REVIEW: Mammalian Hibernation and Regulation of Lipid Metabolism: A Focus on Non-coding RNAs
D. Lang-Ouellette, T. G. Richard, and P. Jr. Morin*
Department of Chemistry and Biochemistry, Université de Moncton, 18 Antonine-Maillet avenue, Moncton, New Brunswick, Canada E1A 3E9; fax: (506) 858-4541; E-mail: pier.morin@umoncton.ca* To whom correspondence should be addressed.
Received May 30, 2014; Revision received June 10, 2014
Numerous species will confront severe environmental conditions by undergoing significant metabolic rate reduction. Mammalian hibernation is one such natural model of hypometabolism. Hibernators experience considerable physiological, metabolic, and molecular changes to survive the harsh challenges associated with winter. Whether as fuel source or as key signaling molecules, lipids are of primary importance for a successful bout of hibernation and their careful regulation throughout this process is essential. In recent years, a plethora of non-coding RNAs has emerged as potential regulators of targets implicated in lipid metabolism in diverse models. In this review, we introduce the general characteristics associated with mammalian hibernation, present the importance of lipid metabolism prior to and during hibernation, as well as discuss the potential relevance of non-coding RNAs such as miRNAs and lncRNAs during this process.
KEY WORDS: hibernation, hypometabolism, lipid synthesis, fatty acid degradation, microRNAs, long non-coding RNAsDOI: 10.1134/S0006297914110030
Abbreviations: lncRNAs, long non-coding RNAs; miRNAs, microRNAs.
Several animals when confronted with environmental challenges such as
cold temperatures or food restriction will enter a hypometabolic state
in which they will remain until favorable conditions return. Natural
strategies of hypometabolism are numerous and include, but are not
limited to, aestivation, diapause, daily torpor, and mammalian
hibernation. Hibernators can sustain cold ambient conditions for
prolonged periods of time, and the changes associated with the process
of hibernation are well characterized. Besides reduced metabolic rate,
mammalian hibernation is also associated with a reduction in heart
rate, blood flow, and oxygen consumption [1].
Significant metabolic changes are observed including a switch from a
carbohydrate-based metabolism to one that leverages stored lipids [2]. At the biochemical and molecular levels, a
plethora of modifications have been documented in hibernating mammals
including reversible protein phosphorylation, differential expression
of selected transcripts and proteins, as well as modulation of
non-coding RNAs [3, 4]. This
review discusses the current knowledge associated with mammalian
hibernation with a particular focus on the potential role of non-coding
RNAs involved in the regulation of lipid metabolism during this
hypometabolic state.
MAMMALIAN HIBERNATION: A “COOL” PROCESS?
Natural models of hypometabolism. Metabolic rate depression is essential for survival of numerous species confronted with extreme environmental conditions and notably underlies aestivation, diapause, and hibernation. A series of concerted physiological, biochemical, and molecular levels are observed during these hypometabolic states. Such processes are usually associated with basal metabolic rate reduction to levels between 5 and 40% of those experienced by resting animals [5]. Aestivation, for example, is a state of aerobic torpor leveraged by species that are usually confronted with arid conditions. Well-characterized aestivating vertebrates notably include amphibians such as the spadefoot toad Scaphiopus couchii and mollusks such as the land snail Otala lactea [6]. An important element for long-term survival of species that aestivate is the accumulation, conservation, and usage of fuel reserves. Aerobic oxidation of lipid reserves is employed by species such as S. couchii as energy source during aestivation [7]. Hypometabolism is also observed in species that undergo diapause such as cold-hardy insects. Diapause is undertaken by species as a developmental response to changing seasons and environmental conditions. This genetically programmed phenomenon allows species to face seasonally recurring environmental stresses as well as to initiate and regulate growth and development in favorable environmental conditions [8]. Although not its exclusive role, diapause often positively influences cold hardiness and overwintering survival in insects. Diapause-associated changes, including synthesis of cryoprotectants and changes in membrane lipid composition, ultimately help with protection against cold stress [9, 10]. Not unlike diapausing insects, mammalian hibernators also have to confront cold temperatures for extended periods of time. Several groups of mammals have been reported to hibernate including marsupials, rodents, bats, and primates [11]. A plethora of changes are undertaken to enter, maintain, and exit the hypometabolic state associated with hibernation. A strong reliance on lipid catabolism as the primary fuel source as well as differential expression of selected, torpor-responsive, transcripts is notably observed [12, 13]. As a well-characterized model of hypometabolism and a particular research interest of our laboratory, the focus of the review will now be directed towards mammalian hibernation.
Physiological changes associated with mammalian hibernation. The changes associated with mammalian hibernation are numerous and well documented. A period of hyperphagia that leads to body fat increase is frequently experienced in animals prior to the hibernating season as significant triglyceride synthesis and storage is undertaken in white adipose tissue [12]. These are essential, as lipid metabolism is an essential fuel source for hibernating species. A typical hibernating season consists in a series of multiple torpor bouts that can last up to several weeks and that are separated by brief arousal periods [14]. The duration of the torpor periods differs between species and is influenced by ambient temperatures [15]. Small mammalian hibernators, such as the thirteen-lined ground squirrel Ictidomys tridecemlineatus or the little brown bat Myotis lucifugus, can reduce vital functions significantly during torpor. Core body temperature is reduced to levels nearing the freezing point while heart rate can fall to 10 beats/min from the 350-400 beats/min measured in euthermic animals [11]. Another fascinating physiological feat observed in hibernating mammals includes a considerable reduction in breathing rate [16]. From a metabolic standpoint, mammalian hibernators can suppress their metabolism by up to 99% during torpor when compared with the basal metabolic rate measured in euthermic animals [11]. It is estimated that hibernators can save up to 90% of the energy they would otherwise require if they were to remain active during the cold season [17].
Molecular switches underlying metabolic depression in torpid animals. Numerous biochemical and molecular changes underlie metabolic rate depression during mammalian hibernation. Energy-consuming cellular processes including transmembrane ion transport and transcription are significantly suppressed in torpid animals [18, 19]. Interestingly, the rapid and reversible physiological changes associated with the torpor-arousal cycles experienced by hibernators must be supported by an equally dynamic system at the molecular level. Reversible protein phosphorylation has emerged over time as a crucial posttranslational mechanism involved in regulating key molecular players involved in metabolic rate depression. Phosphorylation-mediated pyruvate dehydrogenase activity inhibition, leading to carbohydrate catabolism suppression, is well documented in different models of hypometabolism [20, 21]. Na+,K+-ATPase activity is another example of an enzyme with an activity that is strongly influenced by phosphorylation in mammalian hibernation [18]. Despite the importance of reversible protein phosphorylation for metabolic rate suppression, additional posttranslational mechanisms including sumoylation and ubiquitination are modulated to various degrees in hypometabolism [22, 23]. Protein synthesis, an important ATP-consuming process, is significantly inhibited during hibernation. A study performed in brain of torpid and euthermic ground squirrels showed that the translational rate during hibernation could be as low as 0.04% when compared with the one measured in euthermic squirrels [24]. An in vivo study of protein synthesis in hibernating Syrian hamsters Mesocricetus auratus demonstrated significant reduction of this process in several organs [25]. Posttranslational modifications of initiation and elongation factors of protein synthesis, such as eIF2α and eEF2, underlie in part the changes observed in translational rate during torpor [24]. Significant alterations in the activity of the mTOR signaling cascade, a key pathway linked to protein synthesis, have been reported in hibernating skeletal muscle of ground squirrels and could also account for the reduced translational rates observed in torpor [26]. More recently, studies have highlighted the potential importance of the small non-coding RNAs microRNAs (miRNAs) in regulating expression of selected transcripts in different models of hypometabolism providing yet another mean of translational control for hibernating mammals [4]. A particular emphasis will be placed in the final section of this review on the role that miRNAs can play during hibernation.
LIPID METABOLISM AND HIBERNATION
Lipid synthesis and degradation: fueling hibernation. Mammalian hibernators cycle on an annual basis between active and hibernating periods. Energy requirements associated with these two states differ significantly. Active hibernators spend a significant portion of their time eating and accumulating fat stores, primarily as triglycerides, that contribute to a considerable gain in body mass [12]. The hibernating golden-mantled ground squirrels Callospermophilus lateralis can spend up to 57% of their active hours feeding prior to hibernation [27]. Elevated activities of enzymes involved in triglycerides synthesis, such as diacylglycerol acyltransferase, have been reported in white adipose tissue of summer active golden-mantled ground squirrels and of summer active yellow-bellied marmots Marmota flaviventris [28, 29]. These studies further demonstrated that enzymes involved in fatty acid synthesis possess higher activities in selected tissues of summer active animals when compared with torpid animals. In addition, recent work performed in tissues of hibernating golden-mantled ground squirrels showed significant overexpression in torpor of the phosphorylated and inactive form of acetyl-CoA carboxylase, an enzyme that catalyzes an essential step in fatty acid synthesis [30]. While in torpor, hibernators address their energy demands primarily via β-oxidation of fatty acids sourced from white adipose tissue [12]. Acyl-CoA dehydrogenase, the enzyme catalyzing the initial step in β-oxidation, exhibits marked induction in its activity in brown adipose tissue and white adipose tissue of the hibernating jerboa Jaculus orientalis [31]. Increased fatty acid plasma concentration has also been reported in selected hibernating models such as M. flaviventris, suggesting increased lipolysis in torpid animals [32, 33]. In addition to being an important fuel source for hibernators, it is also important to emphasize the role of fatty acids, and in particular polyunsaturated fatty acids, in preserving the fluidity of membrane phospholipids at low body temperatures [34]. Several studies have demonstrated that hibernating mammals or cold-water fish, for example, notably displayed a higher proportion of polyunsaturated fatty acids in their body fats than species in warmer climates [35, 36].
Regulatory roles of lipids during hibernation. Besides being an important source of energy and essential molecules underlying membrane fluidity at low temperatures, lipids can also influence a myriad of torpor-related processes via diverse signaling and regulatory functions. For example, fatty acids can act as ligands and activate the nuclear receptors peroxisome proliferator-activated receptors (PPARs) [37]. These transcription factors regulate lipid metabolism via differential expression of numerous target genes. Target genes such as pyruvate dehydrogenase kinase isozyme-4 and apolipoprotein A-I can impact lipid metabolism as well as ketone body formation, respectively, and are differentially expressed in hibernation [38, 39]. PPAR protein levels are also modulated in several models of hypometabolism including PPARγ in the hibernating bat M. lucifugus, ground squirrel I. tridecemlineatus, and jerboa J. orientalis [40-42]. PPARα is differentially expressed in hibernating J. orientalis and has been put forward as a key controller of torpor [43, 44]. In addition to PPARs, additional nuclear receptors such as the retinoid-related orphan receptor alpha (RORα) and the liver X receptors (LXRs) can regulate expression of genes involved in lipid metabolism and also warrant closer attention for their potential role during torpor. LXRs target genes including the apolipoprotein E and the ATP-binding cassette transporter AI (ABCA1) can affect cholesterol homeostasis [45]. Plasma and tissue cholesterol levels vary significantly during hibernation, and LXR activity is thought to underlie this balance [46, 47]. RORα, a nuclear receptor that can influence circadian rhythms and metabolism through the expression of its target genes, regulate key molecular players involved in mammalian hibernation [48]. Interestingly, cholesterol sulfate, a RORα ligand, is altered in hibernating ground squirrels as they undergo a hibernation cycle [49]. Whether as an integral part of the energy source associated with torpor, as an important component of biological membranes at low temperatures, or as signaling molecules with far-reaching translational impacts, lipids are undoubtedly essential to several facets of mammalian hibernation. Accordingly, molecules that can impact the synthesis, degradation, or signaling processes associated with lipids are of significant interest.
FAMILIES OF NON-CODING RNAs
Cold-associated miRNAs. MiRNAs are examples of molecules with such ubiquitous implications. MiRNAs are short, approximately 21 nucleotides in length, non-coding transcripts that can bind to target mRNAs and repress their translation. MiRNA biogenesis has been extensively described elsewhere and will not be the scope of this review [50]. It is estimated that expression of more than 60% of all protein-coding genes can be regulated by miRNAs, which highlights the far-reaching capabilities of these molecules in modulating different cellular processes [51, 52]. A growing body of evidence has positioned miRNAs as molecules involved in the regulation of hypometabolic models including diapause in the flesh fly Sarcophaga bullata [53], aestivation in the sea cucumber Apostichopus japonicus [54], and cold hardiness in insects [55, 56]. The freeze-tolerant wood frog Rana sylvatica displayed elevated miR-21 levels and reduced miR-16 levels in skeletal muscle during freezing [57]. Subsequent work in liver of frozen R. sylvatica reported elevated levels of miR-26a, miR-126, and miR-217, three miRNAs that can regulate PTEN expression and thus potentially contribute to anti-apoptotic functions via Akt activation during freezing [58]. Differential expression of miRNAs, including miR-1a-1 and miR-34a, was also presented in frozen hepatopancreas and foot muscle of the freeze-tolerant land snail Littorina littorea [59]. Several studies have reported modulated miRNAs during mammalian hibernation. Pioneering work on this topic demonstrated differentially expressed miRNAs in several tissues of the hibernating I. tridecemlineatus [60]. Subsequent reports on miRNA expression in different hibernating species were presented in recent years and have contributed markedly in positioning these molecules as important molecular players in torpor. The little brown bat M. lucifugus notably exhibits elevated levels of eight miRNA species in hibernating pectoral muscle tissue [61]. Interestingly, transcript targets of these miRNAs were shown to code for muscle-specific proteins, including FoxO3a and SMAD7, suggesting a potential relevance for bat muscle preservation during torpor. A follow-up study evaluating miRNA expression in euthermic and hibernating brain of M. lucifugus also demonstrated eight overexpressed miRNAs with probable impact on axon guidance and focal adhesion processes in torpor [62]. Genomic analysis performed in the Arctic ground squirrel Spermophilus parryii revealed numerous miRNAs such as miR-451 and miR-486 that were differentially expressed at different stages of hibernation when compared with control animals [63]. While the exact role of these miRNAs remains to be determined in torpid S. parryii, such molecules likely impact diverse processes including cell growth. Levels of miR-106b, a miRNA that can influence HIF-1α expression, were also shown to be reduced in skeletal muscle of torpid M. lucifugus and I. tridecemlineatus [64]. A microarray-based approach that assessed miRNA levels in the brain of euthermic and hibernating I. tridecemlineatus revealed down-regulation of miRNAs that were part of the miR-182 and miR-200 families of miRNAs [65]. Members of these families, including miR-200a, miR-200b, and miR-200c, as well as miR-182, were all involved in regulating the expression of ubiquitin-like modifiers, which underlie cellular response to various stresses. As evidenced by these studies, a growing list of differentially expressed miRNAs in different animal species that can confront and survive cold temperatures is being built. This family of cold-associated miRNAs, loosely referred to as “CryomiRs” [4], is likely to expand in the near future.
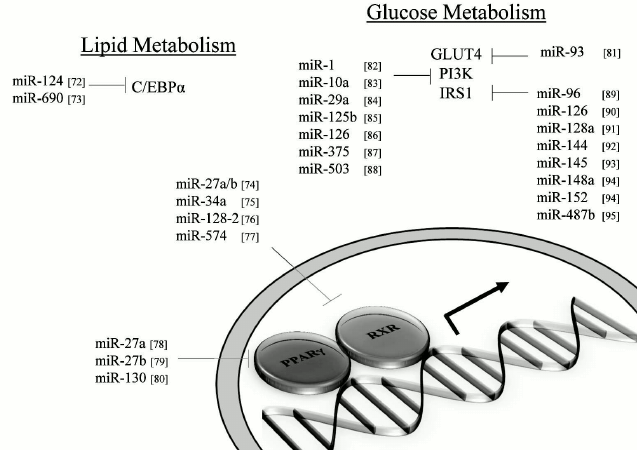
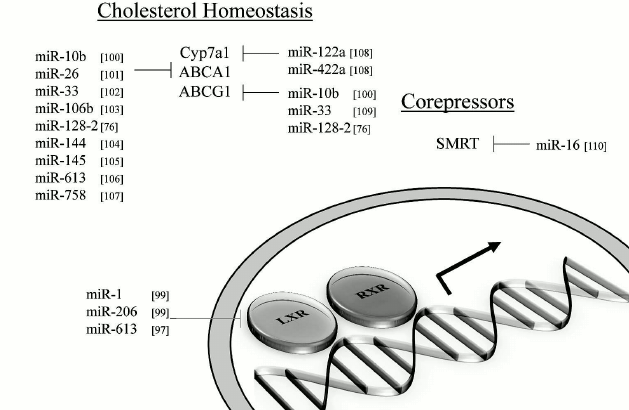
MicroRNAs and impact on lipid metabolism. MiRNAs that can modulate key metabolic cascades of hibernation such as those associated with lipid metabolism are foreseen to join the list of “CryomiRs”. Examples abound, in non-hypometabolic models, of interactions between miRNAs and target transcripts that code for proteins involved in lipid metabolism. For example, miR-33b can target key enzymes involved in fatty acid oxidation such as carnitine palmitoyltransferase 1A and the alpha subunit of AMP kinase (AMPKα) in human hepatic cell lines [66]. AMPKα phosphorylation is also influenced by miR-144 and miR-451 notably via translational regulation of key mediators in this pathway such as MO25α and acetyl-CoA carboxylase [67]. Table 1 presents miRNAs that can target AMPK as well as upstream regulators of this cascade. Interestingly, AMPK and acetyl-CoA carboxylase protein and activity levels are modulated to different extents in hibernating ground squirrels [30, 70], which raises the question of whether or not these miRNAs participate in such control. It is of particular interest to note that miR-144 is strongly expressed in liver tissue of torpid S. parryii [63]. MiR-195, a miRNA that targets fatty acid synthase (FAS) in human osteosarcoma cells, is elevated in hibernating liver tissue of I. tridecemlineatus [71]. The same study demonstrated that protein levels of FAS, an enzyme involved in fatty acid synthesis, were down-regulated under the same conditions, suggesting a potential importance for the miR-195–FAS axis in hibernating ground squirrels. As mentioned above, PPARγ is a key transcription factor modulated in selected models of hibernation and that can regulate the expression of a plethora of target genes involved in lipid metabolism. Numerous miRNAs can regulate the expression of PPARγ and of genes under its control. An overview of the potential miRNA-mediated regulatory nodes associated with PPARγ and its target genes is presented in Fig. 1. MiR-27 and miR-130 for instance can regulate PPARγ expression in mice and human adipocytes, respectively [78-80]. MiR-126 also plays an important role in PPARγ-mediated gene expression via regulation of two PPARγ-associated genes, phosphatidylinositol 3-kinase (PI3K) and insulin receptor substrate-1 (IRS1) [86, 90]. PPARα, also differentially expressed in selected hibernators, is targeted by miR-22 in mouse cardiomyocytes [96]. Expression of the nuclear receptor liver X receptor α (LXRα) is regulated by miRNAs such as miR-1, miR-206, and miR-613 in human and mouse models [97-99]. MiRNAs that can target LXRα and its target genes are presented in Fig. 2. Interestingly and despite the relative lack of expression data on these molecules in hibernating models, the sterol regulatory element-binding proteins (SREBPs) transcription factors are crucial regulators of lipid homeostasis [111]. Multiple evidences of miRNAs that can regulate the expression of SREBPs and their target genes have been put forward. These interactions are summarized in Table 2. MiR-185 and miR-342 have notably been shown to inhibit both SREBP1 and SREBP2 expression, leading to negative regulation of their target genes in human prostate cancer cells [112]. MiR-33 was shown to impact SREBP1 expression in a mouse model and that this regulation had potential implications in obesity [113]. Overall, several transcripts coding for proteins involved in lipid metabolism have been identified as miRNA targets. Validating and understanding the relevance of these miRNA-transcript interactions in natural models of hypometabolism will need to be undertaken next to have a clearer idea of the extent to which miRNAs play a role in such species.
Table 1. MiRNA regulation of AMPK signaling.
MiRNAs with reported transcript targets related to AMPK and upstream
signaling
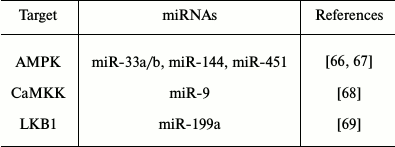
Note: AMPK, AMP-activated protein kinase; CaMKK,
Ca2+/calmodulin-dependent protein kinase kinase; LKB1,
serine/threonine kinase 11.
Fig. 1. PPARγ and related miRNAs. MiRNA-mediated regulation of PPARγ and selected target genes. Note: C/EBPα, CCAAT/enhancer-binding protein alpha; GLUT4, glucose transporter 4; IRS1, insulin receptor substrate-1; PI3K, phosphatidylinositol 3-kinase; PPARγ, peroxisome proliferator-activated receptor gamma; RXR, retinoid X receptor.
Fig. 2. Liver X receptors and related miRNAs. Regulation of LXR and target genes by miRNAs. Note: ABCA1, ATP-binding cassette transporter AI; ABCG1, ATP-binding cassette transporter GI; LXR, liver X receptor; RXR, retinoid X receptor; SMRT, silencing mediator of retinoic acid and thyroid hormone receptor.
Table 2. SREBP-associated miRNAs. MiRNAs
with reported transcript targets associated with SREBPs and target
genes
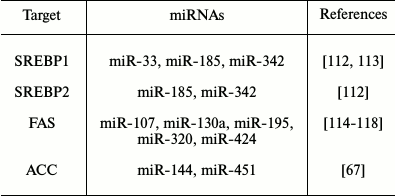
Note: SREBP, sterol regulatory element-binding protein; ACC, acetyl-CoA
carboxylase; FAS, fatty acid synthase.
Long non-coding RNAs: additional “CryomiRs’’? The surface has barely been scratched when it comes to elucidating the impact of miRNAs in torpor, and yet another family of non-coding RNAs emerges as another layer of regulation; the long non-coding RNAs (lncRNAs). LncRNAs are non-coding transcripts that are longer than 200 nucleotides [119]. More than a thousand lncRNAs have been identified to date in mammals and other vertebrates and this number is increasing [120, 121]. Not surprisingly, the list of functions associated with this family of molecules is also expanding. Some of the better known functions associated with lncRNAs include epigenetic regulation of gene expression via recruitment of histone-modifying complexes. The lncRNA Xist can recruit key factors that can contribute to histone methylation and ubiquitination [122]. LncRNAs, including Evf-2, can also act as co-activators of proteins involved in transcriptional regulation [123]. LncRNAs can even affect the subcellular localization of proteins and influence their activity. For example, the lncRNA NRON can affect the cytoplasm-nucleus trafficking of the NFAT transcription factor [124]. With such diverse functions, it is not surprising that lncRNAs can impact different cellular processes including lipid metabolism. A recent study has highlighted a potential role for the lncRNAs colorectal neoplasia differentially expressed (CRNDE) transcripts in colorectal cancer cells [125]. Their findings suggested that such lncRNAs could influence gene expression of key molecules involved in glucose and lipid metabolism. Similarly, work performed in esophageal squamous cell carcinoma identified differentially expressed lncRNAs with the potential to regulate expression of genes implicated in lipid metabolism [126]. APOA1-AS and DYNLRB2-2 are other examples of lncRNAs that can influence the expression of targets involved in lipid homeostasis and metabolism [127, 128]. While the field is still in its infancy, it is reasonable to postulate that lncRNAs play an important part in regulating key molecules involved in lipid metabolism, and other cascades, during mammalian hibernation. Pioneering work in M. lucifugus has revealed decreased levels of the natural antisense long non-coding RNA HIF-1a, known as aHIF, in torpid M. lucifugus skeletal muscle tissue when compared with euthermic samples [64]. The authors proposed a likely correlation between aHIF levels and HIF-1a expression and suggested that the non-coding RNA might have a significant role in influencing transcriptional expression of HIF-1 target genes during torpor. While a clearer knowledge of lncRNA expression and function in mammalian hibernation remains to be performed to better assess the relevance of these molecules in this process, the characterized impact of lncRNAs in non-hibernating models suggest that they will play an important role.
OUTLOOK
Natural models of hypometabolism undergo a series of physiologic, biochemical, and molecular changes to successfully confront environmental challenges that arise. Several metabolic cascades are affected by this hypometabolic state, and this review took a particular interest in discussing the dynamics of lipid metabolism in mammalian hibernators as well as highlighting the potential regulation by miRNAs of key molecular players involved in such pathways. Looking ahead, it is possible to foresee the need to better characterize the miRNA targets, lipid or non-lipid related, underlying mammalian hibernation. In addition, the identification and quantification of additional non-coding RNAs, such as lncRNAs, with potential implications in this process will be of uttermost importance. Ultimately, the far-reaching implications of non-coding RNAs in mammalian hibernation will be elucidated.
This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada awarded to P. J. M.
REFERENCES
1.Lyman, C. P., Willis, J. S., Malan, A., and Wang,
L. C. H. (1982) in Hibernation and Torpor in Mammals and Birds,
Academic Press, New York.
2.Andrews, M. T. (2004) Genes controlling the
metabolic switch in hibernating mammals, Biochem. Soc. Trans.,
32, 1021-1024.
3.Storey, K. B., and Storey, J. M. (2004) Metabolic
rate depression in animals: transcriptional and translational controls,
Biol. Rev. Camb. Philos. Soc., 79, 207-233.
4.Lyons, P. J., Lang-Ouellette, D., and Morin, P. Jr.
(2013) CryomiRs: towards the identification of a cold-associated family
of microRNAs, Comp. Biochem. Physiol. D, 8, 358-364.
5.Guppy, M., and Withers, P. (1999) Metabolic
depression in animals: physiological perspectives and biochemical
generalizations, Biol. Rev. Camb. Philos. Soc., 74,
1-40.
6.Storey, K. B. (2002) Life in the slow lane:
molecular mechanisms of estivation, Comp. Biochem. Physiol. A,
133, 733-754.
7.Jones, R. M. (1980) Metabolic consequences of
accelerated urea synthesis during seasonal dormancy of spadefoot toads,
Scaphiopus couchi and Scaphiopus multiplicatus, J.
Exp. Zool., 212, 255-267.
8.Denlinger, D. L. (2002) Regulation of diapause,
Ann. Rev. Entomol., 47, 93-122.
9.Kostal, V., and Simek, P. (2000) Overwintering
strategy in Pyrrhocoris apterus (Heteroptera): the relations
between life-cycle, chill tolerance and physiological adjustments,
J. Insect Physiol., 46, 1321-1329.
10.Michaud, R. M., and Denlinger, D. L. (2006) Oleic
acid is elevated in cell membranes during rapid cold-hardening and
pupal diapause in the flesh fly, Sarcophaga crassipalpis, J.
Insect Physiol., 52, 1073-1082.
11.Geiser, F. (2004) Metabolic rate and body
temperature reduction during hibernation and daily torpor, Annu.
Rev. Physiol., 66, 239-274.
12.Dark, J. (2005) Annual lipid cycles in
hibernators: integration of physiology and behavior, Annu. Rev.
Nutr., 25, 469-497.
13.Storey, K. B., and Storey, J. M. (2010) Metabolic
rate depression: the biochemistry of mammalian hibernation, Adv.
Clin. Chem., 52, 77-108.
14.Carey, H. V., Andrews, M. T., and Martin, S. L.
(2003) Mammalian hibernation: cellular and molecular responses to
depressed metabolism and low temperature, Physiol. Rev.,
83, 1153-1181.
15.Buck, M. J., and Barnes, B. M. (2000) Effects of
ambient temperature on metabolic rate, respiratory quotient, and torpor
in an arctic hibernator, Am. J. Physiol., 279,
R255-R262.
16.Milsom, W. K., and Jackson, D. C. (2011)
Hibernation and gas exchange, Compr. Physiol., 1,
397-420.
17.Wang, L. C. H., and Wolowyk, M. W. (1988) Torpor
in mammals and birds, Can. J. Zool., 66, 133-137.
18.MacDonald, J. A., and Storey, K. B. (1999)
Regulation of ground squirrel Na+,K+-ATPase
activity by reversible phosphorylation during hibernation, Biochem.
Biophys. Res. Commun., 254, 424-429.
19.Van Breukelen, F., and Martin, S. L. (2002)
Reversible depression of transcription during hibernation, J. Comp.
Physiol. B, 172, 355-361.
20.Brooks, S. P. J., and Storey, K. B. (1992)
Mechanisms of glycolytic control during hibernation in the ground
squirrel Spermophilus lateralis, J. Comp. Physiol.,
162, 23-28.
21.Rider, M. H., Hussain, N., Dilworth, S. M.,
Storey, J. M., and Storey, K. B. (2011) AMP-activated protein
kinase and metabolic regulation in cold-hardy insects, J. Insect
Physiol., 57, 1453-1462.
22.Lee, Y. J., Miyake, S., Wakita, H., McMullen, D.
C., Azuma, Y., Auh, S., and Hallenbeck, J. M. (2007) Protein
SUMOylation is massively increased in hibernation torpor and is
critical for the cytoprotection provided by ischemic preconditioning
and hypothermia in SHSY5Y cells, J. Cereb. Blood Flow Metab.,
27, 950-962.
23.Velickovska, V., and van Breukelen, F. (2007)
Ubiquitylation of proteins in livers of hibernating golden-mantled
ground squirrels, Spermophilus lateralis, Cryobiology,
55, 230-235.
24.Frerichs, K. U., Smith, C. B., Brenner, M.,
DeGracia, D. J., Krause, G. S., Marrone, L., Dever, T. E., and
Hallenbeck, J. M. (1998) Suppression of protein synthesis in brain
during hibernation involves inhibition of protein initiation and
elongation, Proc. Natl. Acad. Sci. USA, 95,
14511-14516.
25.Osborne, P. G., Gao, B., and Hashimoto, M. (2004)
Determination in vivo of newly synthesized gene expression in
hamsters during phases of the hibernation cycle, Jpn. J.
Physiol., 54, 295-305.
26.Wu, C. B., and Storey, K. B. (2012) Regulation of
the mTOR signaling network in hibernating thirteen-lined ground
squirrels, J. Exp. Biol., 215, 1720-1727.
27.Mrosovsky, N., and Boshes, M. (1986) Meal
patterns and food intakes of ground squirrels during circannual cycles,
Appetite, 7, 163-175.
28.Mostafa, N., Everett, D. C., Chou, S. C., Kong,
P. A., Florant, G. L., and Coleman, R. A. (1993) Seasonal changes in
critical enzymes of lipogenesis and triacylglycerol synthesis in the
marmot (Marmota flaviventris), J. Comp. Physiol. B,
163, 463-469.
29.Wang, P., Walter, R. D., Bhat, B. G., Florant, G.
L., and Coleman, R. A. (1997) Seasonal changes in enzymes of
lipogenesis and triacylglycerol synthesis in the golden-mantled ground
squirrel (Spermophilus lateralis), Comp. Biochem. Physiol.
B, 118, 261-267.
30.Healy, J. E., Gearhart, C. N., Bateman, J. L.,
Handa, R. J., and Florant, G. L. (2011) AMPK and ACC change with
fasting and physiological condition in euthermic and hibernating
golden-mantled ground squirrels (Callospermophilus lateralis),
Comp. Biochem. Physiol. A, 159, 322-331.
31.Kabine, M., Clemencet, M. C., Bride, J., El
Kebbaj, M. S., Latruffe, N., and Cherkaoui-Malki, M. (2003) Changes of
peroxisomal fatty acid metabolism during cold acclimatization in
hibernating jerboa (Jaculus orientalis), Biochimie,
85, 707-714.
32.Florant, G. L., Nuttle, L. C., Mullinex, D. E.,
and Rintoul, D. A. (1990) Plasma and white adipose tissue lipid
composition in marmots, Am. J. Physiol., 258,
R1123-1131.
33.Wilson, B. E., Deeb, S., and Florant, G. L.
(1992) Seasonal changes in hormone-sensitive and lipoprotein lipase
mRNA concentrations in marmot white adipose tissue, Am. J.
Physiol., 262, R177-181.
34.Munro, D., and Thomas, D. W. (2004) The role of
polyunsaturated fatty acids in the expression of torpor by mammals: a
review, Zoology (Jena), 107, 29-48.
35.Bell, M. V., Henderson, R. J., and Sargent, J. R.
(1986) The role of polyunsaturated fatty acids in fish, Comp.
Biochem. Physiol. B, 83, 711-719.
36.Frank, C. L. (1991) Adaptations for hibernation
in the depot fats of a ground squirrel (Spermophilus beldingi),
Can. J. Zool., 69, 2707-2711.
37.Wahli, W., and Michalik, L. (2012) PPARs at the
crossroads of lipid signaling and inflammation, Trends Endocrinol.
Metab., 23, 351-363.
38.Martin, S. L., Epperson, L. E., Rose, J. C.,
Kurtz, C. C., Ane, C., and Carey, H. V. (2008) Proteomic analysis of
the winter-protected phenotype of hibernating ground squirrel
intestine, Am. J. Physiol. Regul. Integr. Comp. Physiol.,
295, R316-R328.
39.Andrews, M. T., Squire, T. L., Bowen, C. M., and
Rollins, M. B. (1998) Low-temperature carbon utilization is regulated
by novel gene activity in the heart of a hibernating mammal, Proc.
Natl. Acad. Sci. USA, 95, 8392-8397.
40.Eddy, S. F., and Storey, K. B. (2003)
Differential expression of Akt, PPARγ, and PGC-1 during
hibernation in bats, Biochem. Cell Biol., 81,
269-274.
41.Kabine, M., El Kebbaj, Z., Oaxaca-Castillo, D.,
Clemencet, M. C., El Kebbaj, M. S., Latruffe, N., and Cherkaoui-Malki,
M. (2004) Peroxisome proliferator-activated receptors as regulators of
lipid metabolism; tissue differential expression in adipose tissues
during cold acclimatization and hibernation of jerboa (Jaculus
orientalis), Biochimie, 86, 763-770.
42.Eddy, S. F., Morin, P. Jr., and Storey, K. B.
(2005) Cloning and expression of PPAR-γ and PGC-1α from the
hibernating ground squirrel, Spermophilus tridecemlineatus,
Mol. Cell. Biochem., 269, 175-182.
43.El Kebbaj, Z., Andreoletti, P., Mountassif, D.,
Kabine, M., Schohn, H., Dauca, M., Latruffe, N., El Kebbaj, M. S., and
Cherkaoui-Malki, M. (2009) Differential regulation of peroxisome
proliferator-activated receptor (PPAR)-α1 and truncated
PPARα2 as an adaptive response to fasting in the control of
hepatic peroxisomal fatty acid β-oxidation in the hibernating
mammal, Endocrinology, 150, 1192-1201.
44.Ishida, N. (2009) Role of PPARα in the
control of torpor through FGF21-NPY pathway: from circadian clock to
seasonal change in mammals, PPAR Res., 2009, 412949.
45.Colin, S., Bourguignon, E., Boullay, A. B.,
Tousaint, J. J., Huet, S., Caira, F., Staels, B., Lestavel, S.,
Lobaccaro, J. M., and Delerive, P. (2008) Intestine-specific regulation
of PPARα gene transcription by liver X receptors,
Endocrinology, 149, 5128-5135.
46.Russom, J. M., Guba, G. R., Sanchez, D., Tam, C.
F., Lopez, G. A., and Garcia, R. E. (1992) Plasma lipoprotein
cholesterol concentrations in the golden-mantled ground squirrel
(Spermophilus lateralis): a comparison between pre-hibernators
and hibernators, Comp. Biochem. Physiol. B, 102,
573-578.
47.Kolomiytseva, I. K., Perepelkina, N. I.,
Zharikova, A. D., and Popov, V. I. (2008) Membrane lipids and
morphology of brain cortex synaptosomes isolated from hibernating
Yakutian ground squirrel, Comp. Biochem. Physiol. B, 151,
386-391.
48.Green, C. B., Takahashi, J. S., and Bass, J.
(2008) The meter of metabolism, Cell, 134, 728-742.
49.Nelson, C. J., Otis, J. P., Martin, S. L., and
Carey, H. V. (2009) Analysis of the hibernation cycle using LC-MS-based
metabolomics in ground squirrel liver, Physiol. Genom.,
37, 43-51.
50.Krol, J., Loedige, I., and Filipowicz, W. (2010)
The widespread regulation of microRNA biogenesis, function and decay,
Nat. Rev. Genet., 11, 597-610.
51.Bartel, D. P. (2009) MicroRNAs: target
recognition and regulatory functions, Cell, 136,
215-233.
52.Friedman, R. C., Farh, K. K., Burge, C. B., and
Bartel, D. P. (2009) Most mammalian mRNAs are conserved targets of
microRNAs, Genome Res., 19, 92-105.
53.Reynolds, J. A., Clark, J., Diakoff, S. J., and
Denlinger, D. L. (2013) Transcriptional evidence for small RNA
regulation of pupal diapause in the flesh fly, Sarcophaga
bullata, Insect Biochem. Mol. Biol., 43, 982-989.
54.Chen, M., Zhang, X., Liu, J., and Storey, K. B.
(2013) High-throughput sequencing reveals differential expression of
miRNAs in intestine from sea cucumber during aestivation, PloS
One, 8, e76120.
55.Courteau, L. A., Storey, K. B., and Morin, P. Jr.
(2012) Differential expression of microRNA species in a freeze tolerant
insect, Eurosta solidaginis, Cryobiology, 65,
210-214.
56.Lyons, P. J., Poitras, J. J., Courteau, L. A.,
Storey, K. B., and Morin, P. Jr. (2013) Identification of
differentially regulated microRNAs in cold-hardy insects, Cryo
Lett., 34, 83-89.
57.Biggar, K. K., Dubuc, A., and Storey, K. (2009)
MicroRNA regulation below zero: differential expression of miRNA-21 and
miRNA-16 during freezing in wood frogs, Cryobiology, 59,
317-321.
58.Zhang, J., and Storey, K. B. (2012) Akt signaling
and freezing survival in the wood frog, Rana sylvatica,
Biochim. Biophys. Acta, 1830, 4828-4837.
59.Biggar, K. K., Kornfeld, S. F., Maistrovski, Y.,
and Storey, K. B. (2012) MicroRNA regulation in extreme environments:
differential expression of microRNAs in the intertidal snail
Littorina littorea during extended periods of freezing and
anoxia, Genom. Proteom. Bioinform., 10, 302-309.
60.Morin, P. Jr., Dubuc, A., and Storey, K. B.
(2008) Differential expression of microRNA species in organs of
hibernating ground squirrels: a role in translational suppression
during torpor, Biochim. Biophys. Acta, 1779, 628-633.
61.Kornfeld, S. F., Biggar, K. K., and Storey, K. B.
(2012) Differential expression of mature microRNAs involved in muscle
maintenance of hibernating little brown bats, Myotis lucifugus:
a model of muscle atrophy resistance, Genom. Proteom.
Bioinform., 10, 295-301.
62.Biggar, K. K., and Storey, K. B. (2014)
Identification and expression of microRNA in the brain of hibernating
bats, Myotis lucifugus, Gene, 544, 67-74.
63.Liu, Y., Hu, W., Wang, H., Lu, M., Shao, C.,
Menzel, C., Yan, Z., Li, Y., Zhao, S., Khaitovich, P., Liu, M., Chen,
W., Barnes, B. M., and Yan, J. (2010) Genomic analysis of miRNAs in an
extreme mammalian hibernator, the Arctic ground squirrel, Physiol.
Genom., 42A, 39-51.
64.Maistrovski, Y., Biggar, K. K., and Storey, K. B.
(2012) HIF-1α regulation in mammalian hibernators: role of
non-coding RNA in HIF-1α control during torpor in ground
squirrels and bats, J. Comp. Physiol. B, 182,
849-859.
65.Lee, Y. J., Johnson, K. R., and Hallenbeck, J. M.
(2012) Global protein conjugation by ubiquitin-like-modifiers during
ischemic stress is regulated by microRNAs and confers robust tolerance
to ischemia, PLoS One, 7, e47787.
66.Davalos, A., Goedeke, L., Smibert, P., Ramirez,
C. M., Warrier, N. P., Andreo, U., Cirera-Salinas, D., Rayner, K.,
Suresh, U., Pastor-Pareja, J. C., Esplugues, E., Fisher, E. A.,
Penalva, L. O. F., Moore, K. J., Suarez, Y., Lai, E. C., and
Fernandez-Hermando, C. (2011) miR-33a/b contribute to the regulation of
fatty acid metabolism and insulin signaling, Proc. Natl. Acad. Sci.
USA, 108, 9232-9237.
67.Turczynska, K. M., Bhattachariya, A., Sall, J.,
Goransson, O., Sward, K., Hellstrand, P., and Albinsson, S. (2013)
Stretch-sensitive down-regulation of the miR-144/451 cluster in
vascular smooth muscle and its role in AMP-activated protein kinase
signaling, PLoS One, 8, e65135.
68.Chang, F., Zhang, L. H., Xu, W. P., Jing, P., and
Zhan, P. Y. (2014) microRNA-9 attenuates amyloid-β-induced
synaptotoxicity by targeting calcium/calmodulin-dependent protein
kinase kinase 2, Mol. Med. Rep., 9, 1917-1922.
69.Lee, G. C., Kim, Y. W., Kim, E. H., Meng, Z.,
Huang, W., Hwang, S. J., and Kim, S. G. (2012) Farnesoid X receptor
protects hepatocytes from injury by repressing miR-199a-3p, which
increases levels of LKB1, Gastroenterology, 145,
1206-1217.
70.Belke, D. D., Wang, L. C., and Lopaschuk, G. D.
(1998) Acetyl-CoA carboxylase control of fatty acid oxidation in hearts
from hibernating Richardson’s ground squirrels, Biochim.
Biophys. Acta, 1391, 25-36.
71.Lang-Ouellette, D., and Morin, P. Jr. (2014)
Differential expression of miRNAs with metabolic implications in
hibernating thirteen-lined ground squirrels, Ictidomys
tridecemlineatus, Mol. Cell. Biochem., 394,
291-298.
72.Ponomarev, E. D., Veremeyko, T., Barteneva, N.,
Krichevsky, A. M., and Weiner, H. L. (2011) MicroRNA-124 promotes
microglia quiescence and suppresses EAE by deactivating macrophages via
the C/EBP-α-PU.1 pathway, Nat. Med., 17, 64-70.
73.Hegde, V. L., Tomar, S., Jackson, A., Rao, R.,
Yang, X., Singh, N. P., Nagarkatti, P. S., and Nagarkatti, M. (2014)
Distinct microRNA expression profile and targeted biological pathways
in functional myeloid-derived suppressor cells induced by
Δ9-tetrahydrocannabinol in vivo: regulation of
CCAAT/enhancer-binding protein α by microRNA-690, J. Biol.
Chem., 288, 36810-36826.
74.Ji, J., Zhang, J., Huang, G., Qian, J., Wang, X.,
and Mei, S. (2009) Over-expressed microRNA-27a and 27b influence fat
accumulation and cell proliferation during rat hepatic stellate cell
activation, FEBS Lett., 583, 759-766.
75.Oda, Y., Nakajima, M., Tsuneyama, K., Takamiya,
M., Aoki, Y., Fukami, T., and Yokoi, T. (2014) Retinoid X receptor
α in human liver is regulated by miR-34a, Biochem.
Pharmacol., 90, 179-187.
76.Adlakha, Y. K., Khanna, S., Singh, R., Singh, V.
P., Agrawal, A., and Saini, N. (2013) Pro-apoptotic miRNA-128-2
modulates ABCA1, ABCG1 and RXRα expression and cholesterol
homeostasis, Cell Death Dis., 4, e780.
77.Guerit, D., Philipot, D., Chuchana, P., Toupet,
K., Brondello, J. M., Mathieu, M., Jorgensen, C., and Noel, D. (2013)
Sox9-regulated miRNA-574-3p inhibits chondrogenic differentiation of
mesenchymal stem cells, PLoS One, 8, e62582.
78.Kim, S. Y., Kim, A. Y., Lee, H. W., Son, Y. H.,
Lee, G. Y., Lee, J. W., Lee, Y. S., and Kim, J. B. (2010) miR-27a is a
negative regulator of adipocyte differentiation via suppressing
PPARγ expression, Biochem. Biophys. Res. Commun.,
392, 323-328.
79.Lee, J. J., Drakaki, A., Iliopoulos, D., and
Struhl, K. (2012) MiR-27b targets PPARγ to inhibit growth, tumor
progression and the inflammatory response in neuroblastoma cells,
Oncogene, 31, 3818-3825.
80.Lee, E. K., Lee M. J., Abdelmohsen, K., Kim, W.,
Kim, M. M., Srikantan, S., Martindale, J. L., Hutchison, E. R., Kim, H.
H., Marasa, B. S., Selimyan, R., Egan, J. M., Smith, S. R., Fried, S.
K., and Gorospe, M. (2011) miR-130 suppresses adipogenesis by
inhibiting peroxisome proliferator-activated receptor gamma expression,
Mol. Cell. Biol., 31, 626-638.
81.Chen, Y. H., Heneidi, S., Lee, J. M., Layman, L.
C., Stepp, D. W., Gamboa, G. M., Chen, B. S., Chazenbalk, G., and
Azziz, R. (2013) miRNA-93 inhibits GLUT4 and is overexpressed in
adipose tissue of polycystic ovary syndrome patients and women with
insulin resistance, Diabetes, 62, 2278-2286.
82.Yu, Q. Q., Wu, H., Huang, X., Shen, H., Zhang,
B., Xiang, C. C., Yu, S. M., Guo, R. H., and Chen, L. (2014) MiR-1
targets PIK3CA and inhibits tumorigenic properties of A549 cells,
Biomed. Pharmacother., 68, 155-161.
83.Hu, R., Pan, W., Fedulov, A. V., Jester, W.,
Jones, M. R., Weiss, S. T., Panettieri, R. A. J., Tantisira, K., and
Lu, Q. (2014) MicroRNA-10a controls airway smooth muscle cell
proliferation via direct targeting of the PI3 kinase pathway, FASEB
J., 28, 2347-2357.
84.Pandey, A. K., Verma, G., Vig, S., Srivastava,
S., Srivastava, A. K., and Datta, M. (2010) miR-29a levels are elevated
in the db/db mice liver and its overexpression leads to attenuation of
insulin action on PEPCK gene expression in HepG2 cells, Mol. Cell.
Endocrinol., 332, 125-132.
85.Li, J., You, T., and Jing, J. (2014) MiR-125b
inhibits cell biological progression of Ewing’s sarcoma by
suppressing the PI3K/Akt signaling pathway, Cell. Prolif.,
47, 152-160.
86.Guo, C., Sah, J. F., Beard, L., Willson, J. K.,
Markowitz, S. D., and Guda, K. (2008) The noncoding RNA, miR-126,
suppresses the growth of neoplastic cells by targeting
phosphatidylinositol 3-kinase signaling and is frequently lost in colon
cancers, Genes Chromosomes Cancer, 47, 939-946.
87.Wang, Y., Tang, Q., Li, M., Jiang, S., and Wang,
X. (2014) MicroRNA-375 inhibits colorectal cancer growth by targeting
PIK3CA, Biochem. Biophys. Res. Commun., 444, 199-204.
88.Yang, Y., Liu, L., Zhang, Y., Guan, H., Wu, J.,
Zhu, X., Yuan, J., and Li, M. (2014) MiR-503 targets PI3K p85 and
IKK-β and suppresses progression of non-small cell lung cancer,
Int. J. Cancer, 135, 1531-1542.
89.Jeong, H. J., Park, S. Y., Yang, W. M., and Lee,
W. (2013) The induction of miR-96 by mitochondrial dysfunction causes
impaired glycogen synthesis through translational repression of IRS-1
in SK-Hep1 cells, Biochem. Biophys. Res. Commun., 434,
503-508.
90.Zhou, Y., Feng, X., Liu, Y. L., Ye, S. C., Wang,
H., Tan, W. K., Tian, T., Qiu, Y. M., and Luo, H. S. (2013)
Down-regulation of miR-126 is associated with colorectal cancer cells
proliferation, migration and invasion by targeting IRS-1 via the AKT
and ERK1/2 signaling pathways, PLoS One, 8, e81203.
91.Motohashi, N., Alexander, M. S.,
Shimizu-Motohashi, Y., Myers, J. A., Kawahara, G., and Kunkel, L. M.
(2013) Regulation of IRS1/Akt insulin signaling by microRNA-128a during
myogenesis, J. Cell Sci., 126, 2678-2691.
92.Karolina, D. S., Armugam, A., Tavintharan, S.,
Wong, M. T., Lim, S. C., Sum, C. F., and Jeyaseelan, K. (2011) MicroRNA
144 impairs insulin signaling by inhibiting the expression of insulin
receptor substrate 1 in type 2 diabetes mellitus, PLoS One,
6, e22839.
93.Wang, Y., Hu, C., Cheng, J., Chen, B., Ke, Q.,
Lv, Z., Wu, J., and Zhou, Y. (2014) MicroRNA-145 suppresses
hepatocellular carcinoma by targeting IRS1 and its downstream Akt
signaling, Biochem. Biophys. Res. Commun., 446,
1255-1260.
94.Xu, Q., Jiang, Y., Yin, Y., Li, Q., Jing, Y., Qi,
Y. T., Xu, Q., Li, W., Lu, B., Peiper, S. S., Jiang, B. H., and Liu, L.
Z. (2013) A regulatory circuit of miR-148a/152 and DNMT1 in modulating
cell transformation and tumor angiogenesis through IGF-IR and IRS1,
J. Mol. Cell. Biol., 5, 3-13.
95.Nossent, A. Y., Eskildsen, T. V., Andersen, L.
B., Bie, P., Bronnum, H., Schneider, M., Andersen, D. C., Welten, S.
M., Jeppesen, P. L., Hamming, J. F., Hansen, J. L., Quax, P. H., and
Sheikh, S. P. (2013) The 14q32 microRNA-487b targets the antiapoptotic
insulin receptor substrate 1 in hypertension-induced remodeling of the
aorta, Ann. Surg., 258, 743-751.
96.Gurha, P., Wang, T., Larimore, A. H., Sassi, Y.,
Abreu-Goodger, C., Ramirez, M. O., Reddy, A. K., Engelhardt, S.,
Taffet, G. E., Wehrens, X. H., Entman, M. L., and Rodriguez, A. (2013)
microRNA-22 promotes heart failure through coordinate suppression of
PPAR/ERR-nuclear hormone receptor transcription, PLoS One,
8, e75882.
97.Ou, Z., Wada, T., Gramignoli, R., Li, S., Strom,
S. C., Huang, M., and Xie, W. (2011) MicroRNA hsa-miR-613 targets the
human LXRα gene and mediates a feedback loop of LXRα
autoregulation, Mol. Endocrinol., 25, 584-596.
98.Miller, A. M., Gilchrist, D. S., Nijjar, J.,
Araldi, E., Ramirez, C. M., Lavery, C. A., Fernandez-Hernando, C.,
McInnes, I. B., and Kurowska-Stolarska, M. (2013) MiR-155 has a
protective role in the development of non-alcoholic hepatosteatosis in
mice, PLoS One, 8, e72324.
99.Zhong, D., Huang, G., Zhang, Y., Zeng, Y., Xu,
Z., Zhao, Y., He, X., and He, F. (2013) MicroRNA-1 and microRNA-206
suppress LXRα-induced lipogenesis in hepatocytes, Cell.
Signal., 25, 1429-1437.
100.Wang, D., Xia, M., Yan, X., Li, D., Wang, L.,
Xu, Y., Jin, T., and Ling, W. (2012) Gut microbiota metabolism of
anthocyanin promotes reverse cholesterol transport in mice via
repressing miRNA-10b, Circ. Res., 111, 967-981.
101.Sun, D., Zhang, J., Xie, J., Wei, W., Chen, M.,
and Zhao, X. (2012) MiR-26 controls LXR-dependent cholesterol efflux by
targeting ABCA1 and ARL7, FEBS Lett., 586, 1472-1479.
102.Rayner, K. J., Suarez, Y., Davalos, A.,
Parathath, S., Fitzgerald, M. L., Tamehiro, N., Fisher, E. A., Moore,
K. J., and Fernandez-Hernando, C. (2010) MiR-33 contributes to the
regulation of cholesterol homeostasis, Science, 328,
1570-1573.
103.Kim, J., Yoon, H., Ramirez, C. M., Lee, S. M.,
Hoe, H. S., Fernandez-Hernando, C., and Kim, J. (2012) MiR-106b impairs
cholesterol efflux and increases Aβ levels by repressing ABCA1
expression, Exp. Neurol., 235, 476-483.
104.De Aguiar Vallim, T. Q., Tarling, E. J., Kim,
T., Civelek, M., Baldan, A., Esau, C., and Edwards, P. A. (2013)
MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and
plasma high-density lipoprotein after activation of the nuclear
receptor farnesoid X receptor, Circ. Res., 112,
1602-1612.
105.Kang, M. H., Zhang, L. H., Wijesekara, N., de
Haan, W., Butland, S., Bhattacharjee, A., and Hayden, M. R. (2013)
Regulation of ABCA1 protein expression and function in hepatic and
pancreatic islet cells by miR-145, Arterioscler. Thromb. Vasc.
Biol., 33, 2724-2732.
106.Zhao, R., Feng, J., and He, G. (2014) miR-613
regulates cholesterol efflux by targeting LXRα and ABCA1 in
PPARγ activated THP-1 macrophages, Biochem. Biophys. Res.
Commun., 448, 329-334.
107.Ramirez, C. M., Davalos, A., Goedeke, L.,
Salerno, A. G., Warrier, N., Cirera-Salinas, D., Suarez, Y., and
Fernandez-Hernando, C. (2011) MicroRNA-758 regulates cholesterol efflux
through posttranscriptional repression of ATP-binding cassette
transporter A1, Arterioscler. Thromb. Vasc. Biol., 31,
2707-2714.
108.Song, K. H., Li, T., Owsley, E., and Chiang, J.
Y. (2010) A putative role of micro RNA in regulation of cholesterol
7α-hydroxylase expression in human hepatocytes, J. Lipid
Res., 51, 2223-2233.
109.Marquart, T. J., Allen, R. M., Ory, D. S., and
Baldan, A. (2010) miR-33 links SREBP-2 induction to repression of
sterol transporters, Proc. Natl. Acad. Sci. USA, 107,
12228-12232.
110.Zhou, R., Li, X., Hu, G., Gong, A. Y.,
Drescher, K. M., and Chen, X. M. (2012) miR-16 targets transcriptional
corepressor SMRT and modulates NF-κB-regulated transactivation of
interleukin-8 gene, PLoS One, 7, e30772.
111.Osborne, T. F., and Espenshade, P. J. (2009)
Evolutionary conservation and adaptation in the mechanism that
regulates SREBP action: what a long, strange tRIP it’s been,
Genes Dev., 23, 2578-2591.
112.Li, X., Chen, Y. T., Mukhopadhyay, N. K., Kim,
J., Freeman, M. R., and Huang, W. C. (2013) MicroRNA-185 and 342
inhibit tumorigenicity and induce apoptosis through blockade of the
SREBP metabolic pathway in prostate cancer cells, PLoS One,
8, e70987.
113.Horie, T., Nishino, T., Baba, O., Kuwabara, Y.,
Nakao, T., Nishiga, M., Usami, S., Izuhara, M., Sowa, N., Yahagi, N.,
Shimano, H., Matsumura, S., Inoue, K., Marusawa, H., Nakamura, T.,
Hasegawa, K., Kume, N., Yokode, M., Kita, T., Kimura, T., and Ono, K.
(2013) MicroRNA-33 regulates sterol regulatory element-binding protein
1 expression in mice, Nat. Commun., 4, 2883.
114.Xiao, F., Yu, J., Liu, B., Guo, Y., Li, K.,
Deng, J., Zhang, J., Wang, C., Chen, S., Du, Y., Lu, Y., Xiao, Y.,
Zhang, Z., and Guo, F. (2014) A novel function of microRNA 130a-3p in
hepatic insulin sensitivity and liver steatosis, Diabetes,
63, 2631-2642.
115.Mao, J. H., Zhou, R. P., Peng, A. F., Liu, Z.
L., Huang, S. H., Long, X. H., and Shu, Y. (2012) microRNA-195
suppresses osteosarcoma cell invasion and migration in vitro by
targeting FASN, Oncol. Lett., 4, 1125-1129.
116.Park, J. H., Ahn, J., Kim, S., Kwon, D. Y., and
Ha, T. Y. (2011) Murine hepatic miRNAs expression and regulation of
gene expression in diet-induced obese mice, Mol. Cells,
31, 33-38.
117.Cheng, C., Chen, Z. Q., and Shi, X. T. (2014)
MicroRNA-320 inhibits osteosarcoma cells proliferation by directly
targeting fatty acid synthase, Tumor Biol., 35,
4177-4183.
118.Long, X. H., Mao, J. H., Peng, A. F., Zhou, Y.,
Huang, S. H., and Liu, Z. L. (2013) Tumor suppressive microRNA-424
inhibits osteosarcoma cell migration and invasion via targeting fatty
acid synthase, Exp. Ther. Med., 5, 1048-1052.
119.Rinn, J. L., and Chang, H. Y. (2012) Genome
regulation by long noncoding RNAs, Annu. Rev. Biochem.,
81, 145-166.
120.Guttman, M., Amit, I., Garber, M., French, C.,
Lin, M. F., Feldser, D., Huarte, M., Zuk, O., Carey, B. W., Cassady, J.
P., Cabili, M. N., Jaenisch, R., Mikkelsen, T. S., Jacks, T., Hacohen,
N., Bernstein, B. E., Kellis, M., Regev, A., Rinn, J. L., and Lander,
E. S. (2009) Chromatin signature reveals over a thousand highly
conserved large non-coding RNAs in mammals, Nature, 458,
223-227.
121.Pauli, A., Valen, E., Lin, M. F., Garber, M.,
Vastenhouw, N. L., Levin, J. Z., Fan, L., Sandelin, A., Rinn, J. L.,
Regev, A., and Schier, A. F. (2011) Systematic identification of long
noncoding RNAs expressed during zebrafish embryogenesis, Genome
Res., 22, 577-591.
122.Tavares, L., Dimitrova, E., Oxley, D., Webster,
J., Poot, R., Demmers, J., Bezstarosti, K., Taylor, S., Ura, H., Koide,
H., Wutz, A., Vidal, M., Elderkin, S., and Brockdorff, N. (2012)
RYBP–PRC1 complexes mediate H2A ubiquitylation at polycomb target
sites independently of PRC2 and H3K27me3, Cell, 148,
664-678.
123.Feng, J., Bi, C., Clark, B. S., Mady, R., Shah,
P., and Kohtz, J. D. (2006) The Evf-2 noncoding RNA is transcribed from
the Dlx-5/6 ultraconserved region and functions as a Dlx-2
transcriptional coactivator, Genes Dev., 20,
1470-1484.
124.Willingham, A. T., Orth, A. P., Batalov, S.,
Peters, E. C., Wen, B. G., Aza-Blanc, P., Hogenesch, J. B., and
Schultz, P. G. (2005) A strategy for probing the function of noncoding
RNAs finds a repressor of NFAT, Science, 309,
1570-1573.
125.Ellis, B. C., Graham, L. D., and Molloy, P. L.
(2014) CRNDE, a long non-coding RNA responsive to insulin/IGF
signaling, regulates genes involved in central metabolism, Biochim.
Biophys. Acta, 1843, 372-386.
126.Cao, W., Wu, W., Shi, F., Chen, X., Wu, L.,
Yang, K., Tian, F., Zhu, M., Chen, G., Wang, W., Biddle, F. G., and Gu,
J. (2013) Integrated analysis of long noncoding RNA and coding RNA
expression in esophageal squamous cell carcinoma, Int. J.
Genom., 2013, 480534.
127.Halley, P., Kadakkuzha, B. M., Faghihi, M. A.,
Magistri, M., Zeier, Z., Khorkova, O., Coito, C., Hsiao, J., Lawrence,
M., and Wahlestedt, C. (2014) Regulation of the apolipoprotein gene
cluster by a long noncoding RNA, Cell Rep., 6,
222-230.
128.Hu, Y. W., Yang, J. Y., Ma, X., Chen, Z. P.,
Hu, Y. R., Zhao, J. Y., Li, S. F., Qiu, Y. R., Lu, J. B., Wang, Y. C.,
Gao, J. J., Sha, Y. H., Zheng, L., and Wang, Q. (2014) A
lincRNA-DYNLRB2-2/GPR119/GLP-1R/ABCA1-dependent signal transduction
pathway is essential for the regulation of cholesterol homeostasis,
J. Lipid Res., 55, 681-697.