Identification and Characterization of SlVKOR, a Disulfide Bond Formation Protein from Solanum lycopersicum, and Bioinformatic Analysis of Plant VKORs
Chun-Mei Wan1#, Xiao-Jian Yang1#, Jia-Jia Du1, Ying Lu1, Zhi-Bo Yu1, Yue-Guang Feng2, and Xiao-Yun Wang1,3*
1College of Life Sciences, Shandong Agricultural University, Taian 271018, Shandong, People’s Republic of China; fax: +86-538-8242217; E-mail: xyunwang@sdau.edu.cn; wcm.1121@163.com; xiaojian841@163.com; yuhuashi0633@163.com; enzyme@sdau.edu.cn; bojiezhenmei@163.com2Jinan Academy of Agricultural Sciences, Jinan 250300, Shandong, People’s Republic of China; E-mail: jnnyfeng@163.com
3State Key Laboratory of Crop Biology, Shandong Agricultural University, Taian 271018, Shandong, People’s Republic of China
# These authors contributed equally to this study.
* To whom correspondence should be addressed.
Received December 11, 2013; Revision received January 23, 2014
Homologs of vitamin K epoxide reductase (VKOR) exist widely in plants. However, only VKOR of Arabidopsis thaliana has been the subject of many studies to date. In the present study, the coding region of a VKOR from Solanum lycopersicum (JF951971 in GenBank) was cloned; it contained a membrane domain (VKOR domain) and an additional soluble thioredoxin-like (Trx-like) domain. Bioinformatic analysis showed that the first 47 amino acids in the N-terminus should act as a transit peptide targeting the protein to the chloroplast. Western blot demonstrated that the protein is localized in thylakoid membrane with the Trx-like domain facing the lumen. Modeling of three-dimensional structure showed that SlVKOR has a similar conformation with Arabidopsis and cyanobacterial VKORs, with five transmembrane segments in the VKOR domain and a typical Trx-like domain in the lumen. Functional assay showed that the full-length of SlVKOR with Trx-like domain without the transit peptide could catalyze the formation of disulfide bonds. Similar transit peptides at the N-terminus commonly exist in plant VKORs, most of them targeting to chloroplast according to prediction. Comparison of sequences and structures from different plants indicated that all plant VKORs possess two domains, a transmembrane VKOR domain and a soluble Trx-like domain, each having four conservative cysteines. The cysteines were predicted to be related to the function of catalyzing the formation of disulfide bonds.
KEY WORDS: Solanum lycopersicum, plant VKORs, suborganellar localization, bioinformatic analysis, disulfide bond formationDOI: 10.1134/S0006297914050083
Vitamin K epoxide reductase (VKOR) is an integral membrane protein that is widely present in the endoplasmic reticulum of mammalian animals; it reduces vitamin K to support the carboxylation and consequent activation of vitamin K-dependent proteins involved in blood coagulation, and it is the target of the most commonly used oral anticoagulant warfarin [1].
Homologs of VKOR are present in internal thylakoid membrane system of most photoautotrophic cyanobacteria. Different from animal VKORs, the protein consists of two distinct domains: an N-terminal membrane domain, homologous to the catalytic subunit of mammalian VKOR, and a C-terminal soluble Trx-like domain, homologous to the disulfide bond formation protein DsbA in Escherichia coli [2]. The fused protein was demonstrated to be involved in disulfide bond formation in oxygenic photosynthetic organisms and be required for their optimal photoautotrophic growth [3]. Mutant strains with deficiency of the encoding gene of VKOR showed significant growth impairment under photoautotrophic conditions at normal light intensity [3].
Arabidopsis VKOR has been identified recently; it is encoded by the At4g35760 gene and is similar with VKORs from cyanobacteria, which also comprise an integral domain and a soluble Trx-like moiety [4]. GFP-fusion experiments established that the 45 amino acids from the N-terminus act as a transit peptide targeting the protein to plastids [5, 6]. Through Western blot analysis, the protein was demonstrated to be localized in the thylakoids [6].
As for the topology of Arabidopsis VKOR (AtVKOR-DsbA), two similar models have been proposed. One predicted four transmembrane helices with one helix half-inserted into the membrane, with both the N- and the C-termini of the protein located in the oxidative lumen [6]. The other model implies five transmembrane helices with the N-terminus facing the reductive stroma and the C-terminus in the lumen [7]. Both models suggest that all functional cysteines and the Trx-like domain are located in the oxidative surroundings. These models are consistent with the 3D structure of Synechococcus VKOR revealed by X-ray crystallographic analysis [2].
It has been demonstrated that Arabidopsis VKOR can catalyze disulfide bond formation, similarly to E. coli DsbA and DsbB proteins [6, 7]. Eight conservative cysteines (four in the VKOR domain and four in the Trx-like domain) are involved in disulfide bond formation [6]. The bimodular Arabidopsis VKOR transfers electrons when catalyzing the formation of disulfide bonds. The Trx-like domain can function as an electron donor for its integral VKOR partner, while the fused VKOR or individual VKOR domain are equally effective in mediating dithiothreitol-dependent reduction of phylloquinone and menaquinone into their respective quinol forms [5]. The soluble Trx-like domain of Arabidopsis VKOR was found to have reduction, oxidation, and isomerization activities in vitro, and it can promote the formation of disulfide bonds in lumenal proteins [7, 8].
Homozygous Arabidopsis VKOR mutants have been demonstrated to display severely defective growth [7, 8]. Mutant lines display more severely stunted growth, lighter green, smaller size leaves, and shorter internodes compared with wild type plants [7]. Arabidopsis VKOR is required for the assembly of photosystem II and plays important roles in redox regulation and in homeostasis of reactive oxygen species [8].
Homologs of VKOR are widely found in plants. However, to our best knowledge, only a few studies have focused on VKOR of Arabidopsis thaliana to date. By sequence comparison, the coding sequence of VKOR from tomato (ABA41597) is quite different. Instead of being a two-domain protein, the sequence of ABA41597 only encodes a short transmembrane region according to the NCBI (National Center for Biotechnology Information) data. In the present investigation, we cloned the sequence of ABA41597 but found it to be a truncated version. So we used RACE (Rapid Amplification of cDNA Ends) to clone the full-length cDNA of VKOR from Solanum lycopersicum and further analyzed its function and location. Using bioinformatic methods, we analyzed plant VKORs to get further insight into their function, location, and structure in the plant kingdom.
MATERIALS AND METHODS
Plant materials, strains, and growth conditions. Yellow tomato selfing species (donated by Ms. Xizhen Ai) were grown in soil in a greenhouse under a 16/8 h light/dark cycle (120 µmol·m–2·s–1) at 26°C. Bacterial strains and their relevant genotypes are shown in Table 1. Cultures were grown in LB medium or M63 minimal medium supplemented with glucose and appropriate antibiotics [9].
Table 1. Strains used in this study
Cloning of the SlVKOR gene. Homologs of the VKOR amino acid sequence from tomato were identified through NCBI Blast search against that of Arabidopsis VKOR (At4g35760) registered in the GenBank library. Total RNA extracted from tomato leaf tissues was used as a template to amplify SlVKOR using primers P1 and P2 (Table 2). The sequencing result revealed that the gene had no termination codon. Then we designed the inner and outer primers and expanded SlVKOR 3′-end of the sequence (primers as P3) using RACE technology. Finally the full-length primers P4 and P5 were designed and the full length of SlVKOR was cloned using cDNA as template.
Table 2. Primers used in this study
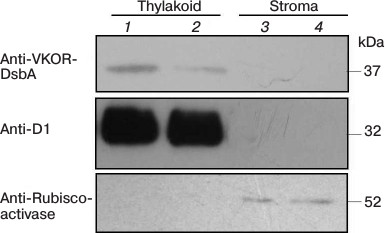
Suborganellar localization of SlVKOR. We first used online software to predict the location of SlVKOR and then separated thylakoid and stroma fractions from tomato plants. Fresh leaves of S. lycopersicum were homogenized in 10 volumes of lysis buffer (20 mM HEPES/KOH, pH 7.5, 10 mM EDTA) and incubated on ice for 30 min. Stroma and thylakoid fractions were obtained from the supernatant and the pellet after centrifugation (42,000g, 30 min, 4°C) [10]; their proteins were separated by SDS-PAGE, transferred to poly(vinylidene difluoride) membrane, and visualized using anti-VKOR (Genscript, China) and anti-Rubisco activase as a control for stromal proteins or anti-D1 (Santa Cruz Biotechnology, USA) as a control for thylakoid proteins.
Functional characterization of SlVKOR. The ability of SlVKOR to catalyze the formation of disulfide bonds was estimated with a motility complementation assay. Plasmid pTrc99a-SlVKOR encoding VKOR without the transit peptide was constructed and introduced into Dsb null strains as represented in Table 1. These strains were stabbed on M63 minimal medium with 0.26% agar and 50 µM isopropyl-β-D-thiogalactopyranoside (IPTG), which was specifically used for motility confirmation as described previously [11]. The strains were incubated at 30°C for 3 days. To determine whether the expression of the target gene can compensate for deficient E. coli disulfide bond formation, the diameters of the strains were measured to characterize colony size.
To further confirm the function of the gene in disulfide bond formation, β-galactosidase activity was tested as a complementation assay. Strains of Dsb null transformed with pTrc99a-SlVKOR plasmid encoding VKOR without transit peptide were streaked on M63 minimal medium with 1.5% agar coated with 20 µg/ml x-gal and 1 mM IPTG and preincubated at 37°C for 2-3 h. The strains were incubated at 37°C for 2 days. Blue color indicated β-galactosidase activity and no disulfide bond formation, whereas white color indicated disulfide bond formation, as described previously [12].
Bioinformatic analysis of plant VKORs. The sequences of plant VKORs were from the NCBI protein databases. Alignments of multiple amino acid sequences were carried out using the DNAMAN software. The location and transit peptides of plant VKORs were predicted using online software including PredSL (http://hannibal.biol.uoa.gr/PredSL/), TargetP1.1 (http://www.cbs.dtu.dk/services/TargetP/), and ChloroP1.1 (http://www.cbs.dtu.dk/services/ChloroP/). The TMHMM server 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and SWISS-MODEL online software [13-15] provided by ExPASy (http://www.expasy.org/) were used to predict the secondary and tertiary structures, respectively. A phylogenetic tree was built using the MEGA 5.2 software.
RESULTS
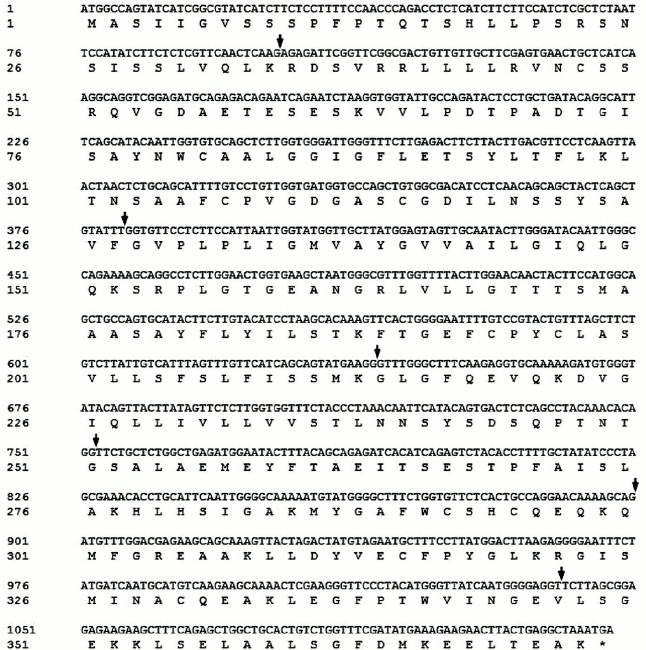
Full-length cDNA of SlVKOR was 1122 bp encoding 373 amino acids. We first used NCBI Blast analysis to search for sequences of plant VKORs. More than 20 sequences from different plants including Arabidopsis, Zea, Sorghum, Oryza, and others were found. Tomato VKOR was unusual because its coding region (ABA41597) was significantly shorter than the other plant VKORs. According to the sequence of ABA41597, up- and down-primers were designed to clone the ABA41597 sequence using the tomato cDNA library. Compared with the sequence of ABA41597, the sequence we cloned lacked an adenine base at position 639, which resulted in a frame shift after the 214th amino acid and no termination codon at the respective position. The results suggested that the gene we cloned might be incomplete. So RACE technology was used to clone the 3′-terminus. The coding sequence with 1122 bp was obtained, and these sequence data have been submitted to GenBank under accession number JF951971. It was named SlVKOR. Compared with the published tomato genome, the gene was located in chromosome 2 of the S. lycopersicum (cultivar Heinz 1706 chromosome 2), which contained seven exons with 1122 bp ORF and encoded 373 amino acids. The sequence of ABA41597 was a truncated version and missannotated, since sequence comparison indicated an extra adenine base in the fourth exon in ABA41597 with chromosome 2 of S. lycopersicum. The coding region of the ORF of SlVKOR and corresponding amino acid sequence are shown in Fig. 1, where arrows mark the positions of the introns.
Fig. 1. SlVKOR cDNA and the corresponding amino acid sequence. Arrows mark the positions of the introns.
A chloroplast transit peptide exists in the N-terminus of SlVKOR, targeting the protein to thylakoids. PredSL, TargetP, and ChloroP software were used to predict the location of SlVKOR. As shown in Table 3, the first 47 or 53 amino acids from the N-terminus should be a transit peptide targeting the protein to chloroplasts, which was similar with the Arabidopsis VKOR. To determine the suborganellar localization of SlVKOR, the fractions of stroma and thylakoid of chloroplast were subjected to Western blot analysis using antibody against Arabidopsis VKOR, and the purity of the fractions was monitored by the thylakoid D1 protein and the stromal rubisco activase. The SlVKOR protein was exclusively detected in the thylakoid fraction (Fig. 2), which demonstrated that SlVKOR is localized in the thylakoids, just like VKOR from A. thaliana [6].
Table 3. Plant VKORs subcellular
localization and signal peptide sequence prediction
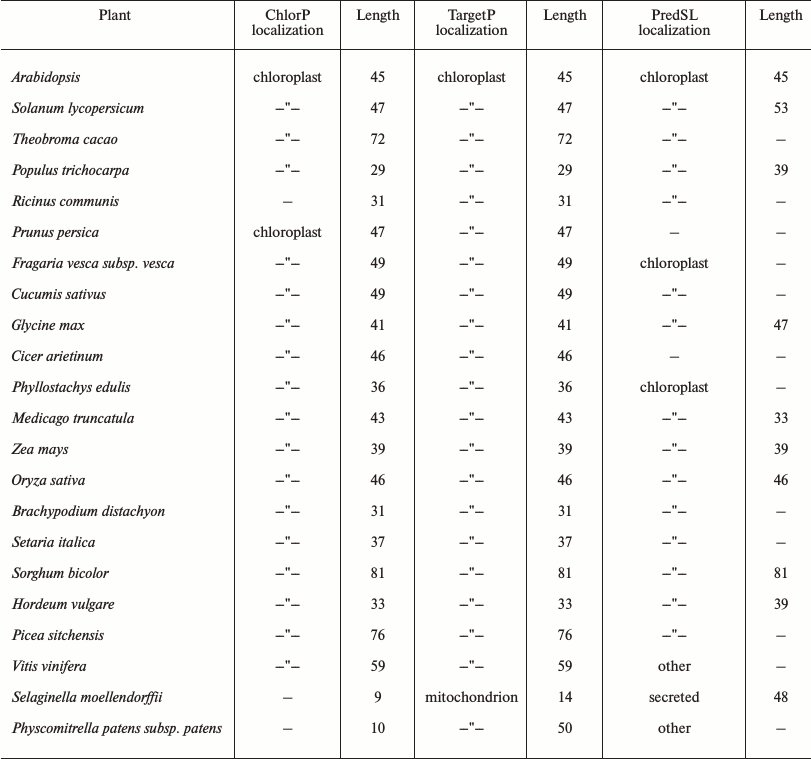
Fig. 2. Suborganellar localization of SlVKOR by Western blotting. Lanes 1 and 2, thylakoid; lanes 3 and 4, stroma.
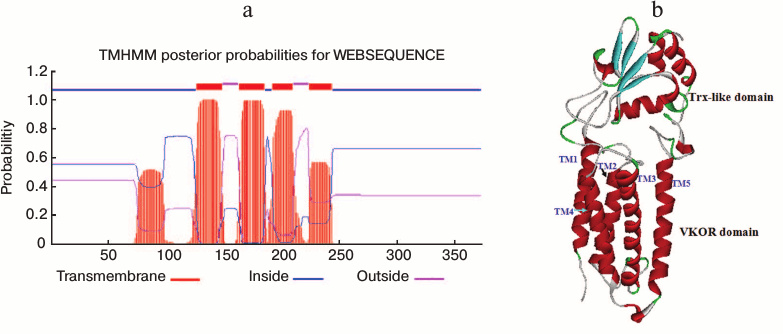
A transmembrane VKOR domain with four or five helices fused with a soluble Trx-like domain in SlVKOR. Online TMHMM software was used to predict the membrane topology of SlVKOR. The results showed that the SlVKOR without transit peptide was a fusion protein containing a transmembrane VKOR domain at the N-terminus and a Trx-like domain at the C-terminus (Fig. 3a). In the VKOR domain, three segments were predicted as transmembrane helices with higher than 90% probabilities: 126-148, 163-185, and 192-209. For the segment 224-243, the probability was higher than 60%. However, the probability of the first segment of the N-terminus, 51-107, to be helical was only about 50%. Arabidopsis VKOR has four transmembrane segments with an additional half inserted segment [6]. So the topology of SlVKOR may resemble that of the Arabidopsis VKOR.
Fig. 3. Membrane topology and three-dimensional structure of SlVKOR. a) Prediction of membrane topology of SlVKOR using TMHMM. The top line shows the predicted topology. The blue and pink curves indicate the probabilities for the inside and outside loops, respectively. The striped region shows the transmembrane helix. b) SlVKOR three-dimensional structure consisting of VKOR and Trx-like domain. Five transmembrane helices (TM1-5) of the VKOR domain are colored in red.
The three-dimensional structure of VKOR from Synechococcus has been determined by X-ray crystallography [2]. We simulated the tertiary structure of SlVKOR via homology modeling through SWISS-MODEL using the crystal three-dimensional structure diagram of VKOR from Synechococcus as a template (the sequences of the two VKORs were 26.2% identical). As shown in Fig. 3b, SlVKOR is an integral protein composing five helices in N-terminus and a Trx-like domain containing a central β-sheet with α-helices linked. This result is consistent with the TMHMM prediction of SlVKOR and the data on its Arabidopsis VKOR counterpart.
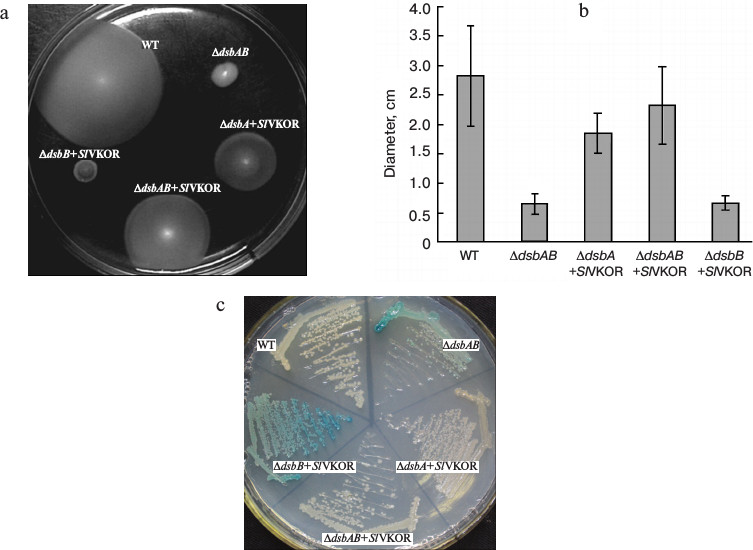
SlVKOR can promote the formation of disulfide bonds in E. coli. To assess the role of SlVKOR in promoting the formation of disulfide bonds, functional complementation assays were performed in disulfide bond formation-deficient strains. In normal E. coli, the formation of protein disulfide bonds is catalyzed by the DsbA and DsbB proteins [16, 17]. When the Dsb system was eliminated in dsbA, dsbB, or double dsbAdsbB mutants, the cell lost its motility on M63 minimal medium due to their inability to introduce a single disulfide bonds critical for proper folding of FlgI, a flagellar motor protein in the periplasmic space [18]. We used the diameter of each colony to quantitate the motility function. Expression of SlVKOR in ΔdsbAB and ΔdsbA mutant strains could restore the motility phenotype as shown in Figs. 4a and 4b. Similarly with Arabidopsis VKOR, SlVKOR could replace the function of membrane protein DsbB and Trx-like protein DsbA in E. coli, which implied that the VKOR domain of SlVKOR can promote the disulfide bond formation in FlgI through oxidizing the Trx-like domain. However, expression of SlVKOR in the strain of ΔdsbB had little effect on the compensation of the motility phenotype. A speculated explanation was that though the Trx-like domain of SlVKOR could oxidize FlgI, as previously described, the reduced E. coli DsbA in ΔdsbB mutant may disturb the effects of the Trx-like domain of SlVKOR.
Fig. 4. Complementation assay of dsb null strains. a) Complementation of the motility phenotype in dsb mutants by SlVKOR. The Δdsb strains (HK361(ΔdsbA), HK320(ΔdsbB), and HK329(ΔdsbAB)) transformed with pTrc99a-SlVKOR were cultured on M63 medium containing 0.26% agar and 0.2% glucose as carbon source. The spots representing different strains after 3 days of incubation at 30°C are shown. b) The diameters of the colonies shown in (a). c) Δdsb strains HK361(ΔdsbA), HK325(ΔdsbB), and Mer600(ΔdsbAB) transformed with pTrc99a-SlVKOR were streaked on a M63 medium X-Gal plate and incubated at 37°C for 2 days. Cells having defects in disulfide bond formation are stained with blue color.
To provide further evidence for the proposed function, we also detected the β-galactosidase activity of Dsb null strains containing β-galactosidase fused to a membrane protein MalF in the genome [12]. When mutant strains grew on minimal M63 medium, β-galactosidase would remain enzymatically active because of deficiency of the formation of disulfide bonds, as indicated by blue color, while a normal E. coli colony was white [19]. As shown in Fig. 4c, the colonies of ΔdsbA and ΔdsbAB mutants with transformed SlVKOR were white, indicating that the expression of SlVKOR could compensate the role in disulfide bond formation. However, in ΔdsbB the colony still showed blue color, which was consistent with the results indicated in the motility assay (Fig. 4a). From these results, we concluded that SlVKOR could catalyze disulfide bond formation in E. coli.
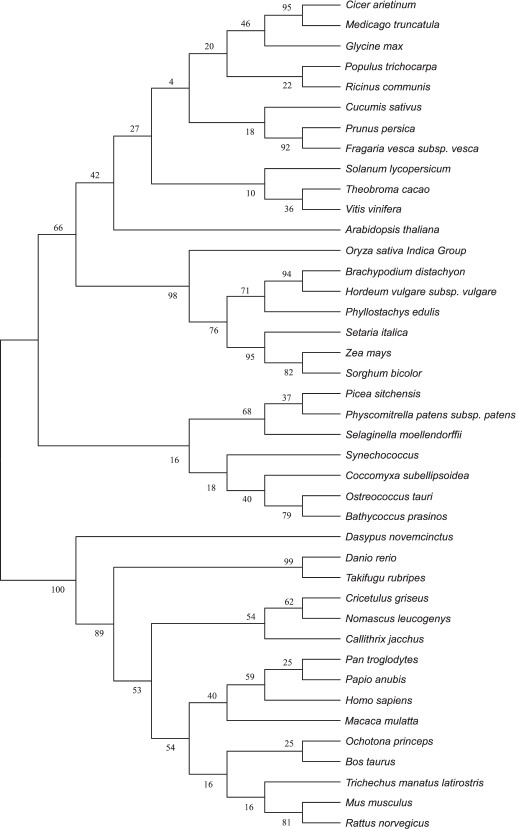
Amino acid sequences and evolutionary tree analysis of plant VKORs. To study the evolutionary relationships among different plant, cyanobacterial, and mammalian VKORs, a phylogenetic tree was constructed using the software MEGA 5.2 though the neighbor-joining method (Fig. 5). The result clearly show that plant VKORs are grouped as a main branch, quite different from the mammalian VKORs, while cyanobacterial VKORs are included in the group of plant VKORs as a subgroup, indicating their close genetic relationship.
Fig. 5. Phylogenetic tree of different VKORs.
Comparing the amino acid sequences of plant VKORs obtained from the NCBI protein database, we found that all these plant VKORs had transit peptides on the N-termini, and most of them targeted to chloroplasts with high probabilities (up to 60%) by predictions using three different methods (Table 3). Three different prediction methods provided the same or similar length of the transit peptides, mostly distributed in the 30-80 amino acids. The longest was VKOR from Sorghum bicolor with 81 amino acids, and that of Selaginella moellendorffii was the shortest one with only 9 amino acids. As to VKOR from S. moellendorffii, the ChlorP software could not give any prediction, while TargetP predicted a transit peptide targeting to chloroplast or mitochondrion with similar probabilities (0.23 and 0.29, respectively). However, PredSL predicted that the transit peptide was a secretion signal sequence. Experimental evidences are further needed to identify the accurate definition of different plant VKORs.
A multiple sequence alignment of the sequences of plant VKORs indicated high sequence identity (Fig. S1; see Supplement to this paper on the website of Biochemistry (Moscow) (http://protein.bio.msu.ru/biokhimiya)). All plant VKORs have a VKOR domain with five transmembrane helices and a soluble Trx-like domain. There are four conservative cysteines in the VKOR domain. The first pair of cysteines was located in a soluble region between two transmembrane helices, and the cysteines are separated by 6-8 amino acids. The second pair of cysteines in the Cys-x-x-Cys motif was located within or near the beginning of the fourth transmembrane helix. In the Trx-like domain, there were also four conservative cysteines with vicinal cysteines in a similar Cys-x-x-Cys motif and two other cysteines separated by about 14 amino acids. The positions of conservative cysteines are marked in Fig. S1. The vicinal cysteines were found in all Trx superfamily members and were incorporated in the active site of an oxidoreductase [20]. It has been experimentally demonstrated that all eight conservative cysteines in Arabidopsis VKOR are essential for the formation of disulfide bonds. Based on the above evidence, we predicted that the eight conservative cysteines in plant VKORs are also involved in an oxidoreduction reaction. In addition to the conservative cysteines, an S or T residue just after the first cysteine pair in plant VKORs might be an active-site residue, which was also conserved in an animal VKOR [4]. Two proline residues (marked in Fig. S1) near the last pair of cysteines in the Trx-like domain of plant VKORs may contribute to the stability and structure of VKOR, since most of the thioredoxin-superfamily members have these highly conserved cis-proline residues that play important roles in their functions and structures [21].
DISCUSSION
VKORs exist almost universally in plants, and most plant VKORs are predicted to be localized in the chloroplast. The phylogenetic tree shows that VKOR can be divided into two categories, animal VKORs and plant VKORs (including cyanobacteria) (Fig. 5). Most VKORs from mammals are transmembrane proteins located in the integral endoplasmic reticulum membrane with three or four predicted transmembrane α-helices [22, 23]. Unlike mammalian VKOR, photoautotrophic cyanobacterial VKORs exist in the internal thylakoid membrane system [2, 3]. Likewise, Arabidopsis VKOR has been confirmed to be localized in the thylakoids of chloroplasts [5, 6]. In this study, it has also been demonstrated that VKOR from tomato also exists in the thylakoids. From the protein database of NCBI, more than 20 plant VKORs have been searched out from different plant species. Bioinformatic analysis showed that all these plant VKORs have transit peptides at the N-termini with about 30-80 amino acids and most plant VKORs are predicted to be localized in the chloroplast (Table 3). Though plant VKORs are likely located in chloroplasts, they are nuclear encoded, synthesized on the ribosomes in the cytoplasm, and imported into the organelle. The transit peptides control the targeting process. On the stroma side of the envelope, the chloroplast-targeting transit peptides are removed. VKORs refold and integrate into the thylakoid membrane. The results of the present study show that the full-length of SlVKOR without its transit peptide is able to catalyze the formation of disulfide bonds in E. coli, consistent with the counterpart of Arabidopsis VKOR. With the transit peptide, SlVKOR and Arabidopsis VKOR do not possess the function of disulfide bond formation [6]. This implies the transit peptides of plant VKORs may affect their conformation and activity.
All plant VKORs may have similar membrane topology and similar residues in the active site. Membrane topology of VKOR from Synechococcus illustrated that the VKOR domain contains five transmembrane helices (TM), and TM5 passes through the membrane to connect with the fused periplasmic Trx-like domain according to the crystal structure [2]. Using the alkaline phosphatase sandwich fusion technique, the topological arrangement of Arabidopsis VKOR was determined. Its integral membrane domain also contains five transmembrane helices at the N-terminus, but the first helix is partly inserted into the thylakoid membrane, which is consistent with the prediction of TMHMM [6, 7]. The SlVKOR sequence is 50% identical to that of Arabidopsis VKOR, and the membrane topology of the VKOR domain of SlVKOR has also been predicted to be similar to that of the Arabidopsis VKOR. Using the TMHMM software to predict plant VKORs, the results showed that most plant VKORs can have a similar membrane topology with that of Arabidopsis (data not shown).
Similar topology in plant VKORs determines the arrangement of conservative cysteines in the corresponding positions. Mutations of any of these cysteines of VKOR from Synechococcus sp. and Arabidopsis led to the loss of VKOR function. These results suggest the conservative cysteines are essential active site residues involved in catalyzing oxidoreduction reactions.
In contrast to animal and bacteria VKORs, an additional Trx-like domain is fused with the VKOR domain in plant VKORs. In the Trx proteins, such as DsbA in E. coli, there is only one pair of cysteines in a Cys-x-x-Cys motif [24]. The Trx-like domains of plant VKORs also share this active site motif. Studies have shown that the Cys-x-x-Cys motif in all the members of the Trx superfamily is necessary for the oxidoreduction reaction. Mutation of either cysteine of this pair in the VKOR from Synechococcus abolished disulfide bridge formation [2]. However, the Trx-like domain of plant VKORs has two additional cysteines in a separated form separated by 14 amino acids. In Arabidopsis, all these four cysteines were found to be essential for promoting disulfide bond formation in vivo [6], but the separated cysteines do not affect the activity of the Trx-like domain in vitro (unpublished data in our lab). Two adjacent active site cysteines (Cys-x-x-Cys) in the Trx-like domain may directly promote electron transfer in a thiol-disulfide exchange reaction, and another pair of separated cysteines probably has a role in maintaining structure [2].
VKORs from S. lycopersicum and A. thaliana can catalyze disulfide bond formation, and all plant VKORs may be involved in electron transfer. Arabidopsis VKOR and SlVKOR have been determined to catalyze the formation of disulfide bonds in vivo. Also, the Trx-like domain of Arabidopsis VKOR has been demonstrated to be active in catalyzing disulfide bond formation in chloroplast proteins in vitro, for example, PsbO and FKBP13. PsbO is critical for PSII assembly and activity [7], and FKBP13 is a thylakoid lumenal peptidyl-prolyl isomerase. Both proteins have disulfide bonds essential for their activities [8]. According to the topology of plant VKORs, the Trx-like domains exist on the lumenal side of the thylakoid, so some lumenal proteins with disulfide bonds may be targets of plant VKORs.
The soluble Trx-like domain of plant VKORs directly catalyzes the formation of disulfide bonds of substrates and functions as an electron donor for its integral VKOR domain in vitro [5, 8]. The VKOR domain or Arabidopsis full-length VKOR can mediate the reduction of phylloquinone and menaquinone into their respective quinol forms [5]. They display strict specificity for the quinone forms of naphthoquinone conjugates [5]. Unlike mammalian VKORs, the Arabidopsis VKOR does not reduce phylloquinone epoxide into phylloquinone, and it is resistant to inhibition by warfarin [5].
The VKOR gene should be an important functional gene in plants since its absence severely affects the growth of Arabidopsis [7, 8]. It has been demonstrated that Arabidopsis VKOR is required for the assembly of PSII and is involved in redox regulation and reactive oxygen species homeostasis. As for plant VKORs, it is an intriguing prospect for future research to understand the mechanism of oxidative folding in the thylakoid lumen.
We thank Dr. Jon Beckwith of Harvard Medical School (Boston, MA, USA) for the generous gift of DHB4 and dsb mutant strains.
This work was supported by the Special Research Fund of Public Welfare of China Agricultural Ministry (201303093). The authors declare no conflict of interests.
REFERENCES
1.Cain, D., Hutson, S. M., and Wallin, R. (1997)
J. Biol. Chem., 272, 29068-29075.
2.Li, W., Schulman, S., Dutton, R. J., Boyd, D.,
Beckwith, J., and Rapoport, T. A. (2010) Nature, 463,
507-512.
3.Singh, A. K., Bhattacharyya-Pakrasi, M., and
Pakrasi, H. B. (2008) J. Biol. Chem., 283,
15762-15770.
4.Goodstadt, L., and Ponting, C. P. (2004) Trends
Biochem. Sci., 29, 289-292.
5.Furt, F., Oostende, C., Widhalm, J. R., Dale, M.
A., Wertz, J., and Basset, G. J. (2010) Plant J., 64,
38-46.
6.Feng, W. K., Wang, L., Lu, Y., and Wang, X. Y.
(2011) FEBS J., 278, 3419-3430.
7.Karamoko, M., Cline, S., Redding, K., Ruiz, N., and
Hamel, P. P. (2011) Plant Cell, 23, 4462-4475.
8.Lu, Y., Wang, H. R., Li, H., Cui, H. R., Feng, Y.
G., and Wang, X. Y. (2013) Plant Cell Rep., 32,
1427-1440.
9.Guzman, L. M., Barondess, J. J., and Beckwith, J.
(1992) J. Bacteriol., 174, 7716-7728.
10.Armbruster, U., Zuhlke, J., Rengstl, B., Kreller,
R., Makarenko, E., Ruhle, T., Schunemann, D., Jahns, P., Weisshaar, B.,
Nickelsen, J., and Leister, D. (2010) Plant Cell, 22,
3439-3460.
11.Jander, G., Martin, N. L., and Beckwith, J.
(1994) EMBO J., 13, 5121-5127.
12.Tian, H., Boyd, D., and Beckwith, J. (2000)
Proc. Natl. Acad. Sci. USA, 97, 4730-4735.
13.Arnold, K., Bordoli, L., Kopp, J., and Schwede,
T. (2006) Bioinformatics, 22, 195-201.
14.Schwede, T., Kopp, J., Guex, N., and Peitsch, M.
C. (2003) Nucleic Acids Res., 31, 3381-3385.
15.Guex, N., and Peitsch, M. C. (1997)
Electrophoresis, 18, 2714-2723.
16.Bardwell, J. C., McGovern, K., and Beckwith, J.
(1991) Cell, 67, 581-589.
17.Bardwell, J. C., Lee, J. O., Jander, G., Martin,
N., Belin, D., and Beckwith, J. (1993) Proc. Natl. Acad. Sci.
USA, 90, 1038-1042.
18.Dailey, F. E., and Berg, H. C. (1993) J.
Bacteriol., 175, 3236-3239.
19.Kadokura, H., Bader, M., Tian, H., Bardwell, J.
C., and Beckwith, J. (2000) Proc. Natl. Acad. Sci. USA,
97, 10884-10889.
20.Goyer, A., Haslekas, C., Miginiac-Maslow, M.,
Klein, U., Le Marechal, P., Jacquot, J. P., and Decottignies, P. (2002)
Eur. J. Biochem., 269, 272-282.
21.Martin, J. L. (1995) Structure, 3,
245-250.
22.Rost, S., Fregin, A., Ivaskevicius, V.,
Conzelmann, E., Hortnagel, K., Pelz, H. J., Lappegard, K., Seifried,
E., Scharrer, I., Tuddenham, E. G., Muller, C. I., Strom, T. M., and
Oldenburg, J. (2004) Nature, 427, 537-541.
23.Oldenburg, J., Bevans, C. G., Muller, C. R., and
Watzka, M. (2006) Antioxid. Redox Signal, 8, 347-353.
24.Kadokura, H., and Beckwith, J. (2010)
Antioxid. Redox Signal, 13, 1231-1246.
25.Kadokura, H., and Beckwith, J. (2002) EMBO
J., 21, 2354-2363.
26.Eser, M., Masip, L., Kadokura, H., Georgiou, G.,
and Beckwith, J. (2009) Proc. Natl. Acad. Sci. USA, 106,
1572-1577.
Supplementary Figure S1 (PDF)