Upstream Open Reading Frames Regulate Translation of the Long Isoform of SLAMF1 mRNA That Encodes Costimulatory Receptor CD150
L. V. Putlyaeva1, A. M. Schwartz1, K. V. Korneev1,2, M. Covic1, L. A. Uroshlev1,3, V. Yu. Makeev1,3, S. E. Dmitriev1,4, and D. V. Kuprash1,2*
1Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, ul. Vavilova 32, 119991 Moscow, Russia; E-mail: kuprash@gmail.com2Faculty of Biology, Lomonosov Moscow State University, 119991 Moscow, Russia
3Vavilov Institute of General Genetics, Russian Academy of Sciences, ul. Gubkina 3, 119991 Moscow, Russia
4Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received June 30, 2014
More than 40% of human genes contain upstream open reading frames (uORF) in their 5′-untranslated regions (5′-UTRs) and at the same time express at least one truncated mRNA isoform containing no uORF. We studied translational regulation by four uORFs found in the 5′-UTR of full-length mRNA for SLAMF1, the gene encoding CD150 membrane protein. CD150 is a member of the CD2 superfamily, a costimulatory lymphocyte receptor, a receptor for measles virus, and a microbial sensor on macrophages. The SLAMF1 gene produces at least two mRNA isoforms that differ in their 5′-UTRs. In the long isoform of the SLAMF1 mRNA that harbors four uORFs in the 5′-UTR, the stop codon of uORF4 overlaps with the AUG codon of the main ORF forming a potential termination–reinitiation site UGAUG, while uORF2 and uORF3 start codons flank a sequence identical to Motif 1 from the TURBS regulatory element. TURBS was shown to be required for a coupled termination–reinitiation event during translation of polycistronic RNAs of some viruses. In a model cell system, reporter mRNA based on the 5′-UTR of SLAMF1 short isoform, which lacks any uORF, is translated 5-6 times more efficiently than the mRNA with 5′-UTR from the long isoform. Nucleotide substitutions disrupting start codons in either uORF2-4 result in significant increase in translation efficiency, while substitution of two nucleotides in TURBS Motif 1 leads to a 2-fold decrease in activity. These data suggest that TURBS-like elements can serve for translation control of certain cellular mRNAs containing uORFs.
KEY WORDS: protein biosynthesis, translation control, uORF, mRNADOI: 10.1134/S0006297914120165
Regulation of gene expression at the posttranscriptional level is one of the key mechanisms for changing of gene expression pattern by a cell in response to internal or external stimuli [1]. Translational control includes a variety of factors functioning at initiation, elongation, and termination stages, and even after its completion. Regulation at the initiation stage could be accomplish by various cis-elements that are present in 5′-untranslated regions (5′-UTRs) of mRNA. One of these elements is an upstream open reading frame (uORF) – sequences flanked by start- and stop-codons in the same frame and localized at a 5′-UTR upstream from the main ORF [2]. This situation is rather common: one or more uORFs upstream from the main ORF are detected in approximately half of human mRNAs [3, 4]. In some cases, a polypeptide synthesized on an uORF is functional [5], but generally the uORFs at 5′-UTRs are used for regulation of expression of a protein encoded by the main ORF [2, 6]. In most of cases, this is a simple and effective way to reduce the expression of the main protein [7], because in accordance with the scanning model [8] the presence of alternative start-codons in a leader sequence leads to reduction in ribosome number at the main ORF start-codon. The level of such reduction directly depends on the start codon context: the more effective is its recognition, the less effective is the translation of the main ORF [9, 10]. However, the presence of one or more uORFs in mRNA could impart resistance to some deficiency of an initiation factor. In this case, the effectiveness of translation increases under certain conditions [6, 11]. Thus, the system of uORFs in mRNA leaders of some stress-response genes allows these transcripts to be actively translated under eIF2 inactivation conditions [12, 13]. Theoretically, any mRNA that contains a few uORFs at the 5′-UTR could have unusual dependence on the availability of eIF2 [12]. The presence of uORFs could also lead to the ability of mRNAs to switch synthesis between two protein isoforms depending on the concentration of active eIF2 and eIF4E [14].
Translation of mRNA harboring an uORF in its 5′-UTR involves a variety of mechanisms. The most important in our case is the mechanism of coupled termination–reinitiation involving the regulatory element TURBS (Termination/Translation Upstream Ribosome Binding Site). This mechanism has been described for genomes of some viruses, e.g. caliciviruses [15, 16] and influenza B virus [17]. It was suggested that the TURBS region with core sequence UGGGA interacts with the complementary region of loop 26 of 18S RNA and/or with the eIF3 factor, leading to ribosome retention near the stop-codon with consequent reinitiation at a start-codon that overlaps the stop-codon or is located near the start [16, 18, 19].
CD150 encoded by the SLAMF1 gene is a 70-kDa transmembrane glycoprotein that in humans and mice is expressed at surfaces of B- and T-lymphocytes (at various differentiation stages), mature dendritic cells, thrombocytes, monocytes, and macrophages [20]. SLAMF1 expression increases under activation of lymphocytes and monocytes [21]. In T-lymphocytes, CD150 stimulates antigen-specific CD28-independent proliferation and induces IFNγ synthesis [22], while in B-lymphocytes it induces (or increases) proliferation and synthesis of immunoglobulins [23]. CD150 could also be a receptor for measles virus [24] and act in the process of recognition of gram-negative bacteria and consequent macrophage activation [25]. It was shown that imbalance in regulation of expression of SLAMF1 can lead to progression of autoimmune diseases and immune deficiency disorders [26]. In studies of some B-cell lymphoma, increased expression of CD150 and its participation in activation of mitogen-activated protein kinases were shown [27, 28].
Some mRNA isoforms of the SLAMF1 gene that differ in 5′-UTR length are annotated. We found that the 5′-UTR of the long isoform contains four uORFs and suggested that it could play some role in regulation of gene expression. It is also interesting that the stop-codon of the last uORF overlaps the start-codon of the main reading frame (UGAUG), and the 5′-UTR of the long isoform contains characteristic for TURBS core sequence UGGGA; this suggests the action of a coupled termination–reinitiation mechanism [15, 17]. Here we have analyzed possible mechanisms of long isoform SLAMF1 mRNA translation in a model system using site-directed mutagenesis and selective inhibition of transcription factors.
MATERIALS AND METHODS
Plasmids. Gene engineering manipulations were done according to standard methods. Enzymes from Fermentas/Thermo Scientific (Lithuania) and SibEnzyme (Russia) were used. For developing the basic construct “long”, the 5′-UTR sequence of SLAMF1 mRNA long isoform (with 5′-terminus corresponding to BC012602 mRNA) was amplified together with the first 10 triplets of the gene coding sequence and cloned into pGL3-control vector (Promega, USA) using endonucleases HindIII and NarI. This sequence was cloned in the same frame with the firefly luciferase gene, Fluc, in which its own start-codon was deleted. As the amplified fragment had the HindIII recognition site, the recognition site of type IIS endonuclease BsaI was added to the sequence of the forward primer into the nucleotide context, so HindIII-compatible sticky end formed after restriction. On the basis of the “long” construction, the mutant variants “mut AUG2/3”, “mut AUG3/4” and “mut AUG4” with substitutions of start-codons AUG of corresponding uORFs to leucine triplet UUG or UAG stop-codon were obtained. The construct “mut UGGGA” contained substitution of two nucleotides in the TURBS core sequence to UGACA. Mutagenesis was made by double-step PCR using nested primers; all constructs obtained were checked by sequencing. The primer sequences are listed in the table.
Oligonucleotide primers used in the study

The m7G-capped and polyadenylated RNAs were obtained and purified as described in [29]. PCR products obtained from corresponding plasmids were used as templates. PCR products “long”, “mut AUG2/3”, “mut AUG3/4”, “mut AUG4”, and “mut UGGGA” were obtained using forward primer T7CDfor. Shorter PCR products “del AUG1” (truncated to the start of uORF 1) and “short” (with 5′-terminus corresponding to AK304237 mRNA) were obtained using forward primers T7CD12 and T7CDsh, respectively. All forward primers contained T7 RNA-polymerase promoter sequence. Reverse primer FLA50, which was used for obtaining all PCR products, corresponded to the distal region of Fluc and contained a poly(T) sequence. RNA transcription in vitro was done using T7 RiboMAX Large Scale RNA Production kit (Promega, USA). For preparation of m7G-capped transcripts, m7G cap analog 3′-O-Me-m7GpppG (New England Biolabs, USA) was added to transcription mixture in the ratio 5 : 1 to GTP. RNAs obtained in this way were purified by precipitation with lithium chloride and checked for integrity by denaturing PAGE.
Cell culture and transfection. HEK293T cells were cultured on DMEM (Gibco, USA) with 10% fetal calf serum. The day before transfection, actively proliferating cells were transferred into 24-well plates. After 12-16 h, when confluence achieved 60-80%, transfection was performed using Unifectin-56 (Unifect Group, Russia). The standard protocol was modified to obtain maximal mRNA transfection efficiency according to [30]: a mixture of 0.2 µg of tested mRNA and 0.01 µg of control reporter mRNA (m7G-capped polyadenylated mRNA of Renilla luciferase, Rluc) [31] was incubated with 0.42 µl of Unifectin in 125 µl DMEM for 15 min and then added to the cells. Two hours later, the cells were lysed, and luciferase activities were measured with a Dual Luciferase Assay kit (Promega). To create active eIF2 deficiency, 10 µM sodium arsenite (Sigma-Aldrich, USA) was added to the medium 15 min before transfection. To create active eIF4E deficiency, 250 nM torin 1 (Tocris Bioscience, GB) was added to the medium 15 min before transfection. At least two independent experiments were performed for each mRNA.
RESULTS
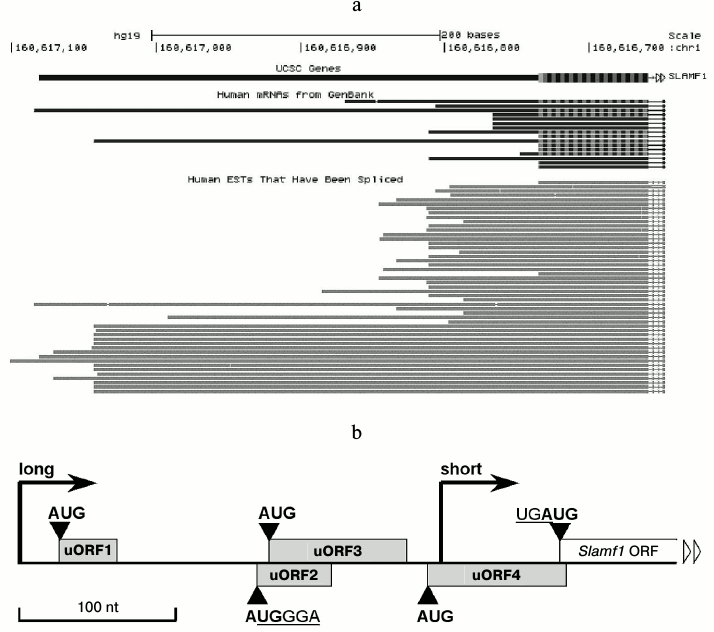
Long isoforms of SLAMF1 mRNA contain few uORFs at 5′-UTR. The GenBank database contains description of ~15 isoforms of SLAMF1 mRNA with different 5′-termini (Fig. 1a). EST analysis shows two dominant transcription starts in SLAMF1 gene (Fig. 1a). Of note, all long transcripts deposited in GenBank are found in tissues that contain no mature lymphoid cells, such as testes and thymus. Our data on SLAMF1 expression obtained on a panel of human B-cell lines also indicate the dominance of SLAMF1 mRNA short isoform in differentiated cells that express CD150 protein, while long isoform is found in CD150-negative pre-B-cells (data not shown). We focused on 5′-UTR of the long isoform of SLAMF1 mRNA. The longest known full length SLAMF1 mRNA has a 5′-UTR of 349 nt length that contains four uORFs, i.e. of 34 nt (1), 48 nt (2), 87 nt (3), and 87 nt (4) lengths, in the order from the 5′-terminus (Fig. 1b). The uORF2 almost completely overlaps uORF3, and the stop-codon of uORF4 crossovers at one nucleotide with the start of the main reading frame of SLAMF1 (UGAUG). Moreover, in the beginning of uORF2 the core sequence of the TURBS element (AUGGGAGAUG) is found. Thus, the structure of SLAMF1 mRNA long isoform contains some features that could reduce its translation efficiency (the presence of uORFs) and others that probably increase it (due to coupled termination–reinitiation). To understand the influence of these features on translation, we obtained a set of reporter mRNAs and analyzed their translation efficiency in a model system.
Fig. 1. Structure of SLAMF1 5′-UTR. a) Abundance of SLAMF1 transcripts in GenBank database according to analysis of genome.ucsc.edu server. Upper part, SLAMF1 5′-UTR and beginning of coding sequence; bottom part, spliced EST. b) Schematic view of SLAMF1 5′-UTR structure. Dominating transcription starts of “long” and “short” isoforms are marked by vertical arrows and ORFs by rectangles. The core sequence UGGGA of the region identical to TURBS Motif 1, which overlaps start-codon of uORF2, as well as stop-start sequence at uORF4 and SLAMF1 ORF junction are also shown.
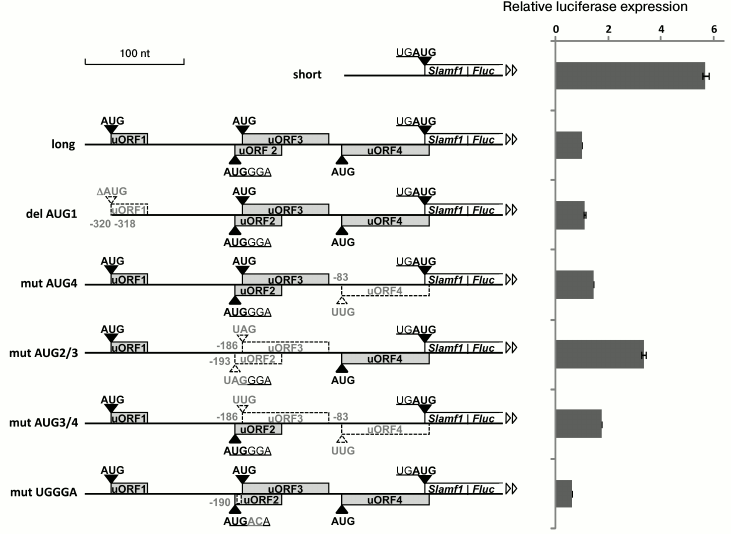
5′-UTR of SLAMF1 mRNA long isoform reduces translation efficiency of reporter gene. To analyze the role of uORFs in regulation of SLAMF1 gene translation, we performed mutagenesis of their start-codons, separately and in combinations. The analysis was done using the method of reporter mRNA transfection to exclude possible impact of nucleotide substitutions on the transcription initiation. Capped and polyadenylated mRNAs, which contained different variants of SLAMF1 5′-UTR fused to coding part of firefly luciferase Fluc, were synthesized by in vitro transcription of PCR-products obtained from corresponding plasmids (see “Materials and Methods”) and then transfected into HEK293T cells. After 2 h, cells were lysed and the reporter activity was measured. Normalized results of measurements of luciferase activity are presented in Fig. 2. We show that the shorter variant of the 5′-UTR without uORFs provides 5-6-fold more effective expression of the reporter gene in comparison to the long form containing four uORFs (Fig. 2). Nucleotide substitutions in separate start-codons of uORFs of long isoform 5′-UTR resulted in increase in luciferase activity, excluding uORF1, whose deletion led to no significant effect. From comparison of “mut AUG3+4” and “mut AUG2+3” activities, it could be suggested that uORF2 makes the largest contribution in the reducing of SLAMF1 mRNA long isoform translation efficiency. The most interesting is the decrease in construction activity as a result of substitutions in the UGGGA sequence (Fig. 2), which violates the hypothetical interaction between mRNA and 18S rRNA and/or eIF3 [16, 19]. These substitutions did not affect the adjacent start-codons of uORF2 and uORF3, and they did not lead to principal changes in key positions of the local nucleotide context of these AUG codons [8]. This result could indicate violation of the termination–reinitiation mechanism in the region between uORF4 and the main reading frame. Especially interesting is the fact that substitutions in the AUG codon of uORF2 and in the UGGGA sequence, being located only a few nucleotides from each other, lead to opposite effects. This observation could indicate complexity of the system of fine regulation of SLAMF1 mRNA translation.
Fig. 2. Upstream ORFs affect translation efficiency of the downstream gene. Short uORFs are denoted by gray rectangles, fused ORF of SLAMF1 and Fluc reporter is designated as white rectangle. Coordinates of changed or deleted (del AUG) start-codons and two nucleotides, which are substituted in UGGGA consensus, are denoted. Start-codons and uORFs disrupted as the result of nucleotide substitutes are denoted by dashed white triangles and rectangles. Nucleotides are enumerated from the main SLAMF1 ORF start-codon.
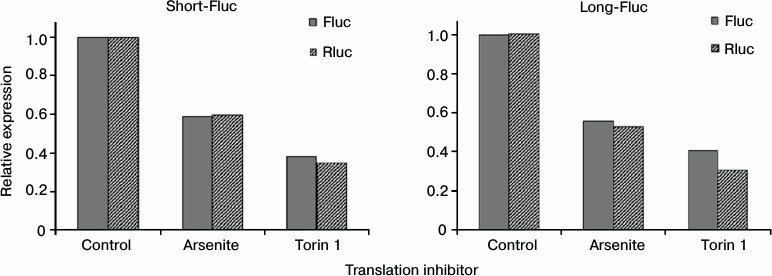
Translation of mRNAs containing both variants of SLAMF1 mRNA 5′-UTR is equally inhibited by deficiency of active eIF2 and eIF4E. The complex structure of the 5′-UTR of SLAMF1 mRNA long isoform could indicate the action of translational regulatory mechanisms that are sensitive to concentrations of particular translation factors. To test this assumption, sodium arsenite or torin 1 were added in addition to HEK293T cells transfected by reporter mRNAs obtained in vitro. Arsenite induces the phosphorylation of initiation factor eIF2, thereby reducing its active concentration in the cell [32, 33]. Torin inhibits mTOR kinase, leading to dephosphorylation and activation of 4E-BP proteins that bind and inactivate eIF4E [34]. Applying of each of the inhibitors led to simultaneous reduction of translation of all constructions studied and the control Rluc mRNA (Fig. 3). Thereby, 5′-UTR of SLAMF1 mRNA long isoform does not have anomalous dependence on the availability of eIF2 and eIF4E factors. This suggests that the translation of SLAMF1 mRNA long isoforms is ineffective independently of cell stress or activation, and regulation of CD150 protein expression occurs on the transcription level from the alternative start.
Fig. 3. Effect of eIF2 and eIF4E translation factor inhibitors on activity of reporter constructs with 5′-UTRs of SLAMF1 mRNA short and long forms. Dark gray denotes the level of Fluc luciferase expression under control of the studied 5′-UTR; diagonal hatching shows Rluc internal control expression. For clarity, expression levels of all constructs are normalized to values obtained in the absence of translation inhibitors. Normalized to internal control signals of short 5′-UTR exceeded signals of long form by 5.4 ± 0.6-fold.
Regulation of expression by switching to synthesis of mRNA short isoform without uORFs might be a widespread phenomenon. The inhibiting effect of uORFs on translation of downstream genes has been repeatedly described in the literature for various organisms (e.g. [35, 36]); this could be the “fast” evolutionary solution of the problem of protein expression regulation without disturbing the complex spatiotemporal regulation at the transcriptional level [37]. To clarify if the situation observed for SLAMF1 gene is characteristic for human genes, we analyzed the data on 15,778 human genes represented in the untranslated sequences database, dbUTR [38]. According to the literature [4], we found that 58% of genes present in the database contain uORFs in the 5′-UTR. Moreover, 72% of these genes (i.e. near 42% of all human genes) also have mRNA isoforms that do not contain uORFs. Thus, the phenomenon of differential transcription and translation of different isoforms described here for SLAMF1 mRNA is rather common. Similar mechanisms of switching to the short mRNA isoform could be widely used by cells for fine regulation of gene expression.
We thank S. P. Sidorenko for fruitful collaboration and useful discussions and O. L. Polyanovskii for discussion of the experimental data.
This work was supported by the Russian Foundation for Basic Research grants 11-04-90484-Ukr_f_a (D. V. K.), 12-04-32203-mol_a (A. M. S.), and 13-04-01121-a (S. E. D.).
REFERENCES
1.Schwanhausser, B., Busse, D., Li, N., Dittmar, G.,
Schuchhardt, J., Wolf, J., Chen, W., and Selbach, M. (2011) Global
quantification of mammalian gene expression control, Nature,
473, 337-342.
2.Barbosa, C., Peixeiro, I., and Romao, L. (2013)
Gene expression regulation by upstream open reading frames and human
disease, PLoS Genet., 9, e1003529.
3.Meijer, H. A., and Thomas, A. A. M. (2002) Control
of eukaryotic protein synthesis by upstream open reading frames in the
5′-untranslated region of an mRNA, Biochem. J.,
367, 1-11.
4.Wethmar, K., Barbosa-Silva, A., Andrade-Navarro, M.
A., and Leutz, A. (2014) uORFdb – a comprehensive literature
database on eukaryotic uORF biology, Nucleic Acids Res.,
42, D60-67.
5.Andrews, S. J., and Rothnagel, J. A. (2014)
Emerging evidence for functional peptides encoded by short open reading
frames, Nat. Rev. Genet., 15, 193-204.
6.Somers, J., Poyry, T., and Willis, A. E. (2013) A
perspective on mammalian upstream open reading frame function, Int.
J. Biochem. Cell Biol., 45, 1690-1700.
7.Calvo, S. E., Pagliarini, D. J., and Mootha, V. K.
(2009) Upstream open reading frames cause widespread reduction of
protein expression and are polymorphic among humans, Proc. Natl.
Acad. Sci. USA, 106, 7507-7512.
8.Kozak, M. (1989) The scanning model for
translation: an update, J. Cell Biol., 108, 229-241.
9.Kozak, M. (2005) Regulation of translation via mRNA
structure in prokaryotes and eukaryotes, Gene, 361,
13-37.
10.Wang, X.-Q., and Rothnagel, J. A. (2004)
5′-Untranslated regions with multiple upstream AUG codons can
support low-level translation via leaky scanning and reinitiation,
Nucleic Acids Res., 32, 1382-1391.
11.Barbosa, C., and Romao, L. (2014) Translation of
the human erythropoietin transcript is regulated by an upstream open
reading frame in response to hypoxia, RNA, 20,
594-608.
12.Hinnebusch, A. G. (2005) Translational regulation
of GCN4 and the general amino acid control of yeast, Annu. Rev.
Microbiol., 59, 407-450.
13.Baird, T. D., and Wek, R. C. (2012) Eukaryotic
initiation factor 2 phosphorylation and translational control in
metabolism, Adv. Nutr., 3, 307-321.
14.Calkhoven, C. F., Muller, C., and Leutz, A.
(2000) Translational control of C/EBPalpha and C/EBPbeta isoform
expression, Genes Dev., 14, 1920-1932.
15.Luttermann, C., and Meyers, G. (2007) A bipartite
sequence motif induces translation reinitiation in feline calicivirus
RNA, J. Biol. Chem., 282, 7056-7065.
16.Poyry, T. A. A., Kaminski, A., Connell, E. J.,
Fraser, C. S., and Jackson, R. J. (2007) The mechanism of an
exceptional case of reinitiation after translation of a long ORF
reveals why such events do not generally occur in mammalian mRNA
translation, Genes Dev., 21, 3149-3162.
17.Powell, M. L., Brown, T. D. K., and Brierley, I.
(2008) Translational termination-reinitiation in viral systems,
Biochem. Soc. Trans., 36, 717-722.
18.Powell, M. L., Leigh, K. E., Poyry, T. A. A.,
Jackson, R. J., Brown, T. D. K., and Brierley, I. (2011) Further
characterization of the translational termination-reinitiation signal
of the influenza B virus segment 7 RNA, PLoS One, 6,
e16822.
19.Luttermann, C., and Meyers, G. (2009) The
importance of inter- and intramolecular base pairing for translation
reinitiation on a eukaryotic bicistronic mRNA, Genes Dev.,
23, 331-344.
20.Cocks, B. G., Chang, C. C., Carballido, J. M.,
Yssel, H., de Vries, J. E., and Aversa, G. (1995) A novel receptor
involved in T-cell activation, Nature, 376, 260-263.
21.Howie, D., Okamoto, S., Rietdijk, S., Clarke, K.,
Wang, N., Gullo, C., Bruggeman, J. P., Manning, S., Coyle, A. J.,
Greenfield, E., Kuchroo, V., and Terhorst, C. (2002) The role of SAP in
murine CD150 (SLAM)-mediated T-cell proliferation and interferon gamma
production, Blood, 100, 2899-2907.
22.Aversa, G., Carballido, J., Punnonen, J., Chang,
C. C., Hauser, T., Cocks, B. G., and De Vries, J. E. (1997) SLAM and
its role in T cell activation and Th cell responses, Immunol. Cell
Biol., 75, 202-205.
23.Punnonen, J., Cocks, B. G., Carballido, J. M.,
Bennett, B., Peterson, D., Aversa, G., and de Vries, J. E. (1997)
Soluble and membrane-bound forms of signaling lymphocytic activation
molecule (SLAM) induce proliferation and Ig synthesis by activated
human B lymphocytes, J. Exp. Med., 185, 993-1004.
24.Sidorenko, S. P., and Clark, E. A. (2003) The
dual-function CD150 receptor subfamily: the viral attraction, Nat.
Immunol., 4, 19-24.
25.Berger, S. B., Romero, X., Ma, C., Wang, G.,
Faubion, W. A., Liao, G., Compeer, E., Keszei, M., Rameh, L., Wang, N.,
Boes, M., Regueiro, J. R., Reinecker, H.-C., and Terhorst, C. (2010)
SLAM is a microbial sensor that regulates bacterial phagosome functions
in macrophages, Nat. Immunol., 11, 920-927.
26.Keszei, M., Latchman, Y. E., Vanguri, V. K.,
Brown, D. R., Detre, C., Morra, M., Arancibia-Carcamo, C. V.,
Arancibia, C. V., Paul, E., Calpe, S., Castro, W., Wang, N.,
Terhorst, C., and Sharpe, A. H. (2011) Auto-antibody production and
glomerulonephritis in congenic Slamf1–/– and
Slamf2–/– [B6.129] but not in
Slamf1–/– and Slamf2–/–
[BALB/c.129] mice, Int. Immunol., 23, 149-158.
27.Mikhalap, S. V., Shlapatska, L. M., Berdova,
A. G., Law, C. L., Clark, E. A., and Sidorenko, S. P. (1999) CDw150
associates with src-homology 2-containing inositol phosphatase and
modulates CD95-mediated apoptosis, J. Immunol., 162,
5719-5727.
28.Yurchenko, M. Y., Kashuba, E. V.,
Shlapatska, L. M., Sivkovich, S. A., and Sidorenko, S. P. (2005) The
role of CD150-SH2D1A association in CD150 signaling in Hodgkin’s
lymphoma cell lines, Exp. Oncol., 27, 24-30.
29.Dmitriev, S. E., Andreev, D. E., Terenin, I. M.,
Olovnikov, I. A., Prassolov, V. S., Merrick, W. C., and Shatsky, I. N.
(2007) Efficient translation initiation directed by the
900-nucleotide-long and GC-rich 5′-untranslated region of the
human retrotransposon LINE-1 mRNA is strictly cap dependent rather than
internal ribosome entry site mediated, Mol. Cell. Biol.,
27, 4685-4697.
30.Andreev, D. E., Dmitriev, S. E., Zinovkin, R.,
Terenin, I. M., and Shatsky, I. N. (2012) The 5′-untranslated
region of Apaf-1 mRNA directs translation under apoptosis conditions
via a 5′-end-dependent scanning mechanism, FEBS Lett.,
586, 4139-4143.
31.Andreev, D. E., Dmitriev, S. E., Terenin, I. M.,
Prassolov, V. S., Merrick, W. C., and Shatsky, I. N. (2009)
Differential contribution of the m7G-cap to the 5′-end-dependent
translation initiation of mammalian mRNAs, Nucleic Acids Res.,
37, 6135-6147.
32.McEwen, E., Kedersha, N., Song, B., Scheuner, D.,
Gilks, N., Han, A., Chen, J.-J., Anderson, P., and Kaufman, R. J.
(2005) Heme-regulated inhibitor kinase-mediated phosphorylation of
eukaryotic translation initiation factor 2 inhibits translation,
induces stress granule formation, and mediates survival upon arsenite
exposure, J. Biol. Chem., 280, 16925-16933.
33.Terenin, I. M., Dmitriev, S. E., Andreev, D. E.,
and Shatsky, I. N. (2008) Eukaryotic translation initiation machinery
can operate in a bacterial-like mode without eIF2, Nat. Struct. Mol.
Biol., 15, 836-841.
34.Thoreen, C. C., Chantranupong, L., Keys, H. R.,
Wang, T., Gray, N. S., and Sabatini, D. M. (2012) A unifying model for
mTORC1-mediated regulation of mRNA translation, Nature,
485, 109-113.
35.Matsui, M., Yachie, N., Okada, Y., Saito, R., and
Tomita, M. (2007) Bioinformatic analysis of posttranscriptional
regulation by uORF in human and mouse, FEBS Lett., 581,
4184-4188.
36.Zhang, Y., Li, W., and Vore, M. (2007)
Translational regulation of rat multidrug resistance-associated protein
2 expression is mediated by upstream open reading frames in the
5′ untranslated region, Mol. Pharmacol., 71,
377-383.
37.Vaughn, J. N., and Arnim, A. G. (2012)
uORF-mediated translational control in eukaryotes, in Encyclopedia
of Systems Biology (Dubitzky, W., Wolkenhauer, O., Cho, K., and
Yokota, H., eds.) Springer Science & Business Media, pp.
2325-2327.
38.Grillo, G., Turi, A., Licciulli, F., Mignone, F.,
Liuni, S., Banfi, S., Gennarino, V. A., Horner, D. S., Pavesi, G.,
Picardi, E., and Pesole, G. (2010) UTRdb and UTRsite (RELEASE 2010): a
collection of sequences and regulatory motifs of the untranslated
regions of eukaryotic mRNAs, Nucleic Acids Res., 38,
D75-80.