
139.Gordon, A. M., Homsher, E., and Regnier, M.
(2000) Physiol. Rev., 80, 853-924.
140.Kobayashi, T., and Solaro, R. J. (2005)
Annu. Rev. Physiol., 67, 39-67.
141.Mckillop, D. F., and Geeves, M. A. (1993)
Biophys. J., 65, 693-701.
142.Maytum, R., Lehrer, S. S., and Geeves, M. A.
(1999) Biochemistry, 38, 1102-1110.
143.Hoffman, R. M., Blumenschein, T. M., and Sykes,
B. D. (2006) J. Mol. Biol., 361, 625-633.
144.Lehrer, S. S. (2011) J. Muscle Res. Cell
Motil., 32, 203-208.
145.Bers, D. M. (2000) Circ. Res.,
87, 275-281.
146.Xu, C., Craig, R., Tobacman, L., Horowitz, R.,
and Lehman, W. (1999) Biophys. J., 77, 985-992.
147.Lindert, S., Kekenes-Huskey, P. M., Huber, G.,
Pierce, L., and Mccammon, J. A. (2012) J. Phys. Chem. B,
116, 8449-8459.
148.Ovaska, M., and Taskinen, J. (1991)
Proteins, 11, 79-94.
149.Dong, W., Rosenfeld, S. S., Wang, C. K.,
Gordon, A. M., and Cheung, H. C. (1996) J. Biol. Chem.,
271, 688-694.
150.Gagne, S. M., Tsuda, S., Li, M. X., Smillie, L.
B., and Sykes, B. D. (1995) Nat. Struct. Biol., 2,
784-789.
151.Slupsky, C. M., and Sykes, B. D. (1995)
Biochemistry, 34, 15953-15964.
152.Spyracopoulos, L., Li, M. X., Sia, S. K.,
Gagne, S. M., Chandra, M., Solaro, R. J., and Sykes, B. D. (1997)
Biochemistry, 36, 12138-12146.
153.Dong, W. J., Xing, J., Villain, M., Hellinger,
M., Robinson, J. M., Chandra, M., Solaro, R. J., Umeda, P. K., and
Cheung, H. C. (1999) J. Biol. Chem., 274,
31382-31390.
154.Vibert, P., Craig, R., and Lehman, W. (1997)
J. Mol. Biol., 266, 8-14.
155.Haselgrove, J. C., and Huxley, H. E. (1973)
J. Mol. Biol., 77, 549-568.
156.Parry, D. A., and Squire, J. M. (1973) J.
Mol. Biol., 75, 33-55.
157.Poole, K. J., Lorenz, M., Evans, G., Rosenbaum,
G., Pirani, A., Craig, R., Tobacman, L. S., Lehman, W., and Holmes, K.
C. (2006) J. Struct. Biol., 155, 273-284.
158.Robinson, J. M., Dong, W. J., Xing, J., and
Cheung, H. C. (2004) J. Mol. Biol., 340, 295-305.
159.Houmeida, A., Heeley, D. H., Belknap, B., and
White, H. D. (2010) J. Biol. Chem., 285, 32760-32769.
160.Xing, J., Jayasundar, J. J., Ouyang, Y., and
Dong, W. J. (2009) J. Biol. Chem., 284, 16432-16441.
161.Hofmann, P. A., and Fuchs, F. (1987) Am. J.
Physiol., 253, C90-C96.
162.Allen, D. G., and Kentish, J. C. (1985) J.
Mol. Cell Cardiol., 17, 821-840.
163.De Tombe, P. P., Mateja, R. D., Tachampa, K.,
Ait Mou, Y., Farman, G. P., and Irving, T. C. (2010) J. Mol. Cell
Cardiol., 48, 851-858.
164.Shiels, H. A., and White, E. (2008) J. Exp.
Biol., 211, 2005-2013.
165.Arteaga, G. M., Palmiter, K. A., Leiden, J. M.,
and Solaro, R. J. (2000) J. Physiol., 526, 541-549.
166.Terui, T., Sodnomtseren, M., Matsuba, D.,
Udaka, J., Ishiwata, S., Ohtsuki, I., Kurihara, S., and Fukuda, N.
(2008) J. Gen. Physiol., 131, 275-283.
167.Konhilas, J. P., Irving, T. C., Wolska, B. M.,
Jweied, E. E., Martin, A. F., Solaro, R. J., and De Tombe, P. P. (2003)
J. Physiol., 547, 951-961.
168.Matsuba, D., Terui, T., Jin, O., Tanaka, H.,
Ojima, T., Ohtsuki, I., Ishiwata, S., Kurihara, S., and Fukuda, N.
(2009) J. Gen. Physiol., 133, 571-581.
169.Tachampa, K., Wang, H., Farman, G. P., and De
Tombe, P. P. (2007) Circ. Res., 101, 1081-1083.
170.Sedlis, S. P. (1992) Cardiovasc. Clin.,
22, 3-18.
171.Di Diego, J. M., and Antzelevitch, C. (2011)
Heart Rhythm, 8, 1963-1968.
172.Crampin, E. J., Smith, N. P., Langham, A. E.,
Clayton, R. H., and Orchard, C. H. (2006) Philos. Trans. A Math.
Phys. Eng. Sci., 364, 1171-1186.
173.Bountra, C., Kaila, K., and Vaughan-Jones, R.
D. (1988) J. Physiol., 398, 341-360.
174.Yan, G. X., and Kleber, A. G. (1992) Circ.
Res., 71, 460-470.
175.Park, C. O., Xiao, X. H., and Allen, D. G.
(1999) Am. J. Physiol., 276, H1581-H1590.
176.Jennings, R. B., and Reimer, K. A. (1991)
Annu. Rev. Med., 42, 225-246.
177.Metzger, J. M., Parmacek, M. S., Barr, E.,
Pasyk, K., Lin, W. I., Cochrane, K. L., Field, L. J., and Leiden, J. M.
(1993) Proc. Natl. Acad. Sci. USA, 90, 9036-9040.
178.Dargis, R., Pearlstone, J. R., Barrette-Ng, I.,
Edwards, H., and Smillie, L. B. (2002) J. Biol. Chem.,
277, 34662-34665.
179.Reiser, P. J., Westfall, M. V., Schiaffino, S.,
and Solaro, R. J. (1994) Am. J. Physiol., 267,
H1589-H1596.
180.Day, S. M., Westfall, M. V., Fomicheva, E. V.,
Hoyer, K., Yasuda, S., La Cross, N. C., D’alecy, L. G., Ingwall,
J. S., and Metzger, J. M. (2006) Nat. Med., 12,
181-189.
181.Pineda-Sanabria, S. E., Robertson, I. M., Li,
M. X., and Sykes, B. D. (2013) Cardiovasc. Res., 97,
481-489.
182.Nosek, T. M., Brotto, M. A., and Jin, J. P.
(2004) Arch. Biochem. Biophys., 430, 178-184.
183.Zhang, P., Kirk, J. A., Ji, W., Dos Remedios,
C. G., Kass, D. A., Van Eyk, J. E., and Murphy, A. M. (2012)
Circulation, 126, 1828-1837.
184.Van Der Velden, J., Papp, Z., Zaremba, R.,
Boontje, N. M., De Jong, J. W., Owen, V. J., Burton, P. B., Goldmann,
P., Jaquet, K., and Stienen, G. J. (2003) Cardiovasc. Res.,
57, 37-47.
185.Zhang, J., Guy, M. J., Norman, H. S., Chen, Y.
C., Xu, Q., Dong, X., Guner, H., Wang, S., Kohmoto, T., Young, K. H.,
Moss, R. L., and Ge, Y. (2011) J. Proteome Res., 10,
4054-4065.
186.Noland, T. A., Jr., Guo, X., Raynor, R. L.,
Jideama, N. M., Averyhart-Fullard, V., Solaro, R. J., and Kuo, J. F.
(1995) J. Biol. Chem., 270, 25445-25454.
187.Noland, T. A., Jr., Raynor, R. L., Jideama, N.
M., Guo, X., Kazanietz, M. G., Blumberg, P. M., Solaro, R. J., and Kuo,
J. F. (1996) Biochemistry, 35, 14923-14931.
188.Kobayashi, T., Yang, X., Walker, L. A., Van
Breemen, R. B., and Solaro, R. J. (2005) J. Mol. Cell Cardiol.,
38, 213-218.
189.Noland, T. A., Jr., Raynor, R. L., and Kuo, J.
F. (1989) J. Biol. Chem., 264, 20778-20785.
190.Buscemi, N., Foster, D. B., Neverova, I., and
Van Eyk, J. E. (2002) Circ. Res., 91, 509-516.
191.Cuello, F., Bardswell, S. C., Haworth, R. S.,
Yin, X., Lutz, S., Wieland, T., Mayr, M., Kentish, J. C., and Avkiran,
M. (2007) Circ. Res., 100, 864-873.
192.You, B., Yan, G., Zhang, Z., Yan, L., Li, J.,
Ge, Q., Jin, J. P., and Sun, J. (2009) Biochem. J., 418,
93-101.
193.Oliveira, S. M., Zhang, Y. H., Solis, R. S.,
Isackson, H., Bellahcene, M., Yavari, A., Pinter, K., Davies, J. K.,
Ge, Y., Ashrafian, H., Walker, J. W., Carling, D., Watkins, H.,
Casadei, B., and Redwood, C. (2012) Circ. Res., 110,
1192-1201.
194.Solis, R. S., Ge, Y., and Walker, J. W. (2011)
Protein Sci., 20, 894-907.
195.Solaro, R. J., Moir, A. J., and Perry, S. V.
(1976) Nature, 262, 615-617.
196.Gautel, M., Zuffardi, O., Freiburg, A., and
Labeit, S. (1995) EMBO J., 14, 1952-1960.
197.Yamasaki, R., Wu, Y., Mcnabb, M., Greaser, M.,
Labeit, S., and Granzier, H. (2002) Circ. Res., 90,
1181-1188.
198.Kruger, M., and Linke, W. A. (2006) J.
Muscle Res. Cell Motil., 27, 435-444.
199.Stull, J. T., Brostrom, C. O., and Krebs, E. G.
(1972) J. Biol. Chem., 247, 5272-5274.
200.Pratje, E., and Heilmeyer, L. M. (1972) FEBS
Lett., 27, 89-93.
201.Strang, K. T., Sweitzer, N. K., Greaser, M. L.,
and Moss, R. L. (1994) Circ. Res., 74, 542-549.
202.Hagemann, D., and Xiao, R. P. (2002) Trends
Cardiovasc. Med., 12, 51-56.
203.Bers, D. M., and Perez-Reyes, E. (1999)
Cardiovasc. Res., 42, 339-360.
204.Van Der Velden, J. (2011) Pflugers
Arch., 462, 155-163.
205.Solaro, R. J., Henze, M., and Kobayashi, T.
(2013) Circ. Res., 112, 355-366.
206.Zhang, R., Zhao, J., Mandveno, A., and Potter,
J. D. (1995) Circ. Res., 76, 1028-1035.
207.Kentish, J. C., Mccloskey, D. T., Layland, J.,
Palmer, S., Leiden, J. M., Martin, A. F., and Solaro, R. J. (2001)
Circ. Res., 88, 1059-1065.
208.Wijnker, P. J., Foster, D. B., Tsao, A. L.,
Frazier, A. H., Dos Remedios, C. G., Murphy, A. M., Stienen, G. J., and
Van Der Velden, J. (2013) Am. J. Physiol. Heart Circ. Physiol.,
304, H260-H268.
209.Mittmann, K., Jaquet, K., and Heilmeyer, L. M.,
Jr. (1992) FEBS Lett., 302, 133-137.
210.Solis, R. S., Ge, Y., and Walker, J. W. (2008)
J. Muscle Res. Cell Motil., 29, 203-212.
211.Chandra, M., Dong, W. J., Pan, B. S., Cheung,
H. C., and Solaro, R. J. (1997) Biochemistry, 36,
13305-13311.
212.Zhang, R., Zhao, J., and Potter, J. D. (1995)
J. Biol. Chem., 270, 30773-30780.
213.Pena, J. R., and Wolska, B. M. (2004)
Cardiovasc. Res., 61, 756-763.
214.Pi, Y., Kemnitz, K. R., Zhang, D., Kranias, E.
G., and Walker, J. W. (2002) Circ. Res., 90, 649-656.
215.Pi, Y., Zhang, D., Kemnitz, K. R., Wang, H.,
and Walker, J. W. (2003) J. Physiol., 552, 845-857.
216.Takimoto, E., Soergel, D. G., Janssen, P. M.,
Stull, L. B., Kass, D. A., and Murphy, A. M. (2004) Circ. Res.,
94, 496-504.
217.De Tombe, P. P., and Stienen, G. J. (1995)
Circ. Res., 76, 734-741.
218.Stelzer, J. E., Patel, J. R., Walker, J. W.,
and Moss, R. L. (2007) Circ. Res., 101, 503-511.
219.Chen, P. P., Patel, J. R., Rybakova, I. N.,
Walker, J. W., and Moss, R. L. (2010) J. Gen. Physiol.,
136, 615-627.
220.Messer, A. E., Jacques, A. M., and Marston, S.
B. (2007) J. Mol. Cell Cardiol., 42, 247-259.
221.Steinberg, S. F. (2012) Physiology
(Bethesda), 27, 130-139.
222.Shin, H. G., Barnett, J. V., Chang, P., Reddy,
S., Drinkwater, D. C., Pierson, R. N., Wiley, R. G., and Murray, K. T.
(2000) Cardiovasc. Res., 48, 285-299.
223.Sumandea, M. P., Rybin, V. O., Hinken, A. C.,
Wang, C., Kobayashi, T., Harleton, E., Sievert, G., Balke, C. W.,
Feinmark, S. J., Solaro, R. J., and Steinberg, S. F. (2008) J. Biol.
Chem., 283, 22680-22689.
224.Sumandea, M. P., Pyle, W. G., Kobayashi, T., De
Tombe, P. P., and Solaro, R. J. (2003) J. Biol. Chem.,
278, 35135-35144.
225.Kooij, V., Boontje, N., Zaremba, R., Jaquet,
K., Dos Remedios, C., Stienen, G. J., and Van Der Velden, J. (2010)
Basic Res. Cardiol., 105, 289-300.
226.Hidalgo, C., Hudson, B., Bogomolovas, J., Zhu,
Y., Anderson, B., Greaser, M., Labeit, S., and Granzier, H. (2009)
Circ. Res., 105, 631-638.
227.Ruse, C. I., Willard, B., Jin, J. P., Haas, T.,
Kinter, M., and Bond, M. (2002) Anal. Chem., 74,
1658-1664.
228.Malhotra, A., Kang, B. P., Cheung, S., Opawumi,
D., and Meggs, L. G. (2001) Diabetes, 50, 1918-1926.
229.Wu, S. C., and Solaro, R. J. (2007) J. Biol.
Chem., 282, 30691-30698.
230.Hinken, A. C., Hanft, L. M., Scruggs, S. B.,
Sadayappan, S., Robbins, J., Solaro, R. J., and Mcdonald, K. S. (2012)
J. Muscle Res. Cell Motil., 33, 439-448.
231.Sakthivel, S., Finley, N. L., Rosevear, P. R.,
Lorenz, J. N., Gulick, J., Kim, S., Vanburen, P., Martin, L. A., and
Robbins, J. (2005) J. Biol. Chem., 280, 703-714.
232.Macgowan, G. A., Du, C., Cowan, D. B., Stamm,
C., Mcgowan, F. X., Solaro, R. J., Koretsky, A. P., and Del Nido, P. J.
(2001) Am. J. Physiol. Heart Circ. Physiol., 280,
H835-H843.
233.Kirk, J. A., Macgowan, G. A., Evans, C., Smith,
S. H., Warren, C. M., Mamidi, R., Chandra, M., Stewart, A. F., Solaro,
R. J., and Shroff, S. G. (2009) Circ. Res., 105,
1232-1239.
234.Burkart, E. M., Sumandea, M. P., Kobayashi, T.,
Nili, M., Martin, A. F., Homsher, E., and Solaro, R. J. (2003) J.
Biol. Chem., 278, 11265-11272.
235.Mathur, M. C., Kobayashi, T., and Chalovich, J.
M. (2008) Biophys. J., 94, 542-549.
236.Dong, X., Sumandea, C. A., Chen, Y. C.,
Garcia-Cazarin, M. L., Zhang, J., Balke, C. W., Sumandea, M. P., and
Ge, Y. (2012) J. Biol. Chem., 287, 848-857.
237.Molnar, A., Borbely, A., Czuriga, D., Ivetta,
S. M., Szilagyi, S., Hertelendi, Z., Pasztor, E. T., Balogh, A.,
Galajda, Z., Szerafin, T., Jaquet, K., Papp, Z., Edes, I., and Toth, A.
(2009) J. Biol. Chem., 284, 1031-1039.
238.Haworth, R. S., Cuello, F., Herron, T. J.,
Franzen, G., Kentish, J. C., Gautel, M., and Avkiran, M. (2004)
Circ. Res., 95, 1091-1099.
239.Haworth, R. S., Roberts, N. A., Cuello, F., and
Avkiran, M. (2007) J. Mol. Cell Cardiol., 43,
686-695.
240.Layland, J., Solaro, R. J., and Shah, A. M.
(2005) Cardiovasc. Res., 66, 12-21.
241.Ayaz-Guner, S., Zhang, J., Li, L., Walker, J.
W., and Ge, Y. (2009) Biochemistry, 48, 8161-8170.
242.Risnik, V. V., and Gusev, N. B. (1984)
Biochim. Biophys. Acta, 790, 108-116.
243.Gusev, N. B., Dobrovolskii, A. B., and Severin,
S. E. (1980) Biochem. J., 189, 219-226.
244.Villar-Palasi, C., and Kumon, A. (1981) J.
Biol. Chem., 256, 7409-7415.
245.He, X., Liu, Y., Sharma, V., Dirksen, R. T.,
Waugh, R., Sheu, S. S., and Min, W. (2003) Am. J. Pathol.,
163, 243-251.
246.Vahebi, S., Kobayashi, T., Warren, C. M., De
Tombe, P. P., and Solaro, R. J. (2005) Circ. Res., 96,
740-747.
247.Pfleiderer, P., Sumandea, M. P., Rybin, V. O.,
Wang, C., and Steinberg, S. F. (2009) J. Muscle Res. Cell
Motil., 30, 67-72.
248.Swiderek, K., Jaquet, K., Meyer, H. E.,
Schachtele, C., Hofmann, F., and Heilmeyer, L. M., Jr. (1990) Eur.
J. Biochem., 190, 575-582.
249.Katoh, N., Wise, B. C., and Kuo, J. F. (1983)
Biochem. J., 209, 189-195.
250.Raggi, A., Grand, R. J., Moir, A. J., and
Perry, S. V. (1989) Biochim. Biophys. Acta, 997,
135-143.
251.Noland, T. A., Jr., and Kuo, J. F. (1991) J.
Biol. Chem., 266, 4974-4978.
252.Jideama, N. M., Noland, T. A., Jr., Raynor, R.
L., Blobe, G. C., Fabbro, D., Kazanietz, M. G., Blumberg, P. M.,
Hannun, Y. A., and Kuo, J. F. (1996) J. Biol. Chem., 271,
23277-23283.
253.Noland, T. A., Jr., and Kuo, J. F. (1993) J.
Mol. Cell Cardiol., 25, 53-65.
254.Sumandea, M. P., Burkart, E. M., Kobayashi, T.,
De Tombe, P. P., and Solaro, R. J. (2004) Ann. NY Acad. Sci.,
1015, 39-52.
255.Zhang, J., Zhang, H., Ayaz-Guner, S., Chen, Y.
C., Dong, X., Xu, Q., and Ge, Y. (2011) Biochemistry, 50,
6081-6092.
256.Gusev, N. B., Barskaya, N. V., Verin, A. D.,
Duzhenkova, I. V., Khuchua, Z. A., and Zheltova, A. O. (1983)
Biochem. J., 213, 123-129.
257.Herve, J. C., and Sarrouilhe, D. (2006)
Prog. Biophys. Mol. Biol., 90, 225-248.
258.Chu, G., and Kranias, E. G. (2006) Novartis
Found. Symp., 274, 156-171.
259.Barefield, D., and Sadayappan, S. (2010) J.
Mol. Cell Cardiol., 48, 866-875.
260.Jideama, N. M., Crawford, B. H., Hussain, A.
K., and Raynor, R. L. (2006) Int. J. Biol. Sci., 2,
1-9.
261.Deshmukh, P. A., Blunt, B. C., and Hofmann, P.
A. (2007) Am. J. Physiol. Heart Circ. Physiol., 292,
H792-H799.
262.Jaquet, K., Thieleczek, R., and Heilmeyer, L.
M., Jr. (1995) Eur. J. Biochem., 231, 486-490.
263.Husberg, C., Agnetti, G., Holewinski, R. J.,
Christensen, G., and Van Eyk, J. E. (2012) Proteomics,
12, 973-978.
264.Ceulemans, H., and Bollen, M. (2004)
Physiol. Rev., 84, 1-39.
265.Nicolaou, P., and Kranias, E. G. (2009)
Front. Biosci., 14, 3571-3585.
266.Kurimchak, A., and Grana, X. (2012)
Gene, 499, 1-7.
267.Zhang, Z., Biesiadecki, B. J., and Jin, J. P.
(2006) Biochemistry, 45, 11681-11694.
268.Feng, H. Z., Biesiadecki, B. J., Yu, Z. B.,
Hossain, M. M., and Jin, J. P. (2008) J. Physiol., 586,
3537-3550.
269.Zhang, Z., Feng, H. Z., and Jin, J. P. (2006)
Arch. Biochem. Biophys., 515, 37-45.
270.Li, M. X., Robertson, I. M., and Sykes, B. D.
(2008) Biochem. Biophys. Res. Commun., 369, 88-99.
271.Geguchadze, R. N., Krylatov, A. V., and Gusev,
N. B. (1990) Biomed. Sci., 1, 37-42.
272.Li, Y., Love, M. L., Putkey, J. A., and Cohen,
C. (2000) Proc. Natl. Acad. Sci. USA, 97, 5140-5145.
273.Pollesello, P., Ovaska, M., Kaivola, J.,
Tilgmann, C., Lundstrom, K., Kalkkinen, N., Ulmanen, I., Nissinen, E.,
and Taskinen, J. (1994) J. Biol. Chem., 269,
28584-28590.
274.Sorsa, T., Heikkinen, S., Abbott, M. B.,
Abusamhadneh, E., Laakso, T., Tilgmann, C., Serimaa, R., Annila, A.,
Rosevear, P. R., Drakenberg, T., Pollesello, P., and Kilpelainen, I.
(2001) J. Biol. Chem., 276, 9337-9343.
275.Kleerekoper, Q., and Putkey, J. A. (1999) J.
Biol. Chem., 274, 23932-23939.
276.Li, M. X., Spyracopoulos, L., Beier, N.,
Putkey, J. A., and Sykes, B. D. (2000) Biochemistry, 39,
8782-8790.
277.Wang, X., Li, M. X., Spyracopoulos, L., Beier,
N., Chandra, M., Solaro, R. J., and Sykes, B. D. (2001) J. Biol.
Chem., 276, 25456-25466.
278.Brixius, K., Reicke, S., and Schwinger, R. H.
(2002) Am. J. Physiol. Heart Circ. Physiol., 282,
H131-H137.
279.Endoh, M. (2003) Expert Opin. Investig.
Drugs, 12, 735-750.
280.Elliott, P., Andersson, B., Arbustini, E.,
Bilinska, Z., Cecchi, F., Charron, P., Dubourg, O., Kuhl, U., Maisch,
B., Mckenna, W. J., Monserrat, L., Pankuweit, S., Rapezzi, C.,
Seferovic, P., Tavazzi, L., and Keren, A. (2008) Eur. Heart J.,
29, 270-276.
281.Towbin, J. A. (2009) Pacing Clin.
Electrophysiol., 32, Suppl. 2, S23-S31.
282.Mohan, S. B., Parker, M., Wehbi, M., and
Douglass, P. (2002) Cleve. Clin. J. Med., 69,
481-487.
283.Ammash, N. M., Seward, J. B., Bailey, K. R.,
Edwards, W. D., and Tajik, A. J. (2000) Circulation, 101,
2490-2496.
284.Willott, R. H., Gomes, A. V., Chang, A. N.,
Parvatiyar, M. S., Pinto, J. R., and Potter, J. D. (2010) J. Mol.
Cell Cardiol., 48, 882-892.
285.Murakami, C., Nakamura, S., Kobayashi, M.,
Maeda, K., Irie, W., Wada, B., Hayashi, M., Sasaki, C., Nakamaru, N.,
Furukawa, M., and Kurihara, K. (2010) Leg. Med. (Tokyo),
12, 280-283.
286.Parvatiyar, M. S., Landstrom, A. P.,
Figueiredo-Freitas, C., Potter, J. D., Ackerman, M. J., and Pinto, J.
R. (2012) J. Biol. Chem., 287, 31845-31855.
287.Chung, W. K., Kitner, C., and Maron, B. J.
(2011) Cardiol. Young, 21, 345-348.
288.Watkins, H., Mckenna, W. J., Thierfelder, L.,
Suk, H. J., Anan, R., O’donoghue, A., Spirito, P., Matsumori, A.,
Moravec, C. S., Seidman, J. G., et al. (1995) N. Engl. J. Med.,
332, 1058-10564.
289.Ashrafian, H., Redwood, C., Blair, E., and
Watkins, H. (2003) Trends Genet., 19, 263-268.
290.Hershberger, R. E., Parks, S. B., Kushner, J.
D., Li, D., Ludwigsen, S., Jakobs, P., Nauman, D., Burgess, D.,
Partain, J., and Litt, M. (2008) Clin. Transl. Sci., 1,
21-26.
291.Moolman, J. C., Corfield, V. A., Posen, B.,
Ngumbela, K., Seidman, C., Brink, P. A., and Watkins, H. (1997) J.
Am. Coll. Cardiol., 29, 549-555.
292.Cummins, B., Auckland, M. L., and Cummins, P.
(1987) Am. Heart J., 113, 1333-1344.
293.Katus, H. A., Looser, S., Hallermayer, K.,
Remppis, A., Scheffold, T., Borgya, A., Essig, U., and Geuss, U. (1992)
Clin. Chem., 38, 386-393.
294.Thygesen, K., Alpert, J. S., and White, H. D.
(2007) J. Am. Coll. Cardiol., 50, 2173-2195.
295.Mendis, S., Thygesen, K., Kuulasmaa, K.,
Giampaoli, S., Mahonen, M., Ngu Blackett, K., and Lisheng, L. (2011)
Int. J. Epidemiol., 40, 139-146.
296.Thygesen, K., Alpert, J. S., Jaffe, A. S.,
Simoons, M. L., Chaitman, B. R., White, H. D., Katus, H. A., Lindahl,
B., Morrow, D. A., Clemmensen, P. M., Johanson, P., Hod, H., Underwood,
R., Bax, J. J., Bonow, R. O., Pinto, F., Gibbons, R. J., Fox, K. A.,
Atar, D., Newby, L. K., Galvani, M., Hamm, C. W., Uretsky, B. F., Steg,
P. G., Wijns, W., Bassand, J. P., Menasche, P., Ravkilde, J., Ohman, E.
M., Antman, E. M., Wallentin, L. C., Armstrong, P. W., Januzzi, J. L.,
Nieminen, M. S., Gheorghiade, M., Filippatos, G., Luepker, R. V.,
Fortmann, S. P., Rosamond, W. D., Levy, D., Wood, D., Smith, S. C., Hu,
D., Lopez-Sendon, J. L., Robertson, R. M., Weaver, D., Tendera, M.,
Bove, A. A., Parkhomenko, A. N., Vasilieva, E. J., and Mendis, S.
(2012) Circulation, 126, 2020-2035.
297.Kavsak, P. A., Macrae, A. R., Yerna, M. J., and
Jaffe, A. S. (2009) Clin. Chem., 55, 573-577.
298.Reichlin, T., Hochholzer, W., Bassetti, S.,
Steuer, S., Stelzig, C., Hartwiger, S., Biedert, S., Schaub, N.,
Buerge, C., Potocki, M., Noveanu, M., Breidthardt, T., Twerenbold, R.,
Winkler, K., Bingisser, R., and Mueller, C. (2009) N. Engl. J.
Med., 361, 858-867.
299.Diercks, D. B., Peacock, W. F. T., Hollander,
J. E., Singer, A. J., Birkhahn, R., Shapiro, N., Glynn, T., Nowack, R.,
Safdar, B., Miller, C. D., Lewandrowski, E., and Nagurney, J. T. (2012)
Am. Heart J., 163, 74-80 e4.
300.Hof, D., Klingenberg, R., and Von Eckardstein,
A. (2013) Methods Mol. Biol., 963, 385-406.
301.Mueller, M., Vafaie, M., Biener, M.,
Giannitsis, E., and Katus, H. A. (2013) Circ. J., 77,
1653-1661.
302.Bima, A., and Sikaris, K. (2012) Intern.
Med. J., 42, Suppl. 5, 16-22.
303.Collinson, P. O. (2012) Postgrad. Med.
J., 88, 348-352.