PPX1 Gene Overexpression Has no Influence on Polyphosphates in Saccharomyces cerevisiae
L. P. Lichko1, M. A. Eldarov2, M. V. Dumina2, and T. V. Kulakovskaya1*
1Skryabin Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, pr. Nauki 5, 142290 Pushchino, Moscow Region, Russia; fax: (495) 956-3370; E-mail: alla@ibpm.pushchino.ru2Center for Bioengineering, Russian Academy of Sciences, pr. 60-letiya Oktyabrya 7/1, 117312 Moscow, Russia
* To whom correspondence should be addressed.
Received June 20, 2014; Revision received June 30, 2014
The role of exopolyphosphatase PPX1 in polyphosphate metabolism in yeasts has been studied in strains of Saccharomyces cerevisiae with inactivated PPX1 and PPN1 genes transformed by the expression vector carrying the yeast PPX1 gene. Exopolyphosphatase activity in transformant strains increased 90- and 40-fold compared to the ΔPPX1 and ΔPPN1 strains, respectively. The purified recombinant exopolyphosphatase PPX1 was similar to the PPX1 of wild strains in its substrate specificity and requirement for divalent metal cations. It was more active with tripolyphosphate and low molecular mass polyphosphates than with high molecular mass polyphosphates and required Mg2+ for its activity. The high level of recombinant PPX1 expression caused no decrease in polyphosphate content in the cells of the transformant. This fact suggests the restricted role of PPX1 in polyphosphate metabolism in yeasts.
KEY WORDS: polyphosphates, exopolyphosphatase, PPX1 gene, Saccharomyces cerevisiae, overexpressionDOI: 10.1134/S000629791411008X
Abbreviations: polyP, inorganic polyphosphates.
Dedicated to the memory of Prof. I. S. Kulaev
Inorganic polyphosphates (polyP) are biopolymers that perform numerous
regulatory functions in living cells [1-5]. It is necessary to investigate the enzymes of
polyP metabolism in order to understand their functions.
In the yeast Saccharomyces cerevisiae, the exopolyphosphatases (EC 3.6.1.11) cleaving Pi at the end of the polymer chain are encoded by two nonhomologous genes, PPX1 (http://www.uniprot.org/uniprot/P38698) and PPN1 (http://www.uniprot.org/uniprot/Q04119) [6-8]. These enzymes differ in their substrate specificity, requirement for divalent cations, cellular localization, and, evidently, their role in polyP metabolism [9]. One of the approaches to investigation of their function in polyP metabolism is to use strains with mutations in the genes mentioned above. These mutations were shown to have different effects on polyP content and chain length in different compartments of S. cerevisiae cells [8]. Inactivation of the PPX1 gene did not influence the content and chain lengths of polyP in all compartments of S. cerevisiae, whereas PPN1 inactivation resulted in increased polyP chain lengths in the cytoplasm, mitochondria, and vacuoles [8]. These facts suggest that the function of PPX1 is not confined only to polyP hydrolysis. BLAST demonstrates a high sequence similarity between the yeast exopolyphosphatase PPX1 and the human protein h-prune, which is a partner of the metastatic suppressor nm23-H1 and shows an exopolyphosphatase activity [10].
Another approach to elucidation of exopolyphosphatase function is generation of strains with overexpression of these enzymes. Recently we showed that an S. cerevisiae strain with 10-fold higher expression of exopolyphosphatase PPN1 compared to the parent strain exhibits a 3-fold decrease in polyP amount [11].
The aims of this work were to obtain S. cerevisiae strains with overexpression of the PPX1 gene, to study the properties of the recombinant enzyme, and to assess the effect of this overexpression on polyP under different culture conditions.
MATERIALS AND METHODS
Yeast strains and culture conditions. The parent strain CRY and the strains CRX with inactivated PPX1 gene (ΔPPX1) and CRN with inactivated PPN1 gene (ΔPPN1) were obtained from A. Kornberg’s laboratory [6, 7]. The transformant strains were obtained by the method described earlier [11]. The creation of the pMB1 expression vector with an “expression cassette” consisting of a strong constitutive promoter of glyceraldehyde-3-phosphate dehydrogenase (PKG1) and a terminator of phosphoglycerate kinase (TDH) of S. cerevisiae has been described earlier [11]. The fragment with the PPX1 exopolyphosphatase coding sequence was obtained by PCR, taking advantage of the known data on the open reading frame (ORF) sequence of the S. cerevisiae YHR201c gene (GenBank access number NM_001179332.1). The 1200-bp PCR fragment corresponding to PPX1 ORF was obtained with the DNA template of the CRY strain using primers PPX1_F 5′-ggtacATGTCGCCTTTGAGAAAGACGGTT-3′ and PPX1_R 5′-ggtctcgagTCACTCTTCCAGGTTTGAGTACGCT-3′. The 5′-flanking sequences of these primers with PciI and XhoI sites, respectively, were used for subsequent cloning of the PCR fragment. The fragment was gel-purified, treated with BspHI and XhoI, and inserted into the NcoI/XhoI-cut vector pMB1. The PPX1 gene insert in the selected clones was sequenced, and a clone lacking nonspecific PCR mutations was selected and designated as pMB1/PPX1 Sc. All DNA manipulations were performed using kits and enzymes produced by SibEnzyme (Russia), Fermentas MBI (Lithuania), and Stratagene (USA), in accordance with manufacturers’ instructions. DNA was sequenced using an ABI BigDye 3.1 sequencing kit (Applied Biosystems, USA) according to the manufacturer’s protocol in an ABI 3730 DNA analyzer.
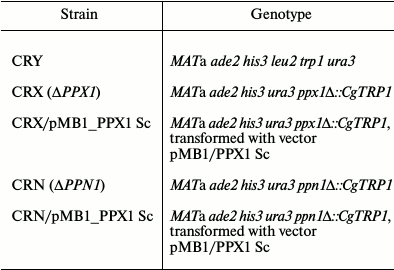
The strains CRX and CRN were transformed by the pMB1/PPX1 Sc plasmid using competent cells treated with lithium acetate [11]. The transformed clones were selected on plates with minimal medium without uracil, using complementation by the URA3 plasmid gene of the ura3 mutations in this strain. The resultant transformants were characterized by PCR to confirm the presence of the vector in the selected strains. The data on strain genotypes are presented in Table 1.
Table 1. Strains of Saccharomyces
cerevisiae used in the work
The parent yeast CRY, CRN and CRX strains were maintained on agarized YPD medium and strain CRX/pMB1_PPX1 Sc was maintained on agarized YNB medium without uracil [11]. The yeast strains were grown on a synthetic minimal YNB medium containing (per liter) 1.7 g of bacto-yeast nitrogen bases (Difco, USA), 20 g of glucose, 20 mg of L-tryptophane, 20 mg L-histidine, 20 mg L-methionine, 20 mg adenine, and 60 mg of L-leucine. Uracil (20 mg/liter) was added during the cultivation of strains CRY, CRN and CRX.
The cultures were grown at 29°C in flasks with 250 ml of the YNB medium on a shaker at 145 rpm. The cells of all strains were grown to the stationary growth phase for 24 h. The biomass was harvested by centrifugation at 3000g and twice washed with cold distilled water.
For study of polyP accumulation under phosphate surplus, the strains CRX and CRX/pMB1_PPX1 Sc were used. After 24-h cultivation in YNB medium containing 8.2 mM Pi, the cells were placed in the YNB containing 0.02 mM Pi (1 ml of inoculate per 250 ml medium). Pi-deficient medium was prepared according the method described earlier [12]. After 24-h cultivation, the cells were separated by centrifugation, placed in the YNB medium with 8.2 mM Pi (~1 g wet biomass per 250 ml), and cultivated for 2 h.
Obtaining of spheroplasts and cytosol. To obtain spheroplasts, the cells were washed in 0.8 M mannitol in 5% Na-citrate, pH 6.5, and incubated in the same buffer with lyophilized snail gut juice (150 mg per gram of wet biomass) and dithiothreitol (10 mg per gram of wet biomass) for 70 min at 30°C. A cytosol fraction was obtained as described earlier [8]. The spheroplasts were washed with the same solution without snail gut juice and dithiothreitol and lysed in a glass homogenizer with Teflon pestle in 25 mM Tris-HCl buffer, pH 7.2, with 0.1 M sorbitol and 0.5 mM phenylmethylsulfonyl fluoride (Sigma, USA) at 4°C followed by centrifugation at 15,000g for 60 min. The supernatant was defined as cytosol fraction.
Recombinant exopolyphosphatase purification. All steps of enzyme purification were carried out at 4°C. The cytosol preparation from ~3 g of wet biomass (CRX/pMB1_PPX1 Sc) in 25 mM Tris-HCl buffer, pH 7.2, was supplemented with ammonium sulfate to 50% saturation. After 30-min with stirring, the precipitate formed was removed by centrifugation (9000g, 30 min). Ammonium sulfate was added to the supernatant to 85% saturation. In 1 h the precipitate was collected by centrifugation under the same conditions and dissolved in 25 mM Tris-HCl, pH 7.2, with 0.1% Triton X-100 (buffer A). The preparation was dialyzed in an Amicon cell (YM-10 membrane) with five volumes of the same buffer. Then it was loaded onto a column (1.6 × 7 cm) with DEAE-Toyopearl 650 M (Toson, Japan) equilibrated with buffer A. The column was washed with 100 ml of buffer A, and the exopolyphosphatase was eluted at a flow rate of 30 ml/h with increasing KCl concentrations (0-0.4 M) in buffer A. The gradient volume was 200 ml.
The fractions with exopolyphosphatase activity were pooled and loaded into a column (1.6 × 7 cm) with heparin-agarose (Sigma). After washing the column with 50 ml of buffer A, the enzyme was eluted at a flow rate of 15 ml/h with a step gradient, using 0.4, 0.5, 0.6, and 0.7 M KCl solutions in buffer A (10 ml of each). The fractions with exopolyphosphatase activity (0.5 and 0.6 M KCl) were pooled and subjected to ultrafiltration using the Amicon cell (YM-10 membrane).
Exopolyphosphatase assay. Exopolyphosphatase activity was assayed at 30°C by the rate of Pi formation. The incubation medium contained 50 mM Tris-HCl, pH 7.2, 2.5 mM MgSO4, and 2 mM (as Pi) polyP with an average chain length of 208 phosphate residues (polyP208) (Monsanto, USA). PolyP was prepurified from pyrophosphate and orthophosphate as described previously [13]. The amount of the enzyme forming 1 μmol of Pi per minute was taken as a unit of enzyme activity (U).
Protein concentration was assayed with BSA used as a standard [14].
Polyphosphate assay. The acid-soluble polyphosphate was extracted by twofold treatment of biomass samples with 0.5 N HClO4 at 0°C for 15 min under stirring [15]. The amount of the acid-insoluble polyphosphate was estimated by Pi content after treatment of residual biomass with 0.5 N HClO4 at 100°C for 20 min. Pi was assayed colorimetrically according the method described in [8].
RESULTS AND DISCUSSION
Exopolyphosphatase activity and polyphosphate under PPX1 overexpression. The strains under study have shown no difference during their growth in the YNB medium (Table 1). No matter what mutant strain (ΔPPX1 or ΔPPN1) was used as a recipient of the expression vector carrying the PPX1 gene, exopolyphosphatase activity in the cytosol fractions increased by more than 10-fold compared to the initial strains (Table 2). It exceeded the exopolyphosphatase activity in the cytosol of the parent strain CRY by more than 20-fold (Table 2).
Table 2. Exopolyphosphatase activity in the
cytosol (U/mg protein) and polyP content in biomass (μmol/g wet
biomass) of the strains of S. cerevisiae
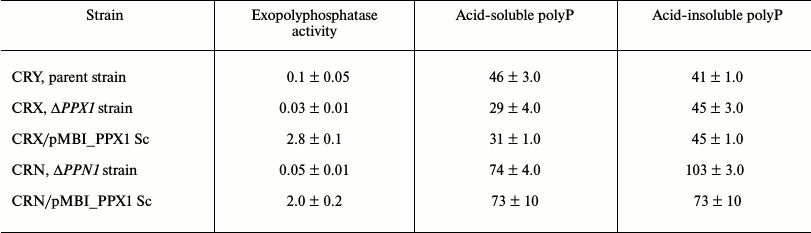
The amounts of relatively short-chain acid-soluble polyP and longer-chain acid-insoluble polyP were determined in the strains under study. In spite of the fact that the ΔPPX1 and ΔPPN1 strains differed from the parent strain in the amount of polyP, PPX1 expression in the respective transformants had no effect on the level of these polymers. Only a small decrease in the level of acid-insoluble polyP can be mentioned in the strain CRN/pMB1_PPX1 Sc (Table 2). In the ΔPPN1 strain transformed by the same vector carrying the PPN1 gene, a considerable decrease in acid-soluble and acid-insoluble polyP was observed as indicated earlier [11].
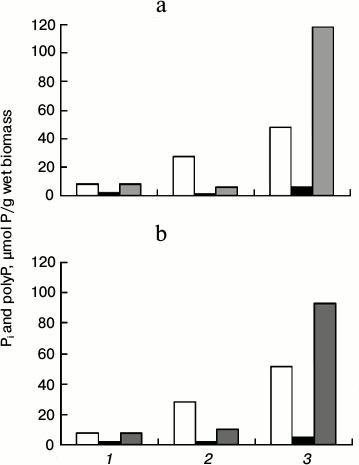
When yeast cells subjected to Pi starvation are placed in a Pi-rich medium, they rapidly accumulate polyP (surplus conditions) [15]. This method of cultivation is suitable for the estimation of polyP synthesis by the cells. The figure shows the results of such experiment for the ΔPPX1 strain and its transformant CRX/pMB1_PPX1 Sc. Both strains equally utilized polyP under Pi deficiency and both of them accumulated considerable amounts of polyP in 2 h of cultivation under the surplus conditions. The accumulated polyP was represented for the most part by the acid-insoluble fraction (figure). Such distribution among the fractions is probably accounted for by peculiarities of the strains. The level of polyP accumulation in the strain with PPX1 overexpression was lower by 20% compared to the ΔPPX1 strain. Thus, PPX1 overexpression does not prevent polyP accumulation under surplus conditions.
Content of Pi (1), acid-soluble polyP (2), and acid-insoluble polyP (3) in cells of S. cerevisiae: a) strain ΔPPX1; b) strain CRX/pMB1_PPX1 Sc with overexpression of PPX1. White columns, cultivation in medium with 8.2 mM Pi for 24 h; black columns, cultivation in medium with 0.02 mM Pi for 24 h; gray columns, the cells were re-inoculated from Pi-deficient medium into the medium with 8.2 mM Pi and cultivated for 2 h
Purification and properties of recombinant PPX1. The results of recombinant enzyme purification are shown in Table 3. The preparation was not contaminated with pyrophosphatase (Table 4) but showed several minor protein bands under PAGE. The substrate specificity and the need for divalent metal cations of the purified recombinant exopolyphosphatase PPX1 were very similar to those of PPX1 of the parent and wild strains of S. cerevisiae studied earlier [9]. The enzyme was more active with tripolyphosphate and short-chain polyP than with long-chain polyP (Table 4). It was inactive in the absence of divalent cations, which is characteristic of PPX1 in contrast to PPN1. Magnesium or cobalt ions were needed for manifestation of the enzyme activity. Ammonium ions had an additional stimulating effect. The specific activity of the enzyme was 150 U/mg protein in the presence of 2.5 mM Mg2+ but increased to 360 U/mg protein on addition of 200 mM NH4Cl. Specific activity of the enzyme increased to 500 U/mg protein when magnesium sulfate was replaced by 0.1 mM cobalt sulfate in the presence of ammonium salt. In the presence of 2.5 mM Mg2+, heparin (1 mg/ml) inhibited the enzyme activity by 80%. This is in agreement with data obtained earlier for PPX1 [8].
Table 3. Purification of exopolyphosphatase
from strain CRX/pMB1_PPX1 Sc
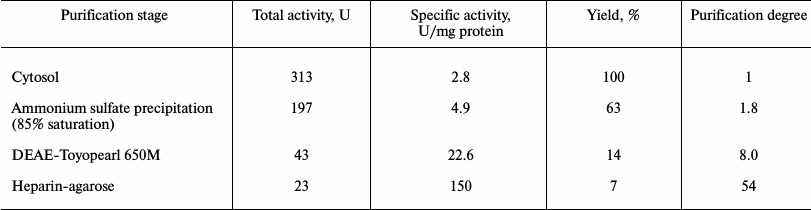
Table 4. Substrate specificity of purified
recombinant exopolyphosphatase
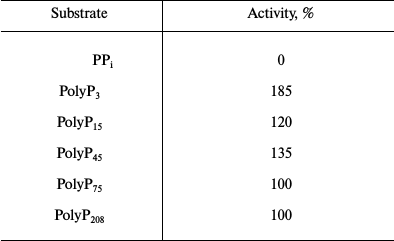
Note: Specific activity taken as 100% was 110 U/mg protein.
Thus, two strains of S. cerevisiae with PPX1 overproduction have been obtained. The cytosol fractions of both strains possessed high exopolyphosphatase activities, with notable changes in polyP contents compared to the initial strains used for transformation. The cytosol of S. cerevisiae is known to contain a substantial portion of the cellular polyP pool [8]. These polymers seem to be somehow protected from hydrolysis by PPX1 in the intact cell. However, in the cytosol preparations polyP is rapidly hydrolyzed even in strains with mutations in the gene of the other exopolyphosphatase, PPN1 [16].
PPX1 expression in heterologous systems results in a marked decrease in polyP content. For example, overproduction of this enzyme in Pseudomonas bacteria leads to elimination of almost 95% of polyP [17]. The expression of the yeast PPX1 in the mitochondria of human cell lines was usefully employed to reduce the level of polyP and to study the role of these polymers in bioenergetic processes and calcium transport in these organelles [18].
The absence of pronounced effect of PPX1 overexpression on polyP content in the yeast raises questions about the function of this enzyme and its participation in polyP metabolism. PPX1 exopolyphosphatase is a constitutive enzyme, and its presence in the cytosol does not prevent polyP accumulation in this compartment [19]. Its participation in polyP degradation is observed under special conditions: during the growth of the ΔPPN1 mutant in a Pi-deficient medium [16]. PPX1 hydrolyzes adenosine-5′-tetraphosphate and guanosine-5′-tetraphosphate [20]. Its functions may be elucidated by revealing new substrates for this enzyme.
This study was supported by the Russian Foundation for Basic Research (grant No. 14-04-00515).
REFERENCES
1.Kulaev, I. S., Vagabov, V. M., and Kulakovskaya, T.
V. (2004) The Biochemistry of Inorganic Polyphosphates,
Wiley.
2.Rao, N. N., Gomez-Garcia, M. R., and Kornberg, A.
(2009) Inorganic polyphosphate: essential for growth and survival,
Ann. Rev. Biochem., 78, 605-647.
3.Pavlov, E., Aschar-Sobbi, R., Campanella, M.,
Turner, R. J., Gomez-Garcia, M. R., and Abramov, A. Y. (2010) Inorganic
polyphosphate and energy metabolism in mammalian cells, J. Biol.
Chem., 285, 9420-9428.
4.Kulakovskaya, T. V. (2012) Inorganic
polyphosphates: Jack of all trades, Biochemistry and Physiology:
Open access, 1, 1000e107.
5.Omelon, S., Ariganello, M., Bonucci, E., Grynpas,
M., and Nanci, A. (2013) A review of phosphate mineral nucleation in
biology and geobiology, Calcif. Tissue Int., 93,
382-396.
6.Wurst, H., Shiba, T., and Kornberg, A. (1995) The
gene for a major exopolyphosphatase of Saccharomyces cerevisiae,
J. Bacteriol., 177, 898-906.
7.Sethuraman, A., Rao, N. N., and Kornberg, A. (2001)
The endopolyphosphatase gene: essential in Saccharomyces
cerevisiae, Proc. Natl. Acad. Sci. USA, 98,
8542-8547.
8.Lichko, L., Kulakovskaya, T., Pestov, N., and
Kulaev, I. (2006) Inorganic polyphosphates and exopolyphosphatases in
cell compartments of the yeast Saccharomyces cerevisiae under
inactivation of PPX1 and PPN1 genes, Biosci.
Rep., 26, 45-54.
9.Lichko, L. P., Andreeva, N. A., Kulakovskaya,
T. V., and Kulaev, I. S. (2003) Exopolyphosphatases of the yeast
Saccharomyces cerevisiae, FEMS Yeast Res., 3,
233-238.
10.Tammenkoski, M., Koivula, K., Cusanelli, E.,
Zollo, M., Steegborn, C., Baykov, A. A., and Lahti, R. (2008) Human
metastasis regulator protein H-prune is a short-chain
exopolyphosphatase, Biochemistry, 47, 9707-9713.
11.Eldarov, M. A., Baranov, M. V., Dumina, M. V.,
Shgun, A. A., Andreeva, N. A., Trilisenko, L. V., Kulakovskaya, T. V.,
Ryasanova, L. P., and Kulaev, I. S. (2013) Polyphosphates and
exopolyphosphatase activities in the yeast Saccharomyces cerevisiae
under overexpression of homologous and heterologous PPN1
genes, Biochemistry (Moscow), 78, 946-953.
12.Rubin, G. M. (1973) The nucleotide sequence of
Saccharomyces cerevisiae 5.8 S ribosomal ribonucleic acid, J.
Biol. Chem., 11, 3860-3875.
13.Andreeva, N. A., and Okorokov, L. A. (1993)
Purification and characterization of highly active and stable
polyphosphatase from Saccharomyces cerevisiae cell envelope,
Yeast, 9, 127-139.
14.Bensandoun, A., and Weinstein, D. (1976) Assay of
protein in the presence of interfering materials, Anal.
Biochem., 70, 241-250.
15.Vagabov, V. M., Trilisenko, L. V., and Kulaev, I.
S. (2000) Dependence of inorganic polyphosphate chain length on the
orthophosphate content in the culture medium of the yeast
Saccharomyces cerevisiae, Biochemistry (Moscow),
65, 349-355.
16.Lichko, L. P., Kulakovskaya, T. V., Kulakovskaya,
E. V., and Kulaev, I. S. (2008) Inactivation of PPX1 and PPN1
genes encoding exopolyphosphatases of Saccharomyces cerevisiae
does not prevent utilization of polyphosphates as phosphate
reserve, Biochemistry (Moscow), 73, 985-989.
17.Chavez, F. P., Mauriaca, C., and Jerez, C. A.
(2009) Constitutive and regulated expression vectors to construct
polyphosphate deficient bacteria, BMC Res. Notes, 2, 50;
doi: 10.1186/1756-0500-2-50.
18.Abramov, A. Y., Fraley, C., Diao, C. T.,
Winkfein, R., Colicos, M. A., Duchen, M. R., French, R. J., and Pavlov,
E. (2007) Targeted polyphosphatase expression alters mitochondrial
metabolism and inhibits calcium-dependent cell death, Proc. Natl.
Acad. Sci. USA, 104, 18091-18096.
19.Kulakovskaya, T. V., Andreeva, N. A., Trilisenko,
L. V., Suetin, S. V., Vagabov, V. M., and Kulaev, I. S. (2005)
Accumulation of polyphosphates and expression of high molecular weight
exopolyphosphatase in the yeast Saccharomyces cerevisiae,
Biochemistry (Moscow), 70, 980-985.
20.Kulakovskaya, T. V., Andreeva, N. A., and Kulaev,
I. S. (1997) Adenosine-5′-tetraphosphate and
guanosine-5′-tetraphosphate – new substrates of the cytosol
exopolyphosphatase of Saccharomyces cerevisiae, Biochemistry
(Moscow), 62, 1180-1184.