REVIEW: Flagella of Halophilic Archaea: Differences in Supramolecular Organization
A. S. Syutkin, M. G. Pyatibratov*, and O. V. Fedorov*
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: mikhailpyatibratov@gmail.com; bratov@vega.protres.ru; fedorov@vega.protres.ru* To whom correspondence should be addressed.
Received September 25, 2014
Archaeal flagella are similar functionally to bacterial flagella, but structurally they are completely different. Helical archaeal flagellar filaments are formed of protein subunits called flagellins (archaellins). Notwithstanding progress in studies of archaeal flagella achieved in recent years, many problems in this area are still unsolved. In this review, we analyze the formation of these supramolecular structures by the example of flagellar filaments of halophilic archaea. Recent data on the structure of the flagellar filaments demonstrate that their supramolecular organization differs considerably in different haloarchaeal species.
KEY WORDS: archaea, flagella, archaella, flagellin, supramolecular structure, halophilicDOI: 10.1134/S0006297914130033
One of the most important discoveries in biology of the previous century is the classification of Archaea as one of the three domains of life, along with Eukarya and Bacteria. They were recognized as a separate branch of prokaryotes by Carl Woese in 1977 based on comparative analysis of 16S rRNA [1]. Many members of this group of organisms are extremophiles, i.e. they occupy ecological niches with extreme temperatures (thermophiles and psychrophiles), pH (acidophiles and alkalophiles), salinity (halophiles), etc. To date, archaea have been found in soil, oceans, and digestive systems of humans and other animals. Archaea possess both characteristics similar to those of Bacteria and Eukarya and properties unique to this group [2]. One of the unique systems of archaea is the flagellar motility apparatus. Its extracellular part is similar to the bacterial analog and is an extended helical filament. However, these structures are fundamentally different in architecture, assembly mechanism, and origin and the archaeal flagella share a number of properties of bacterial type IV pili. There is now no doubt that the archaeal flagella are a unique system of biological motility, and in a recent study it was proposed to call them “archaella” [3]. In this regard, the question arises: how do such different organelles as bacterial and archaeal flagella form similar supramolecular structures?
The mechanism of bacterial flagella spiralization was studied in detail on enterobacteria, in which a helical flagellar filament is composed of only one protein subunit named flagellin. During the co-assembly folding process [4], this protein can have two distinct conformations called the L- and R-conformations. Each longitudinal row of protofilaments consists of flagellin in only one of these conformations. Since the length of protofilaments composed of flagellin in the L- and R-conformation is slightly different, twisting forces leading to flagella coiling arise on their interaction [5].
It is of interest that about 45% of the known genomes of bacteria with a flagellar motility system have multiple copies of flagellin genes, which might be indicative of a selective advantage of additional flagellins [6]. The number of flagellin genes in such microorganisms ranges from 2 (Salmonella enterica, serovar Typhimurium) to 15 (Magnetococcus sp. MC-1).
Studies on Salmonella typhimurium, which has two flagellin genes, fliC and fljB, revealed that its flagellar filaments can be constructed from the product of one of these genes. To change the flagella protein composition, this bacterium uses the phase variation system [7] that includes, along with the flagellin genes, a gene encoding transcription repressor protein of the fliC gene and an invertase gene, which controls the flagellin phase variation. The recombination mechanism (site-specific inversion of the promoter-containing segment) allows the expression of only one of the flagellins at a given time. The fljB gene is located in the same operon with the gene encoding the transcription repressor of the fliC gene. Thus, in different phases a S. typhimurium flagellum consists of different flagellins, which helps pathogens avoid host immune response by changing the antigenic determinants.
The genome of another microorganism, Caulobacter crescentus, has six flagellin genes (fljK, L, M, N, O, J), wherein the protein products of each gene are contained in the wild-type flagella. An interesting property of these flagella is that different parts of the filament are composed of different flagellins. Using deletion analysis, it was shown that each of the flagellins (except FljJ) could alone build functional filaments. Furthermore, it was found that there are no strict requirements to the order of flagellin incorporation into the growing flagellum, and the observed distribution of flagellins along a flagellin filament rather reflects regulation of flagellin secretion [6]. Thus, this organism has high redundancy of flagellin genes, the significance being not yet clear.
Currently archaeal flagella, as compared with bacterial analogs, are not well understood in terms of both their structure and function. However, analysis of genomic sequences shows that in contrast to bacteria, the presence of a single flagellin gene in the genome of archaea (particularly, halophilic archaea) is an exception, and multiplicity of flagellin genes is widespread among Archaea.
In this review, we consider the results of studies on the role of flagellin gene multiplicity that have been conducted mainly on halophilic archaea. The data suggest that the mechanism of formation of the helical flagellar filaments can be different even among related haloarchaea, wherein multiple flagellins can play both a structural role (when each of them is required for functional filament formation) and an adaptive part (when the flagellins may substitute for each other on environmental changes).
FLAGELLAR MOTILITY OF ARCHAEA
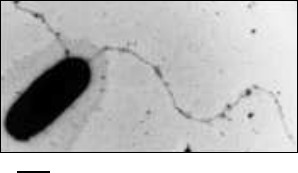
General characteristics. So far, in the Archaea domain flagella based motility has been described only with flagellar filaments (Fig. 1) functioning as in bacteria, like a propeller creating hydrodynamic forces [8].
Fig. 1. Electron micrograph of a Halobacterium salinarum cell with flagella. The scale bar is 500 nm.
Initially, it was believed that the archaeal flagellar motility apparatus was identical to bacterial ones, but detailed studies revealed fundamental differences between them. It was shown that: 1) archaeal flagella are substantially thinner than bacterial flagella and have no internal channel, which indicates fundamental differences in their assembly mechanisms; 2) the protein components of the archaeal flagellar motility apparatus have no homology with components of the bacterial flagellar motility system, but some of them have homology with proteins of bacterial type IV pili; 3) the main structural protein of archaeal flagella – flagellin – is synthesized as a precursor protein with an N-terminal signal peptide. It is now clear that bacterial and archaeal flagella are functional analogs, but they have a different origin and are a striking example of biological convergence. Based on the known facts, it was concluded that the archaeal flagella are a unique biological motility device, and recently the term “archaella” (from archaea and flagella) was proposed for them [3], which has not yet become generally accepted [9].
The location of archaeal flagella on the cell surface can be unipolar, bipolar, or peritrichous. Bundles of flagella located at the cell poles are characteristic of halophilic archaea [8, 10]. Strictly, a flagellar filament is a helical strand with thickness of 10-22 nm and length of several micrometers. Unlike bacterial flagella, filaments of which can be both right- and left-handed, only right-handed filaments have been described in Archaea. The velocity of archaeal cells varies over a very wide range – from 3 µm/s for halophilic archaea to about 600 µm/s for some methanogens. It is interesting that if we consider the velocity of the organisms with respect to their linear dimensions, the methanogenic archaea Methanocaldococcus villosus currently hold the record for speed among all known organisms (500 body lengths per second) [11].
In addition to its primary role in providing motility, archaeal flagella can mediate other functions such as adhesion to abiotic surfaces, formation of cell–cell contacts, and possibly intercellular communication [12, 13].
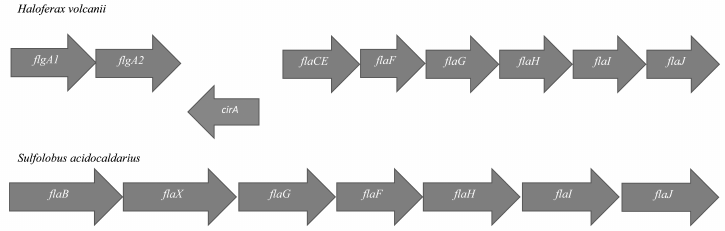
In Archaea, most genes encoding proteins of the motility apparatus are located in the so-called fla-locus (Fig. 2). This locus usually starts with one or more gene copies encoding the main structural component of the flagellum – flagellin (FlaA/B or FlgA/B). The flagellin genes can be transcribed either individually or in association with fla-associated genes lying below. In general, Archaea (especially Euryarchaeota) are characterized by the presence multiple flagellin gene copies in the genome, which can be arranged into one or several operons. Some of the flagellin genes can be located distantly from the fla-locus (Halobacterium salinarum) and even on other replicons (Haloarcula marismortui).
Fig. 2. Structure of the fla-locus in Haloferax volcanii and Sulfolobus acidocaldarius.
In addition to flagellin genes, the fla-locus includes a set of so-called fla-associated genes that are transcribed together. Experiments on the deletion of these genes showed that each of them is critically needed for motility [14-16]. In Euryarchaeota and Crenarchaeota (two main phyla of Archaea), the set of fla-associated genes is slightly different (see Fig. 2).
The flaC and flaE genes can be either individual or fused together (Haloferax volcanii); the products of these genes are membrane proteins. The flaD gene can be either in the composition or at a distance (as in H. volcanii) from the fla-locus. It has been shown on H. salinarum that gene products of the flaCE and flaD genes can interact with components of the chemotaxis system, and thus they participate in switching the rotational direction of the motor [17]. In Crenarchaeota, the flaC, D, E genes are absent, but there is a flaX gene that is absent in Euryarchaeota [18]. It was shown that the FlaX protein can form an oligomeric annular structure due to the C-terminal α-helices [19]. Moreover, it forms a stable complex with the FlaI protein and might be involved in the generation of torque by the flagellar motor. The functions of the flaF and flaG gene products are not yet clear.
The products of flaH, I, and J genes are believed to form a secretory complex involved in flagellum assembly. Theoretical analysis of protein FlaH showed that it contains a typical Walker A motif and an incomplete Walker B motif, which is evidence of possible ATPase activity. In the structure of protein FlaJ, several transmembrane regions are predicted, suggesting that it is an integral membrane protein. Furthermore, two highly charged cytoplasmic loops are present in its structure [18, 20]. This protein is homologous to conserved membrane protein PilC of the Pseudomonas aeruginosa type IV pili system and probably interacts directly with protein FlaI [18]. The latter is an ATPase of the secreting ATPase superfamily and possesses ATPase activity in vitro, which is stimulated in the presence of archaeal membrane lipids. It is assumed that the protein provides the energy to the flagellum assembly system [21].
Another component of the motion system is the signal (leader) peptidase (FlaK or PibD) located on the membrane. Its gene is usually located distantly from the fla-locus. The role of the signal peptidase is to cleave the N-terminal signal peptide from the flagellin molecule, which is synthesized as a precursor protein similarly to type IV pilins [22-25]. The length of the flagellin signal peptide is 6-16 amino acid residues [26]. Genetic studies have shown that the removal of the signal peptide is a critical step in the formation of the motility apparatus [24, 27].
Regulation of assembly of the archaeal flagellar motility apparatus. Flagella assembly and functioning are extremely energy-intensive processes for the cell, and, therefore they are strictly controlled by regulatory mechanisms. The first example of the influence of environmental conditions on flagella synthesis in Archaea was the discovery of the fact that in Methanocaldococcus jannaschii and Methanococcus maripaludis flagella synthesis is induced at low hydrogen concentrations [28, 29]. It should be mentioned that these organisms are methanogens and use molecular hydrogen as an electron donor in the process of methanogenesis, which is an energy source for them. Thus, starvation is one of the factors causing flagella synthesis. A similar effect was observed on another archaeal species, Sulfolobus solfataricus, in which the level of flagellin gene transcription strongly increased when reaching the stationary growth phase or under conditions of nitrogen starvation [30].
The study of the Sulfolobus acidocaldarius fla-operon revealed the presence of two promoters [16]. One promoter is located upstream from the flagellin gene (flaB), and the second is before the flaX gene, from which the flaX-J genes are transcribed. For this organism, as well as for the related S. solfataricus, the phenomenon of flagella synthesis induction under starvation conditions was discovered. Under these conditions, the activation of the promoter located upstream from the flaB gene was increased, but the second promoter had no such effect. These results indicate that the S. acidocaldarius genes encoding the basal body proteins are under control of a constitutive promoter located upstream from the flaX gene, and thus these gene products are constantly present in the cell. Therefore, to make the cell motile, only synthesis of flagellin, whose gene is under control of the promoter activated under starvation, is required. This regulation system allows the cell to complete forming the motility apparatus quickly and to seek conditions that are more favorable. In subsequent studies of this system, several proteins regulating the biosynthesis of the motility apparatus were identified [31, 32].
INVESTIGATION OF ARCHAEAL FLAGELLA STRUCTURE
Mechanisms of assembly and function of archaeal flagellar filaments. The structure of the archaeal flagellar motility apparatus is not as well studied as the bacterial analog. The bacterial flagellum consists of a rigid helical filament that acts as a propeller, a motor producing rotary motion, and a flexible hook junction transmitting torque from the motor to the filament. In Archaea, structures morphologically resembling the bacterial hook have been identified only in methanogens. This area looks like a thickening near the end of the flagellar filament [33].
Archaeal basal bodies have been seen by electron microscopy of cells and membrane fractions treated with the nonionic detergent Triton X-114. In methanogens Methanospirillum hungatei [34] and Methanococcus thermolithotrophicus [35], structures representing two rings connected with a rod, similar to those of gram-positive bacteria, have been described. Furthermore, in work with H. salinarum, the flagella bundles attached to the polar cap-structure have been isolated [36]. On ultra-thin serial cell sections, it corresponds to the discoid lamellar structure (DLS) [37]. It was found that the flagella bundle passes through the plasma membrane and abuts the DLS – a complex electron dense structure 250-300 nm in length located at a distance of 20 nm from the plasma membrane [38, 39]. It is assumed that due to different structure of the cell wall in Archaea, additional structures should hold the flagellum. Currently, the exact structure and mechanism of action of the motor generating rotational motion remain unknown. For H. salinarum, it was shown that the archaeal motor rotation is driven by energy of ATP, unlike the bacterial motor that uses the energy of proton or sodium ion gradients [40].
The proteins of the archaeal basal body are now being actively studied on the model organism Sulfolobus acidocaldarius. It was shown that protein FlaI has ATPase activity in vitro, and this activity is specifically stimulated by adding archaeal membrane lipids [21]. Furthermore, ATP-dependent FlaI oligomerization accompanied by formation of a hexamer with diameter of 14 nm was found [41]. These studies led to the conclusion that protein FlaI is responsible for assembly and rotation of the flagellar filament. Studies of another component of the basal body, protein FlaX, showed that in vitro FlaXc (a truncated variant of FlaX) is capable of forming oligomeric rings with diameter of 30 nm. Moreover, it was found that FlaX through its C-terminal region interacts with FlaI [19]. Further experiments demonstrated that proteins FlaXc, FlaI, and FlaH interact with each other to form a ternary complex [42]. Based on these data, a model of the basal body was proposed according to which the polytopic transmembrane protein FlaJ acts as a motor assembly platform, around which FlaX forms an oligomeric ring with diameter of 30 nm. Then, a hexamer consisting of FlaI is inserted into this ring. Furthermore, the FlaI hexamer is able to form additional connections with protein FlaJ. This complex can then bind protein FlaH that, probably accomplishes nucleotide binding and regulation of motility.
The main part of the flagellum is a helical filament consisting of multiple copies of one or several flagellin proteins. A typical archaeal filament is 10-14 nm in diameter, and a few micrometers in length. Models of H. salinarum and Sulfolobus shibatae flagellar filaments were constructed based on electron microscopic images [43-45]. In both cases, the surface flagellin domain forms a three-start helical line wrapped around the core, which has no internal channel. In both organisms, the inner flagellin core is conservative in size and shape, and its thickness is about 5 nm. This core is apparently formed by hydrophobic helices of the α-N-terminal part of flagellin. Since the archaeal flagellins have a highly conserved region of 30-40 amino acids at the N-terminus [22, 46, 47], it is suggested that a similar structure of the filament core is common for all flagellated archaea. In contrast to the conservative inner domain, the outer part of the filament in these two organisms is different, which explains differences in flagella thickness: 14 nm in S. shibatae and 10 nm in H. salinarum. Differences in the external filament part may reflect adaptation of these organisms to specific environmental conditions [43, 45].
It should be noted that archaeal flagella are rather resistant to dissociating agents and, unlike bacterial analogs, are not capable of self-assembly from individual subunits in vitro. Besides, there is still no resolved spatial structure of archaeal flagellin. Indirect methods of analysis have been used to investigate the archaeal flagellin structure. Our group studied the domain organization of flagellins in the H. salinarum filaments using scanning microcalorimetry, which showed only one heat absorption peak corresponding to a cooperative melting domain, suggesting that all five flagellins of this organism have similar thermodynamic properties [48]. The thermal unfolding of flagellin domains was irreversible, but it did not lead to dissociation of the flagellar filament. Melting of the filaments was observed only in experiments with filament solutions at lower NaCl concentrations (10% or less); at higher salt concentrations (20% NaCl), which are natural for this microorganism, the heat absorption peak was not found in the scanned area (up to 130°C), indicating high thermal stability of the filaments. Interesting results for Natronomonas (previously Natronobacterium) pharaonis flagellins were obtained using scanning microcalorimetry [49]. The flagella of this organism depolymerized into individual subunits in the presence of the nonionic detergent Triton X-100, while the flagellins preserved their tertiary structure and melted cooperatively, although the melting curves of monomeric flagellins and filaments were different in shape, which might be due to destabilization of the tertiary structure of flagellins after dissociation. These results indicated the importance of hydrophobic interactions in forming the flagella structure. As in the case of H. salinarum flagella, the melting of flagellin domains in N. pharaonis was irreversible: reheating the samples did not reveal refolding processes [49].
Glycosylation of archaeal flagellins. One of the characteristic features of the archaeal flagellins is N-glycosylation. For a long time, N-glycosylation was considered as a hallmark of eukaryotes [50]. However, after the discovery of N-glycosylation of the surface (S)-layer protein and flagellins in halophilic archaea H. salinarum [51, 52] and subsequent studies, it was shown that glycosylation is widespread among Archaea [50].
Experiments on violation of N-glycosylation have shown that this process is not vital for some archaea (for H. volcanii only a growth retardation was observed), but it is critical for the life of other archaea such as S. acidocaldarius [53, 54]. It is believed that this posttranslational modification allows proteins to function in severe environments characteristic of archaea [50]. In this connection, it is interesting to note that the number of sites where N-glycosylation occurs, as well as the attached glycan structure, can vary in a cell culture with environmental changes [55]. These results suggest a role of N-glycosylation in adapting to environmental changes.
All flagellins of M. voltae, M. maripaludis, H. salinarum, and H. volcanii are glycoproteins with N-type glycosylation [52, 53, 56-58]. N-glycosylation in Archaea is a multi-step process. In the first stage, the N-glycan is assembled in the cytoplasm on a lipid carrier through sequential addition of carbohydrate monomers by appropriate glycosyltransferases. Then the glycan is transferred across the cytoplasmic membrane by a still unknown enzyme. The N-glycan is then entirely transferred to the flagellin molecule and attached to the asparagine residue in the glycosylation site (N-X-S/T, where X is any amino acid except proline) by AglB oligosaccharyl transferase [59, 60]. The length and structure of the attached glycan can differ in different species.
The analysis of mutant M. voltae and M. maripaludis strains with deletions of the agl genes (encoding proteins of the glycosylation system), which led to a shortening of the glycan chains, showed that the nonglycosylated flagellins and flagellins bearing glycans consisting of only one sugar do not form flagella [59, 60]. In cases when the glycans consisted of two-three sugars, the cells had flagella, but their motility on semisolid agar decreased in comparison with the wild-type cells carrying a normal-length carbohydrate chain (consisting of four monomers). Most of the known amino acid sequences of archaeal flagellins contain several N-glycosylation sites, but not all of these sites are connected with oligosaccharides. For example, despite the presence of six N-X-S/T sites in the H. volcanii major flagellin FlgA1, glycosylation of only three of them has been confirmed experimentally. Using site-directed mutagenesis, it was demonstrated that all three sites are important for the assembly or functioning of the flagellum, since violation of each of the sites causes the loss of motility on semisolid agar plates [53]. Thus, all of the currently available data indicate that N-glycosylation is a posttranslational modification crucial for proper assembly of the archaeal flagellum.
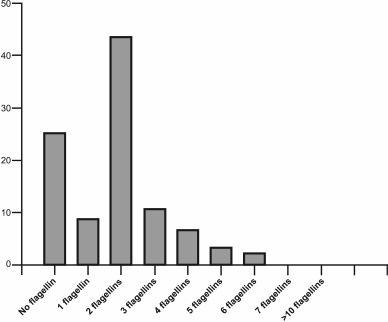
Study of the role of flagellin gene multiplicity in Archaea. The number of flagellin genes in Archaea varies widely, with the multiplicity of these genes in this domain being more widespread than in Bacteria. For example, among 90 haloarchaea with known genomic sequences, about 25% have no flagellin gene, 9% contain a single flagellin gene, 43% have two such genes, and other genomes contain three to six flagellin genes (Fig. 3). The presence of several flagellins in the structure of flagella was confirmed in earlier studies of the archaeal motility apparatus [61, 62]. To determine the role of each of the multiple flagellins in filament formation, experiments on inactivation of the appropriate genes were conducted on some model organisms using both insertions and deletions. Among halophilic archaea, H. salinarum was the first used in these experiments. The genome of this organism contains six flagellin genes (flgA1, A2 forming the A operon, flgB1, B2, B3 forming the B operon, and the separately located flgX gene), however only the products of the first five genes were found as part of the flagella [62]. The first experiments devoted to elucidation of the role of each flagellin were performed using inactivation with an insert; thus, the flgA2-gene inactivation resulted in a significant decrease in cell motility, and only straight flagella were formed [63]. In addition, it was shown that flgB-operon transcription was stopped in the mutant strain. Further studies were carried out using a deletion approach; the results showed that only straight filaments were observed in strains with the flgA1 or flgA2 deletion, as in the case of inactivation [64]. As mentioned above, a bacterial flagellum is helical due to the ability of a single flagellin to accept two conformational states – L or R. The results of H. salinarum flagellin gene inactivation indicate that in this organism the FlgA1 and FlgA2 flagellins might be analogs of two conformational states of the single bacterial flagellin. It was suggested that this principle of supramolecular organization of flagellar filaments might be common for all archaea. This is supported by the results of other experiments on the flagellar filament of the haloalkaliphilic archaea Natrialba magadii, composed of four different flagellins, when two immunologically distinct protofilaments types were found [65].
Fig. 3. Distribution of flagellin genes in haloarchaeal genomes. The analysis was conducted for 91 haloarchaeal species with known genomic sequences.
The role of H. salinarum flgB-operon genes was studied also using a deletion approach. Short curved filaments composed of the FlgB2-flagellin were observed on the cell surface of a mutant strain with deletion of the flgA operon (ΔflgA). However, the analysis of mutant strains in which only the flgB1- or flgB3-flagellin genes were present showed no similar curved structures on the surface of these cells. It was concluded that in the absence of the FlgB2-flagellin, the FlgB1 and FlgB3 cannot form short curved structures [66]. Based on these results, it was suggested that the H. salinarum FlgB2-flagellin forms a hook region. However, using immunodetection of specific peptide loops alternately introduced to the surface of FlgA1, FlgA2 and FlgB2 flagellins, it was recently shown that the FlgB2-flagellin, similarly to FlgA1 and FlgA2 flagellins, is uniformly distributed along the filament length and is not localized near the flagellum base, as could be expected for a hook protein [67, 68].
The role of multiplicity of flagellin genes was also studied on the methanogenic archaea Methanococcus maripaludis. In experiments performed in the laboratory of Prof. K. Jarrell (Canada), it was shown that inactivation of flagellin genes led to complete loss of motility or its abnormalities [15]. A general conclusion from these experiments was that every flagellin gene product is required for normal motility. However, the role of multiplicity of flagellin genes remains unclear. In this connection, it is interesting that representatives of some groups of Archaea, such as the Sulfolobus genus (phylum Crenarchaeota), have only one flagellin gene and are motile [30, 69].
Of special interest for studying the archaeal flagella formation is the halophilic archaea Haloarcula marismortui. This is explained by the following reasons: 1) the flagellin genes are located on different replicons – the chromosome and plasmid; 2) the mass of flagellins calculated from nucleotide sequences is significantly greater than the flagellin mass of other archaea [70]. These facts suggest that the principles of flagella assembly and flagellin biosynthesis regulation in this organism differ from those for the previously studied archaeal motility devices. Haloarcula marismortui was isolated from the water of the Dead Sea; its genome contains a full set of motility genes [70], notwithstanding that originally it was described as having no pronounced motility [71]. In addition to the fla-associated gene cluster, three flagellin genes were annotated in its genome: flaB is located on chromosome I near the fla-locus, flaA1 is on chromosome II, and flaA2 is present on the pNG100 plasmid. It should be noted that a similar arrangement of flagellin genes, when each of them is located on a separate replicon, is extremely rare, and typically all flagellin genes are located on the main chromosome. Putative protein products of the flaA2 and flaB genes have lengths of about 450 amino acid residues and characteristic archaeal flagellin features, such as the presence of a signal peptide (12 amino acids) and highly conserved N-termini. Haloarcula marismortui FlaA2 and FlaB flagellins are homologous to each other (56% identity) and have two conserved regions in the N- and C-termini. The central flagellin part includes highly variable regions containing insertions and deletions, and it is apparently not involved in the formation of intersubunit contacts but rather determines surface filament properties. The hypothetical protein product of the flaA1 gene has length of 84 amino acid residues only, does not contain a signal peptide, and therefore is unable to be secreted and to form a flagellar filament. Thus, in the H. marismortui genome there are only two complete flagellin genes.
During cultivation of the H. marismortui collection strain, two strains (phenotypes) were identified in which the flagellar filaments had different protein composition [72]. These differences are seen on electrophoregrams of isolated filaments: for each of the two strains, a single protein band was observed having a similar, but not identical electrophoretic mobility.
Mass spectrometry of the respective protein bands showed that the filaments isolated from one strain were composed of the FlaA2 flagellin, while filaments isolated from the other strain were formed of the FlaB flagellin. The molecular weights of FlaA2 and FlaB flagellins calculated from their amino acid sequences are 47.4 (45.9) and 46.8 (45.4) kDa, respectively (the weight given in brackets is for a mature protein without a signal peptide). However, the flagellin molecular weights experimentally determined from the electrophoretic mobility were about 70 kDa. Anomalous electrophoretic mobility of haloarchaeal flagellins was observed earlier; it is caused by the high content of negatively charged amino acids in flagellins [73] and due to the posttranslational modifications, particularly glycosylation [50, 56]. The resulting strains were named the FlaA2 strain and FlaB strain. When cultured in a rich culture medium, they were stable and did not move to each other [72].
The existence of two strains differing in the flagellar protein composition resembles the phenomenon of phase variation described for enterobacteria, which use this mechanism for protection from the immune response of the host [74]. It was necessary to clarify how the protein composition of the H. marismortui filaments is regulated. Since the flaA2 gene is located on the pNG100 plasmid, it was suggested that the appearance of the FlaB strain is a result of plasmid loss. To test this hypothesis, the PCR (polymerase chain reaction) was carried out using two pairs of primers for two different and distant from each other sections of the pNG100 plasmid. The experimental results showed that no PCR products were synthesized when the FlaB strain total DNA was used as the template, while upon using the FlaA2 strain DNA the PCR products were accumulated. Thus, the strains FlaA2 and FlaB are a wild-type strain and a strain that has lost the pNG100 plasmid, respectively.
The consequence of these results is that the flagella synthesized by the FlaB strain are composed entirely of FlaB flagellin. It was interesting to check whether FlaB flagellin synthesized in the FlaA2 strain or its flagella consist of the FlaA2 flagellin only. Immunoblotting with FlaB flagellin antibodies demonstrated that the flagella isolated from the FlaA2 strain contained a minor quantity of the FlaB flagellin [72]. It can be assumed that the pNG100 plasmid contains a gene of repressor protein suppressing (but not completely) the FlaB flagellin biosynthesis in the FlaA2 strain. Thus, the protein composition of the H. marismortui flagella is determined by the plasmid, this being the first known case of such type in the domain Archaea [75].
Electron microscopy of preparations of the FlaA2 and FlaB filaments showed that they had a distinct helical shape. The thickness of the FlaA2 and FlaB filaments was 20-22 and 16-18 nm, respectively, i.e. they were much thicker than the previously studied archaeal flagella. The electron microscopy data combined with the fact that the flagella isolated from cells of the FlaB strain consist exclusively of the FlaB-flagellin demonstrated for the first time that in Euryarchaeota a helical flagellum could be constructed from a single type of subunits. This fact is particularly noteworthy because previous studies performed on some representatives of Euryarchaeota showed that they need at least two flagellins to form functional helical flagella [15, 63]. Thus, regardless of the fact that the flagellins in Euryarchaeota are closely related proteins, they can use different principles of flagellar filament formation.
For comparison of their motility, the FlaA2 and FlaB strains were plated on a semisolid agar medium. It was found that both strains formed spots around the inoculation points, which means that both strains are motile, wherein the spot diameter of the FlaB strain was about twice as large as that of the other strain. Since the growth rate of the cells of both strains in liquid medium is approximately the same, it was concluded that under these conditions the cells of the FlaB strain have higher motility [72].
For more information on the structure of filaments consisting of FlaA2 and FlaB flagellins, we used differential scanning microcalorimetry in native saline conditions. On the melting curves, two heat absorption peaks were observed at temperatures of 72oC (peak 1) and 80oC (peak 2) for FlaA2 filaments and 55oC (peak 1) and 84oC (peak 2) for FlaB filaments. Thus, FlaB filaments are more sensitive to higher temperatures than FlaA2.
The results on the melting of H. marismortui filaments differ from the data for H. salinarum, where all five flagellins have similar thermodynamic properties and the melting curve looks like a single heat absorption peak corresponding to one domain [48]. Repeated heating of the partially (peak 1) and fully (peak 2) denatured H. marismortui flagella samples showed complete irreversibility of heat denaturation. Like previously investigated haloarchaeal filaments, the H. marismortui filaments do not dissociate after heating.
Further studies using limited proteolysis demonstrated that under native saline conditions, the H. marismortui filaments are resistant to trypsin attack. Trypsin treatment of the samples heated to 90°C immediately leads to almost complete cleavage of the flagellin with the formation of a number of products of proteolysis. For partially denatured (completion of the first melting peak of heat absorption) FlaB filaments, a somewhat different picture was observed: a predominant part of total flagellin (~80%) is cleaved in a few minutes, while some part remains intact even at long times of incubation. This result indicates that two forms of flagellin FlaB – FlaB-r and FlaB-s – differing in the availability to trypsin attack are present in the filament composition. Comparison of the electrophoretic mobility did not reveal any differences between FlaB-r and FlaB-s, but the results of anion-exchange chromatography showed that under denaturing conditions these forms are not identical. The elution profiles of the two forms are complicated and apparently can be subdivided into several subforms [76]. The observed differences are apparently the result of posttranslational modification of the flagellin. Archaeal flagellins typically are glycoproteins with N-glycosylation; however, neither FlaA2 and FlaB flagellin contains canonical N-glycosylation sites N-X-S(T). Despite this, the data of specific staining with Schiff’s reagent and CHN-analysis confirmed that H. marismortui flagellins are glycoproteins [76]. Here either N-glycosylation by a noncanonical site or some other glycosylation type (e.g. O-glycosylation) takes place. Recently, publications appeared showing cases of noncanonical N-glycosylation in eukaryotes [77], but for archaea similar cases have not been described.
Study of molecular organization of Halorubrum lacusprofundi flagellar filaments. The study of the H. marismortui motility apparatus demonstrated for the first time that functional helical flagella of halophilic archaea could be formed of a single flagellin only. This result showed that the previous principle when the formation of helical filaments required at least two different flagellins is not universal for Euryarchaeota. However, is H. marismortui in this respect a unique organism, or can other halophilic archaea also form a functional flagellum of subunits of a single protein? In recent years, the number of known archaeal genomes increased dramatically, and among them there are several genomes containing only one flagellin gene. One of these organisms, Halorubrum lacusprofundi, was chosen for study. Halorubrum lacusprofundi was isolated from the hypersaline Deep Lake in Antarctica. According to the original description [78], this organism is nonmotile and does not synthesize flagella, while a complete set of fla-associated genes and the single flagellin gene flaB are present in the genome (http://www.ncbi.nlm.nih.gov/genome/1202). The predicted protein product of this gene has size of 243 amino acids and contains a signal peptide.
The collection H. lacusprofundi culture was subjected to several rounds of selection on a semisolid agar medium, whereby cells having notable motility were derived. These cells were used to isolate flagella by the standard procedure. SDS-PAGE of the isolated flagella showed the presence of a single protein band corresponding to (according to mass spectrometry data) the H. lacusprofundi flaB gene product. Electron microscopy of flagella preparations showed that most of the filaments have a distinct helical shape. It is somewhat surprising that polymorphic filaments with a different amplitude and pitch of the helix, including circular shaped filaments, were observed. The thickness of the filaments was 10-11 nm. Subsequent study of the H. lacusprofundi filaments using anion-exchange chromatography allowed, as in the case of H. marismortui, the identification of multiple flagellin forms different in their charge characteristics. It was found that the filaments contain at least two major flagellin forms. The relative content of the flagellin forms in the filament composition was estimated by comparison of the peak areas in the chromatograms. The share of the major flagellin form was about 40% of the total protein [76]. The reason for the flagellin heterogeneity, as in the case of H. marismortui, might be the presence of posttranslational modifications. Unlike H. marismortui flagellins, the H. lacusprofundi flagellin contains six canonical N-glycosylation sites. Staining of the flagellin with Schiff’s reagent gave a positive result [76]. These results are further evidence that the helical flagella of haloarchaea can be constructed of only one flagellin, and the principle observed for H. marismortui is not unique.
Based on these data, and on the general principle according to which for the formation of a superhelix the filament subunits must be nonidentical, it can be assumed that the spiralization of H. marismortui and H. lacusprofundi flagellar filaments are due to conformational lability of flagellins of these organisms. An indirect confirmation of this is the fact that in the filament composition these flagellin subunits are not identical in their accessibility to trypsin attack (for the H. marismortui FlaB-flagellin) and charge characteristics. The difference in the charge characteristics might be a result of varying degrees of posttranslational modifications (glycosylation) of flagellin subunits due to their different conformations.
Role of flagellin gene redundancy in H. marismortui. The study of the H. marismortui motility apparatus showed that the helical and functional flagellum of this organism can be built entirely of FlaB flagellin subunits. Thus, the biological role of the second flagellin gene (flaA2) located on the plasmid remained unclear. In this regard, of interest are studies that explored the role of the redundancy of H. marismortui 16S rRNA genes. The H. marismortui genome contains three 16S rRNA genes (rrnA, rrnB, and rrnC) [70]. The rrnA and rrnC genes are almost identical, and rrnB varies significantly in nucleotide sequence. Studies have shown that the rrnB gene is not vital [79] and, thus, the H. marismortui genome has redundancy of 16S rRNA genes. Investigation of the role of the redundancy showed that the presence of the rrnB gene is a mechanism of adaptation to changes in environmental temperature [80]. With an increase in cultivation temperature, the transcription level of the rrnB gene increased, and its gene product replaced rrnA/C products. Theoretical analysis has shown that the 16S rRNA encoded by rrnB possesses higher thermal stability, and thus, the presence of the rrnB gene increases the rate of the organism’s reaction. For genes whose products perform the same function in different environmental conditions, the term “ecoparalogs” has been proposed [81]. Further analysis revealed that the H. marismortui genome has significant redundancy level: about a hundred genes have been suggested as candidates for ecoparalogs [81]. It should be noted that H. marismortui flagellin genes were not included in this list, possibly because the flagellin gene multiplicity is widespread among Archaea. The flagellin gene redundancy might be a mechanism of adaptation, which is indirectly confirmed by the data of filament melting demonstrating that FlaA2 filaments are more thermally stable than the filaments formed of FlaB flagellin [72].
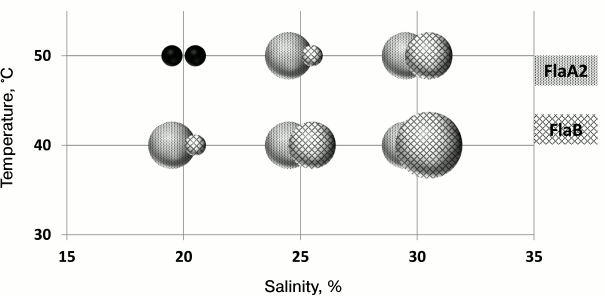
It is known that the stability of halophilic proteins depends on the salinity of the environment. To test the hypothesis that the redundancy of H. marismortui flagellin genes is a mechanism of adaptation, the motilities of FlaA2 and FlaB strains were compared after growth on a semisolid agar medium at different temperatures (40 and 50°C) and salinity (20, 25, and 30%) (Fig. 4). Under 20% salinity and 50°C, neither strain grew. As seen from the results, the cells of the FlaB strain, compared to those of the FlaA2 strain, easily lose their motility when the temperature increases or the salinity decreases. However, with an increasing salt concentration, the FlaB strain motility was restored. It is interesting that under conditions of 30% salt and 40°C the cell motility of the FlaB strain was significantly higher than that of the FlaA2 strain. The reduced cell motility of the FlaB strain under conditions of low salt/high temperature may be caused by disruption of the flagella assembly of the FlaB flagellin. To test this hypothesis, the flagella were isolated from cells growing in the liquid medium. The preparations were analyzed by SDS-PAGE. The results showed that under conditions when the cells of the FlaB strain were nonmotile on a semisolid agar medium, no flagella could be isolated from the cells growing in liquid media. The unexpected result was that under conditions when cell motility of both strains on a semisolid agar medium was comparable, the FlaB flagella yield from culture growing in liquid medium was significantly lower than the yield of FlaA2 flagella (25% salt and 40°C) or was absent (30% salt and 50°C). This result might indicate that the flagellin genes are differently regulated in cells growing in liquid and on semisolid media [75]. This issue should be studied further.
Fig. 4. Range of motility of H. marismortui strains under different cultivation conditions (temperature, salinity).
As mentioned previously, the flagella preparations isolated from the FlaA2 strain contain a minor amount of FlaB flagellin that can be detected by immunoblotting [72]. Flagella preparations isolated from the FlaA2 strain grown under different conditions were tested for the presence of FlaB flagellin. Admixture of FlaB flagellin was not detected under the same conditions when FlaB flagella cannot be isolated from the FlaB strain. It was unclear whether the lack of FlaB flagellin resulted in negative regulation of its biosynthesis or in the fact that under certain conditions FlaB flagellin cannot form a flagellum. Professor F. Rodriguez-Valera’s group (Spain) found that the transcription level of FlaB flagellin in the FlaA2 strain at 50°C is three-fold higher compared to that at 27°C (the medium salinity in both cases was 25%). Using immunoblotting, it was shown that flagella preparations isolated from the FlaA2 strain grown at 27°C contain an admixture of FlaB flagellin, and it was not detected in preparations isolated from the FlaA2 strain grown at 50°C. These results indicate that the absence of FlaB flagellin in flagella preparations is the result of its inability to form flagella under conditions of high temperature/low salinity and is not due to suppression of its biosynthesis [75].
As mentioned above, H. marismortui was isolated from the water of the Dead Sea, which is an endorheic salt lake. An interesting property of this lake is that due to heavy rains in the wintertime, the salinity of the water near the surface can be significantly reduced [82]. Flagellar filaments are extracellular structures, and hence highly susceptible to environmental changes. Under such conditions (sudden salinity reduction), the cells having the plasmid (FlaA2 strain) will have selective advantages. The pNG100 plasmid carrying the flaA2 gene may play the role of a “switch”, determining the protein composition of flagellar filaments. The transfer of this plasmid in a cell population can occur due to horizontal transfer [75]. In this regard, it should be noted that as shown in a recent study, haloarchaea can fuse with each other to form intra- and interspecific hybrids, which considerably facilitates the exchange of DNA between cells [83].
Study of Haloferax volcanii flagellins. Another confirmation of the variety of mechanisms used by various archaea to assemble functional flagella and regulate this process are recent data obtained in the group of Prof. M. Pohlschroder (USA) when studying the role of Haloferax volcanii flagellins. Haloferax volcanii is widely used as a model organism for studying the biology of halophilic archaea and producing halophilic proteins in preparative amounts for structural analyses [84]. Its genome contains two flagellin genes, flgA1 and flgA2, organized in one operon wherein the flgA1 stop codon is separated from the flgA2 start codon by 11 bp. To study the role of each of the flagellins in flagella formation, a series of experiments was performed on strains with deletion of flagellin genes [10, 27, 53]. Studies of the flagella isolated from wild-type cells showed that flagellin FlgA1 is the major component of the filaments, and FlgA2 is present in a minor amount [53]. The distribution of FlgA2 along the wild-type filaments, unfortunately, was not determined [10]. Deletion of flgA1 resulted in the loss of cell motility. In this case, according to electron microscopy, the cells retained the ability to form filaments of FlgA2, but their number was greatly reduced. The flagella were found in only 2% of the investigated cells compared to 40% in the control samples. An interesting result was obtained in the study of the strain with flgA2 deletion. This strain unexpectedly was hypermotile compared to the parental strain. When it is grown on semisolid agar, the diameter of the spot formed around the point of seeding was twice greater than that of the parental strain (at comparable rates of cell growth). The cells of a strain with flgA2 deletion had more flagella (3-5 per cell compared to 1-2 in control), and the flagella length was almost three-fold longer than in the parental strain. Interestingly, when the flgA2 gene integrated into the plasmid with an inducible tryptophan promoter was expressed in this strain, its hypermotility was preserved. This result might indicate that a regulatory role of FlgA2 depends on the relative content of FlgA1. Furthermore, in experiments where a mutant strain with a deletion of the entire flagellin operon was complemented by a plasmid containing only the flgA1 gene or both flagellin genes, the cell motility was recovered only when both genes were expressed. The results obtained on H. volcanii demonstrated that functional helical filaments of this microorganism, as in the case of H. marismortui and H. lacusprofundi, can be formed from the product of only one FlgA1 flagellin gene, and FlgA2 plays a structural and regulatory role in the flagellar motility. The authors suggest that under certain conditions FlgA2 can act as a negative regulator of the filament assembly, although not directly, but through a mediator binding to an unknown positive regulator of the flagellin operon [10].
CONCLUSION
Flagella of bacteria and archaea are fundamentally different structures that are similar only in appearance. This external similarity is caused by the fact that bacterial and archaeal flagella are functional analogs, and it should be helical to function as a propeller. The spiralization mechanism of bacterial filaments has been studied in detail, and it is concluded that for coiling the flagellin subunits that form a flagellar filament must be structurally not identical. However, the archaeal flagellar motility apparatus is inadequately studied, and it is interesting how these widely differing organelles are able to form similar supramolecular structures. Previous studies of the motility apparatus of haloarchaea H. salinarum showed that at least two different flagellins are required for this microorganism to form helical flagellar filaments. It was concluded that in H. salinarum two flagellins might be analogs of the two conformational states of a single flagellin in enterobacteria. Since the multiplicity of flagellin genes is widespread among Archaea, it has been suggested that this principle can be universal for this domain.
In recent studies performed on H. volcanii, H. marismortui, and H. lacusprofundi, it has been found that in these archaea the helical and functional flagella can be formed of only one type of flagellin subunits [10, 75, 76] and, thus, mechanisms of formation of flagella helicity in haloarchaea are more diverse than it was thought previously. Results have been obtained that greatly expanded the preexisting views on the role of multiple archaeal flagellins. For the first time, it was shown that the multiplicity of flagellin genes in some of archaea might be a mechanism of adaptation to environmental changes used in the regulation of flagellar assembly [10, 75].
The mechanism for the formation of the helical supramolecular structure of archaeal filaments is still undetermined. However, based on the existing data, as well as on the general principle that the filament forming subunits should not be identical to form a supercoiled structure, we can assume that the spiralization of flagellar filaments in H. volcanii, H. marismortui, and H. lacusprofundi can be due to the existence of different flagellin conformations. Because of the lack of data on a detailed spatial structure of archaeal flagellins in the flagella composition, the question of an exact mechanism of filament helix formation remains open.
Elucidation of the principles of formation of supramolecular structures is one of the most important problems in current biology. Bacterial and archaeal flagella are probably the most convenient objects for studying this problem. As seen from this review, the assembly of these external cell structures occurs differently, but leads to the same result – forming a rigid helical structure that perfectly executes its mission. Identification of general and specific features of the assembly mechanism will allow the fullest understanding of this molecular–biological problem and make useful conclusions. For example, the property to have highly conserved terminal regions of the polypeptide chain common for bacterial and archaeal flagellins, in the case of bacterial flagellin led to the description of co-assembly protein folding, and in Archaea to the identification of the flagellin domain forming the outer flagellum surface. The understanding of the relationship between the characteristics of individual subunits and the supramolecular structure formed by them will allow purposeful change in the properties of the natural ensembles of subunits. Prokaryotic flagella are considered as one of the most promising objects for the creation of artificial nanofibers with desired properties [67, 85]. In this context, Archaea are of particular interest because these organisms live under extreme conditions, and their formed structures have a high resistance to dissociating effects.
This study was supported by the Russian Foundation for Basic Research (grant Nos. 14-04-01604-a and 14-04-31621-mol_a).
REFERENCES
1.Woese, C. R., and Fox, G. E. (1977) Phylogenetic
structure of the prokaryotic domain: the primary kingdoms, Proc.
Natl. Acad. Sci. USA, 74, 5088-5090.
2.Vorob’eva, L. I. (2007) Archaea [in
Russian], Akademkniga, Moscow.
3.Jarrell, K. F., and Albers, S. V. (2012) The
archaellum: an old motility structure with a new name, Trends
Microbiol., 20, 307-312.
4.Fedorov, O. V. (1998) Protein co-assembly folding
as a mechanism of supramolecular structure formation, Uspekhi Biol.
Khim., 38, 239-256.
5.Calladine, C. R. (1978) Change of waveform in
bacterial flagella: the role of mechanics at the molecular level, J.
Mol. Biol., 118, 457-479.
6.Faulds-Pain, A., Birchall, C., Aldridge, C., Smith,
W. D., Grimaldi, G., Nakamura, S., Miyata, T., Gray, J., Li, G., Tang,
J. X., Namba, K., Minamino, T., and Aldridge, P. D. (2011) Flagellin
redundancy in Caulobacter crescentus and its implications for
flagellar filament assembly, J. Bacteriol., 193,
2695-2707.
7.Silverman, M., Zieg, J., Hilmen, M., and Simon, M.
(1979) Phase variation in Salmonella: genetic analysis of a
recombinational switch, Proc. Natl. Acad. Sci. USA, 76,
391-395.
8.Alam, M., and Oesterhelt, D. (1984) Morphology,
function and isolation of halobacterial flagella, J. Mol. Biol.,
176, 459-475.
9.Wirth, R. (2012) Response to Jarrell and Albers:
seven letters less does not say more, Trends Microbiol.,
20, 511-512.
10.Tripepi, M., Esquivel, R. N., Wirth, R., and
Pohlschroder, M. (2013) Haloferax volcanii cells lacking the
flagellin FlgA2 are hypermotile, Microbiology, 159,
2249-2258.
11.Herzog, B., and Wirth, R. (2012) Swimming
behavior of selected species of Archaea, Appl. Environ. Microb.,
78, 1670-1674.
12.Nather, D. J., Rachel, R., Wanner, G., and Wirth,
R. (2006) Flagella of Pyrococcus furiosus: multifunctional
organelles, made for swimming, adhesion to various surfaces, and
cell–cell contacts, J. Bacteriol., 188,
6915-6923.
13.Jarrell, K. F., Ding, Y., Nair, D. B., and Siu,
S. (2013) Surface appendages of archaea: structure, function, genetics
and assembly, Life, 3, 86-117.
14.Patenge, N., Berendes, A., Engelhardt, H.,
Schuster, S. C., and Oesterhelt, D. (2001) The fla gene cluster
is involved in the biogenesis of flagella in Halobacterium
salinarum, Mol. Microbiol., 41, 653-663.
15.Chaban, B., Ng, S. Y., Kanbe, M., Saltzman, I.,
Nimmo, G., Aizawa, S. I., and Jarrell, K. F. (2007) Systematic deletion
analyses of the fla genes in the flagella operon identify
several genes essential for proper assembly and function of flagella in
the archaeon, Methanococcus maripaludis, Mol. Microbiol.,
66, 596-609.
16.Lassak, K., Neiner, T., Ghosh, A., Klingl, A.,
Wirth, R., and Albers, S. (2012) Molecular analysis of the crenarchaeal
flagellum, Mol. Microbiol., 83, 110-124.
17.Schlesner, M., Miller, A., Streif, S.,
Staudinger, W. F., Muller, J., Scheffer, B., and Oesterhelt, D. (2009)
Identification of Archaea-specific chemotaxis proteins which interact
with the flagellar apparatus, BMC Microbiol., 9, 56.
18.Ghosh, A., and Albers, S. V. (2011) Assembly and
function of the archaeal flagellum, Biochem. Soc. Transact.,
39, 64-69.
19.Banerjee, A., Ghosh, A., Mills, D. J., Kahnt, J.,
Vonck, J., and Albers, S. V. (2012) FlaX, a unique component of the
crenarchaeal archaellum, forms oligomeric ring-shaped structures and
interacts with the motor ATPase FlaI, J. Biol. Chem.,
287, 43322-43330.
20.Thomas, N. A., and Jarrell, K. F. (2001)
Characterization of flagellum gene families of methanogenic archaea and
localization of novel flagellum accessory proteins, J.
Bacteriol., 183, 7154-7164.
21.Ghosh, A., Hartung, S., van der Does, C., Tainer,
J. A., and Albers, S. V. (2011) Archaeal flagellar ATPase motor shows
ATP-dependent hexameric assembly and activity stimulation by specific
lipid binding, Biochem. J., 437, 43-52.
22.Kalmokoff, M. L., and Jarrell, K. F. (1991)
Cloning and sequencing of a multigene family encoding the flagellins of
Methanococcus voltae, J. Bacteriol., 173,
7113-7125.
23.Bardy, S. L., and Jarrell, K. F. (2002) FlaK of
the archaeon Methanococcus maripaludis possesses preflagellin
peptidase activity, FEMS Microbiol. Lett., 208,
53-59.
24.Bardy, S. L., and Jarrell, K. F. (2003) Cleavage
of preflagellins by an aspartic acid signal peptidase is essential for
flagellation in the archaeon Methanococcus voltae, Mol.
Microbiol., 50, 1339-1347.
25.Szabo, Z., Albers, S. V., and Driessen, A. J. M.
(2006) Active-site residues in the type IV prepilin peptidase homologue
PibD from the archaeon Sulfolobus solfataricus, J.
Bacteriol., 188, 1437-1443.
26.Ng, S. Y., Chaban, B., and Jarrell, K. F. (2006)
Archaeal flagella, bacterial flagella and type IV pili: a comparison of
genes and posttranslational modifications, J. Mol. Microbiol.
Biotechnol., 11, 167-191.
27.Tripepi, M., Imam, S., and Pohlschroder, M.
(2010) Haloferax volcanii flagella are required for motility but
are not involved in PibD-dependent surface adhesion, J.
Bacteriol., 192, 3093-3102.
28.Mukhopadhyay, B., Johnson, E. F., and Wolfe, R.
S. (2000) A novel pH2 control on the expression of flagella in the
hyperthermophilic strictly hydrogenotrophic methanarchaeaon
Methanococcus jannaschii, Proc. Natl. Acad. Sci. USA,
97, 11522-11527.
29.Hendrickson, E. L., Liu, Y., Rosas-Sandoval, G.,
Porat, I., Soll, D., Whitman, W. B., and Leigh, J. A. (2008) Global
responses of Methanococcus maripaludis to specific nutrient
limitations and growth rate, J. Bacteriol., 190,
2198-2205.
30.Szabo, Z., Sani, M., Groeneveld, M., Zolghadr,
B., Schelert, J., Albers, S. V., and Driessen, A. J. (2007) Flagellar
motility and structure in the hyperthermoacidophilic archaeon
Sulfolobus solfataricus, J. Bacteriol., 189,
4305-4309.
31.Reimann, J., Lassak, K., Khadouma, S., Ettema, T.
J., Yang, N., Driessen, A. J., Klingl, A., and Albers, S. V. (2012)
Regulation of archaella expression by the FHA and von Willebrand
domain-containing proteins ArnA and ArnB in Sulfolobus
acidocaldarius, Mol. Microbiol., 86, 24-36.
32.Lassak, K., Peeters, E., Wrobel, S., and Albers,
S. V. (2013) The one-component system ArnR: a membrane-bound activator
of the crenarchaeal archaellum, Mol. Microbiol., 88,
125-139.
33.Thomas, A. N., Bardy, B. L., and Jarrell, K. F.
(2001) The archaeal flagellum: a different kind of prokaryotic motility
structure, FEMS Microbiol. Rev., 25, 147-174.
34.Kalmokoff, M. L., Jarrell, K. F., and Koval, S.
F. (1988) Isolation of flagella from the archaebacterium
Methanococcus voltae by phase separation with Triton X-114,
J. Bacteriol., 170, 1752-1758.
35.Cruden, D., Sparling, R., and Markovetz, A. J.
(1989) Isolation and ultrastructure of the flagella of Methanococcus
thermolithotrophicus and Methanospirillum hungatei, Appl.
Environ. Microbiol., 55, 1414-1419.
36.Kupper, J., Marwan, W., Typke, D., Grunberg, H.,
Uwer, U., Gluch, M., and Oesterhelt, D. (1994) The flagellar bundle of
Halobacterium salinarium is inserted into a distinct polar cap
structure, J. Bacteriol., 176, 5184-5187.
37.Bakeeva, L. E., Metlina, A. L., Novikova, T. M.,
and Speransky, V. V. (1992) The ultrastructure of the flagellar
apparatus of Halobacterium salinarium, Doklady Akad.
Nauk, 326, 914-915.
38.Metlina, A. L. (2001) Prokaryotic flagella as
biological motility system, Uspekhi Biol. Khim., 41,
229-282.
39.Metlina, A. L. (2004) Bacterial and archaeal
flagella as prokaryotic motility organelles, Biochemistry
(Moscow), 69, 1203-1212.
40.Streif, S., Staudinger, W. F., Marwan, W., and
Oesterhelt, D. (2008) Flagellar rotation in the archaeon
Halobacterium salinarum depends on ATP, J. Mol. Biol.,
384, 1-8.
41.Reindl, S., Ghosh, A., Williams, G. J., Lassak,
K., Neiner, T., Henche, A. L., Albers, S. V., and Tainer, J. A. (2013)
Insights into FlaI functions in archaeal motor assembly and motility
from structures, conformations, and genetics, Mol. Cell,
49, 1069-1082.
42.Banerjee, A., Neiner, T., Tripp, P., and Albers,
S. V. (2013) Insights into subunit interactions in the Sulfolobus
acidocaldarius archaellum cytoplasmic complex, FEBS J.,
280, 6141-6149.
43.Cohen-Krausz, S., and Trachtenberg, S. (2002) The
structure of the archeabacterial flagellar filament of the extreme
halophile Halobacterium salinarum R1M1 and its relation to
eubacterial flagellar filaments and type IV pili, J. Mol. Biol.,
321, 383-395.
44.Trachtenberg, S., Galkin, V. E., and Egelman, E.
H. (2005) Refining the structure of the Halobacterium salinarum
flagellar filament using the iterative helical real space
reconstruction method: insights into polymorphism, J. Mol.
Biol., 346, 665-676.
45.Cohen-Krausz, S., and Trachtenberg, S. (2008) The
flagellar filament structure of the extreme acidothermophile
Sulfolobus shibatae B12 suggests that archeabacterial flagella
have a unique and common symmetry and design, J. Mol. Biol.,
375, 1113-1124.
46.Kalmokoff, M. L., Karnauchow, T. M., and Jarrell,
K. F. (1990) Conserved N-terminal sequences in the flagellins of
archaebacterial, Biochem. Biophys. Res. Commun., 167,
154-160.
47.Bardy, S. L., Eichler, J., and Jarrell, K. F.
(2003) Archaeal signal peptides — a comparative survey at the
genome level, Protein Sci., 12, 1833-1843.
48.Tarasov, V. Y., Kostyukova, A. S., Tiktopulo, E.
I., Pyatibratov, M. G., and Fedorov, O. V. (1995) Unfolding of tertiary
structure of Halobacterium halobium flagellins does not result
in flagella destruction, J. Protein Chem., 14, 27-31.
49.Pyatibratov, M. G., Kostyukova, A. S., Tarasov,
V. Yu., and Fedorov, O. V. (1996) Some principles of formation of the
haloalkaliphilic archaeal flagellar structure, Biochemistry
(Moscow), 61, 1056-1062.
50.Calo, D., Kaminski, L., and Eichler, J. (2010)
Protein glycosylation in Archaea: sweet and extreme,
Glycobiology, 20, 1065-1076.
51.Mescher, M. F., and Strominger, J. L. (1976)
Purification and characterization of a prokaryotic glucoprotein from
the cell envelope of Halobacterium salinarium, J. Biol.
Chem., 251, 2005-2014.
52.Sumper, M. (1987) Halobacterial glycoprotein
biosynthesis, Biochim. Biophys. Acta (BBA) – Rev.
Biomembr., 906, 69-79.
53.Tripepi, M., You, J., Temel, S., Onder, O.,
Brisson, D., and Pohlschroder, M. (2012) N-glycosylation of
Haloferax volcanii flagellins requires known Agl proteins and is
essential for biosynthesis of stable flagella, J. Bacteriol.,
194, 4876-4887.
54.Meyer, B. H., and Albers, S. V. (2014) AglB
catalyzing the oligosaccharyl transferase step of the archaeal
N-glycosylation process is essential in the thermoacidophilic
crenarchaeon Sulfolobus acidocaldarius, MicrobiologyOpen,
3, 531-543.
55.Guan, Z., Naparstek, S., Calo, D., and Eichler,
J. (2012) Protein glycosylation as an adaptive response in Archaea:
growth at different salt concentrations leads to alterations in
Haloferax volcanii S-layer glycoprotein N-glycosylation,
Environ. Microbiol., 14, 743-753.
56.Wieland, F., Paul, G., and Sumper, M. (1985)
Halobacterial flagellins are sulfated glycoproteins, J. Biol.
Chem., 260, 15180-15185.
57.Voisin, S., Houliston, R. S., Kelly, J., Brisson,
J. R., Watson, D., Bardy, S. L., Jarrell, K. F., and Logan, S. M.
(2005) Identification and characterization of the unique N-linked
glycan common to the flagellins and S-layer glycoprotein of
Methanococcus voltae, J. Biol. Chem., 280,
16586-16593.
58.Kelly, J., Logan, S. M., Jarrell, K. F., Vandyke,
D. J., and Vinogradov, E. (2009) A novel N-linked flagellar glycan from
Methanococcus maripaludis, Carbohydr. Res., 344,
648-653.
59.Chaban, B., Voisin, S., Kelly, J., Logan, S. M.,
and Jarrell, K. F. (2006) Identification of genes involved in the
biosynthesis and attachment of Methanococcus voltae N-linked
glycans: insight into N-linked glycosylation pathways in Archaea,
Mol. Microbiol., 61, 259-268.
60.Vandyke, D. J., Wu, J., Logan, S. M., Kelly, J.
F., Mizuno, S., Aizawa, S. I., and Jarrell, K. F. (2009) Identification
of genes involved in the assembly and attachment of a novel flagellin
N-linked tetrasaccharide important for motility in the archaeon
Methanococcus maripaludis, Mol. Microbiol., 72,
633-644.
61.Gerl, L., and Sumper, M. (1988) Halobacterial
flagellins are encoded by a multigene family. Characterization of five
flagellin genes, J. Biol. Chem., 263, 13246-13251.
62.Gerl, L., Deutzmann, R., and Sumper, M. (1989)
Halobacterial flagellins are encoded by a multigene family
Identification of all five gene products, FEBS Lett.,
244, 137-140.
63.Tarasov, V. Y., Pyatibratov, M. G., Tang, S. L.,
Dyall-Smith, M., and Fedorov, O. V. (2000) Role of flagellins from A
and B loci in flagella formation of Halobacterium salinarum,
Mol. Microbiol., 35, 69-78.
64.Tarasov, V. Y., Pyatibratov, M. G., Beznosov, S.
N., and Fedorov, O. V. (2004) On the supramolecular organization of the
flagellar filament in Archaea, Doklady Biochem. Biophys.,
396, 203-205.
65.Pyatibratov, M. G., Leonard, K., Tarasov, V. Y.,
and Fedorov, O. V. (2002) Two immunologically distinct types of
protofilaments can be identified in Natrialba magadii flagella,
FEMS Microbiol. Lett., 212, 23-27.
66.Beznosov, S. N., Pyatibratov, M. G., and Fedorov,
O. V. (2007) On the multicomponent nature of Halobacterium
salinarum flagella, Microbiology (Moscow), 76,
435-441.
67.Beznosov, S. N., Pyatibratov, M. G., and Fedorov,
O. V. (2009) Archaeal flagella as matrices for new nanomaterials,
Nanotechnol. Russia, 4, 373-378.
68.Beznosov, S. N., Pyatibratov, M. G., Veluri, P.
S., Mitra, S., and Fedorov, O. V. (2013) A way to identify archaellins
in Halobacterium salinarum archaella by FLAG-tagging, Centr.
Europ. J. Biol., 8, 828-834.
69.Lewus, P., and Ford, R. M. (1999)
Temperature-sensitive motility of Sulfolobus acidocaldarius
influences population distribution in extreme environments, J.
Bacteriol., 181, 4020-4025.
70.Baliga, N. S., Bonneau, R., Facciotti, M. T.,
Pan, M., Glusman, G., Deutsch, E. W., and Ng, W. V. (2004) Genome
sequence of Haloarcula marismortui: a halophilic archaeon from
the Dead Sea, Genome Res., 14, 2221-2234.
71.Oren, A., Ginzburg, M., Ginzburg, B. Z.,
Hochstein, L. I., and Volcani, B. E. (1990) Haloarcula
marismortui (Volcani) sp. nov., nom. rev., an extremely halophilic
bacterium from the Dead Sea, Int. J. System. Bacteriol.,
40, 209-210.
72.Pyatibratov, M. G., Beznosov, S. N., Rachel, R.,
Tiktopulo, E. I., Surin, A. K., Syutkin, A. S., and Fedorov, O. V.
(2008) Alternative flagellar filament types in the haloarchaeon
Haloarcula marismortui, Canad. J. Microbiol., 54,
835-844.
73.Matagne, A., Joris, B., and Frere, J. M. (1991)
Anomalous behavior of a protein during SDS/PAGE corrected by chemical
modification of carboxylic groups, Biochem. J., 280,
553-556.
74.Ikeda, J. S., Schmitt, C. K., Darnell, S. C.,
Watson, P. R., Bispham, J., Wallis, T. S., and O’Brien, A. D.
(2001) Flagellar phase variation of Salmonella enterica serovar
Typhimurium contributes to virulence in the murine typhoid infection
model but does not influence Salmonella-induced
enteropathogenesis, Infect. Immun., 69, 3021-3030.
75.Syutkin, A. S., Pyatibratov, M. G., Galzitskaya,
O. V., Rodriguez-Valera, F., and Fedorov, O. V. (2014) Haloarcula
marismortui archaellin genes as ecoparalogs, Extremophiles,
18, 341-349.
76.Syutkin, A. S., Pyatibratov, M. G., Beznosov, S.
N., and Fedorov, O. V. (2012) Various mechanisms of flagella helicity
formation in haloarchaea, Microbiology (Moscow), 81,
573-581.
77.Valliere-Douglass, J. F., Eakin, C. M., Wallace,
A., Ketchem, R. R., Wang, W., Treuheit, M. J., and Balland, A. (2010)
Glutamine-linked and non-consensus asparagine-linked oligosaccharides
present in human recombinant antibodies define novel protein
glycosylation motifs, J. Biol. Chem., 285,
16012-16022.
78.Franzmann, P. D., Stackebrandt, E., Sanderson,
K., Volkman, J. K., Cameron, D. E., Stevenson, P. L., and Burton, H. R.
(1988) Halobacterium lacusprofundi sp. nov., a halophilic
bacterium isolated from Deep Lake, Antarctica, System. Appl.
Microbiol., 11, 20-27.
79.Tu, D., Blaha, G., Moore, P. B., and Steitz, T.
A. (2005) Gene replacement in Haloarcula marismortui:
construction of a strain with two of its three chromosomal rRNA operons
deleted, Extremophiles, 9, 427-435.
80.Lopez-Lopez, A., Benlloch, S., Bonfa, M.,
Rodriguez-Valera, F., and Mira, A. (2007) Intragenomic 16S rDNA
divergence in Haloarcula marismortui is an adaptation to
different temperatures, J. Mol. Evol., 65, 687-696.
81.Sanchez-Perez, G., Mira, A., Nyiro, G., Pasic,
L., and Rodriguez-Valera, F. (2008) Adapting to environmental changes
using specialized paralogs, Trends Genet., 24,
154-158.
82.Bodaker, I., Sharon, I., Suzuki, M. T.,
Feingersch, R., Shmoish, M., Andreishcheva, E., Sogin, M. L.,
Rosenberg, M., Maguire, M. E., Belkin, S., Oren, A., and Beja, O.
(2010) Comparative community genomics in the Dead Sea: an increasingly
extreme environment, ISME J., 4, 399-407.
83.Williams, D., Gogarten, J. P., and Papke, R. T.
(2012) Quantifying homologous replacement of loci between haloarchaeal
species, Genome Biol. Evol., 4, 1223-1244.
84.Allers, T., Barak, S., Liddell, S., Wardell, K.,
and Mevarech, M. (2010) Improved strains and plasmid vectors for
conditional overexpression of His-tagged proteins in Haloferax
volcanii, Appl. Environ. Microbiol., 76,
1759-1769.
85.Deutscher, L., Renner, L. D., and Cuniberti, G.
(2014) Flagella – templates for the synthesis of metallic
nanowires, IFMBE Proc., 41, 860-863.