Features of Hydrolysis of Specific and Nonspecific Globular Proteins and Oligopeptides by Antibodies against Viral Integrase from Blood of HIV-Infected Patients
E. S. Odintsova1, P. S. Dmitrenok2, S. V. Baranova1, A. M. Timofeeva1, V. N. Buneva1,3, and G. A. Nevinsky1,3*
1Institute of Chemical Biology and Fundamental Medicine, Siberian Division of the Russian Academy of Sciences, pr. Akademika Lavrentieva 8, 630090 Novosibirsk, Russia; fax: (383) 363-2653; E-mail: nevinsky@niboch.nsc.ru2Pacific Institute of Bioorganic Chemistry, ul. 100-let Vladivostoku 159, Far East Division of the Russian Academy of Sciences, 690022 Vladivostok, Russia
3Novosibirsk State University, pr. Pirogova 10, 630090 Novosibirsk, Russia; fax: (383) 330-3255; E-mail: nsu@nsu.ru
* To whom correspondence should be addressed.
Received June 30, 2014; Revision received September 18, 2014
It was shown previously that, as differentiated from canonical proteases, abzymes against myelin basic protein (MBP) from blood of patients with multiple sclerosis and systemic lupus erythematosus effectively cleaved only MBP, while antibodies (ABs) against integrase (IN) from blood of HIV-infected patients specifically hydrolyzed only IN. In this work, all sites of effective hydrolysis by anti-IN antibodies (IgG and IgM) of 25-mer oligopeptide (OP25) corresponding to MBP were identified using reversed-phase and thin-layer chromatographies and MALDI mass spectrometry. It was found that amino acid sequences of OP25 and other oligopeptides hydrolyzed by anti-MBP abzymes were partially homologous to some fragments of the full sequence of IN. Sequences of IN oligopeptides cleavable by anti-IN abzymes were homologous to some fragments of MBP, but anti-MBP abzymes could not effectively hydrolyze OPs corresponding to IN. The common features of the cleavage sites of OP25 and other oligopeptides hydrolyzed by anti-MBP and anti-IN abzymes were revealed. The literature data on hydrolysis of specific and nonspecific proteins and oligopeptides by abzymes against different protein antigens were analyzed. Overall, the literature data suggest that short OPs, including OP25, mainly interact with light chains of polyclonal ABs, which had lower affinity and specificity to the substrate than intact ABs. However, it seems that anti-IN ABs are the only one example of abzymes capable of hydrolyzing various oligopeptides with high efficiency (within some hours but not days). Possible reasons for the efficient hydrolysis of foreign oligopeptides by anti-IN abzymes from HIV-infected patients are discussed.
KEY WORDS: HIV-infected patients, catalytic antibodies against viral integrase, integrase, myelin basic protein, hydrolysis of peptides of myelin basic proteinDOI: 10.1134/S0006297915020054
Abbreviations: AB, antibody; FM, final reaction mixture; HIV, human immunodeficiency virus; HSA, human serum albumin; IAS, immunogenic amino acid sequence; IgGmix and IgMmix, mixtures of individual antibodies from blood of HIV-infected patients; IN, HIV integrase; MBP, myelin basic protein; MCA, 4-methylcoumaryl-7-amine; MS, multiple sclerosis; OP, oligopeptide; RPC, reverse-phase chromatography; SLE, systemic lupus erythematosus; X, fluorescent residue 6-O-(carboxymethyl)fluorescein ethyl ester.
Artificial catalytically active antibodies, or abzymes, against
transition states of different chemical reactions have been described
in the literature in recent years [1]. Natural
abzymes hydrolyzing DNA, RNA, polysaccharides, oligopeptides (OPs), and
proteins from blood of patients with different autoimmune diseases
(systemic lupus erythematosus (SLE), Hashimoto’s thyroiditis,
polyarthritis, multiple sclerosis (MS), asthma, rheumatoid arthritis,
etc.) and some viral infections are accompanied by disorders in immune
status (hepatitis, HIV-infection, tick-borne encephalitis) [2-6]. In healthy donors and
patients with many other diseases accompanied by poorly pronounced
autoimmune reactions, abzymes are not produced, the generated
antibodies (ABs) have low catalytic activities, and such ABs cannot be
reliably detected by current methods [2-6]. In addition to classic functions of ABs, abzymes
can play both positive and negative roles inducing specific processes
and clinical manifestations under various autoimmune conditions,
including HIV-infection [2-6].
HIV-1 is an etiological agent of a very dangerous human disease – acquired immunodeficiency syndrome (AIDS) [7]. The cycle of HIV-1 replication includes a reverse transcription of viral RNA with subsequent integration of the generated DNA into the host’s genome [8]. The reverse transcription is catalyzed by retroviral RNA-dependent DNA polymerase (reverse transcriptase (RT)) [9, 10]. Replication of retroviruses depends on the efficiency of integration of the retroviral genome double-stranded DNA into the nuclear genome of the host’s cell [8]. The integration stage is catalyzed by the viral enzyme integrase (IN) [11]. Both IN and RT are important “targets” for suppression of the development of HIV.
Blood of HIV-infected patients contains ABs to many viral proteins including RT and IN as well as to human proteins [5, 12, 13]. The first report of the presence of DNA-hydrolyzing ABs in blood of HIV-infected patients lacked proof by testing the known strict parameters of the activity belonging directly to immunoglobulins [14]. Later a study revealed in detail that the DNase activity was indeed an intrinsic feature of antibodies of HIV-infected patients [15]. It was shown recently that IgG and IgM preparations from blood of HIV-infected patients upon affinity chromatography on sorbents containing immobilized RT, IN, HSA, and casein could hydrolyze only the corresponding substrate but not other proteins [16-20]. Long-term incubation of IN with IgG- or IgM-abzymes possessing high proteolytic activity leads to generation of many peptides with different lengths including short OPs [19]. Forty cleavage sites in IN by abzymes from HIV-infected patients detected by MALDI mass spectrometry are localized mainly in seven known immunodominant sequences of IN [19]. Two 20-mer OPs corresponding to immunogenic sequences of IN contained 9-10 clustered major, moderate, and minor cleavage sites [19, 20]. In the combined pool of IgG and IgM from blood of HIV-infected patients, only specific anti-IN ABs could hydrolyze intact globular molecules of the viral integrase.
Multiple sclerosis (MS) is a chronic autoimmune demyelinating disease of the central nervous system leading to various nervous and mental disorders [21]. Its etiology is still unclear, and the adopted theory of its pathogenesis considers inflammation associated with autoimmune reactions to be mainly responsible for the destruction of myelin [21]. Recent studies revealed that the pathogenesis of MS is significantly contributed to by B-cells and auto-ABs against autoantigens of myelin, including myelin basic protein (MBP) [21-23]. Recently the MBP-hydrolyzing activity was shown to be the intrinsic property of IgG, IgM, and IgA preparations from blood of patients with MS [24-26], and specific sites of cleavage of MBP by abzymes were determined [27].
The polyetiologic and polysyndrome character of an autoimmune diffuse disease, SLE, leads to different biochemical, immunological, and clinical manifestations [28]. Note that there is a similarity in the development of medical, biochemical, and immunological manifestations of SLE and MS. Thus, neurological and mental disorders, which lead to the negative prognosis, develop in 50% of patients with SLE [21]. It seems that SLE affects the central nervous system and the brain stronger than any other systemic inflammatory disease and causes mental disorders [21]. Production of various auto-ABs can be associated with impaired functioning of apoptotic cells.
The AB-mediated destruction of nervous cells and rheological disorders suggest two fundamental mechanisms of tissue damage [21]. The interaction between these processes, main hereditary factors, their modification by hormones, as well as complications by many secondary factors can explain a broad spectrum of features in the development of MS and SLE. Some common manifestations specific for both SLE and MS have been revealed, e.g. appearance of patches in the brain [21].
Recently, it was shown that titers of ABs against MBP were 4.2-fold higher in patients with SLE than in healthy donors, but 2.1-fold lower than in patients with MS [4]. However, catalytic activities of abzymes of blood of patients with SLE and MS were similar. IgG and/or IgM in SLE and MS effectively hydrolyzed DNA, RNA, and polysaccharides [1-4] as well as MBP [24-27]. Electrophoretically and immunologically homogenous preparations of IgG from blood of patients with SLE and MS upon affinity chromatography on Sepharose with immobilized MBP (MBP-Sepharose) specifically hydrolyzed only MBP, but not other tested proteins [29]. Anti-MBP abzymes of blood from patients with SLE and MS hydrolyzed MBP by the same sites [27, 30-34]. Anti-MBP abzymes in SLE and MS can attack MBP of the myelin-proteolipid envelope of axons. A standard drug used in MS, Copaxone, is a specific inhibitor of the MBP-hydrolyzing activity of abzymes [27]. Consequently, MBP-hydrolyzing abzymes can play an important negative role in pathogenesis of MS and SLE.
Earlier, anti-IN abzymes of HIV-infected patients were shown to hydrolyze effectively a 21-mer oligopeptide corresponding to one of four immunogenic sequences of MBP [33]. But it was unclear whether it was an occasional phenomenon, a specific feature of ABs against IN, or a consequence of a partial homology of amino acid sequences of these proteins, and whether the partial homology was sufficient for the effective hydrolysis of oligopeptides by anti-IN abzymes of HIV-infected patients. To answer this question, in the present work hydrolysis of the 25-mer oligopeptide (OP25) of MBP (corresponding to another immunogenic sequence of MBP) by anti-IN abzymes from blood of HIV-infected patients was analyzed. The analysis was conducted using MALDI mass spectrometry and reverse-phase (RP), thin-layer (TL), and affinity chromatographies. Moreover, the homology of the amino acid sequences of HIV integrase and MBP was analyzed, and data on hydrolysis of human intact globular MBP and HIV IN as well as on hydrolysis of their oligopeptides by anti-MBP and anti-IN abzymes were compared.
MATERIALS AND METHODS
Reagents, donors, and patients. All reagents were from Sigma (USA) or Pharmacia (Sweden). Homogenous integrase HIV-1 was prepared as described previously [35]. IN-Sepharose was prepared using BrCN-Sepharose according to the standard protocol of the producer.
Preparations from blood of 10 healthy donors and of 24 HIV-infected patients (18-40-year-old men and women) including 16 patients in the pre-AIDS stage and eight patients in the generalized lymphadenopathy stage, diagnosed according to classification of the Centers for Disease Control and Prevention (USA, http://www.cdc.gov/hiv/), were used in this study. The specific IN-hydrolyzing activity of all the AB preparations was tested according to [17, 18]. In the present work, some earlier described preparations of catalytically active IgG and IgM from HIV-infected patients were used to study hydrolysis of MBP and its peptides. Protocols of taking blood samples were in accordance with recommendations of the Regional Committee on Human Ethics of Novosibirsk State Medical University in correspondence with recommendations of the Helsinki Committee on ethics.
Purification of antibodies. Electrophoretically homogenous preparations of IgG and IgM were obtained from blood of healthy donors and HIV-infected donors using affinity chromatography of blood plasma proteins on protein A-Sepharose and subsequent highly effective gel filtration on a column with Superdex 200 HR 10/30 (GE Healthcare, USA) as described in [17, 18].
To prepare blood plasma, blood samples (5 ml) were supplemented with 4% sodium citrate (1/4 of the blood sample volume) and the cells were removed by centrifugation. Blood plasma was applied onto a column with protein A-Sepharose (5 ml) equilibrated with TBS buffer (0.15 M NaCl, 20 mM Tris-HCl (pH 7.5)). Then the column was washed with the same buffer to zero optical density of the eluate. Nonspecifically adsorbed proteins were eluted with the same buffer supplemented with 1% Triton X-100 and 0.5 M NaCl. Then the column was washed with TBS, and the ABs (total fraction of IgG + IgA + IgM) were eluted with 50 mM glycine-HCl buffer (pH 2.6). The total fraction of ABs was collected into a cooled tube that contained 50 µl of 0.5 M Tris-HCl (pH 9.0) and then the solution was adjusted to neutral pH value and dialyzed against 10 mM Tris-HCl (pH 7.5) containing 0.1 M NaCl.
IgM was separated from IgA and IgG by high-performance gel filtration of the total AB fraction on a column with Superdex 200 HR 10/30 equilibrated with 50 mM Tris-HCl (pH 7.5) buffer supplemented with 0.3 M NaCl according to [17, 18]. Then the proteolytic activities of the IgG and IgM preparations were tested. The specific hydrolysis of IN only by abzymes from blood of HIV-infected patients used in this work was analyzed as described earlier [17, 18].
For analyzing averaged characteristics of proteolytic polyclonal ABs, an equimolar mixture was prepared of electrophoretically homogenous IgG (IgGmix) and IgM (IgMmix) with different but a relatively high activity from blood of seven HIV-infected patients. Preparations of individual IgG and IgM used for preparing IgGmix and IgMmix were described earlier [33], and some of their characteristics are presented below. In the control experiments, a mixture of equal quantities of IgG (hd-IgGmix) and IgM (hd-IgMmix) preparations from blood of seven healthy donors was used.
The IgGmix and IgMmix preparations from HIV-infected patients were subjected to affinity chromatography on a column with IN-Sepharose (1 ml) equilibrated with 50 mM Tris-HCl (pH 7.5) buffer containing 0.1 M NaCl as described earlier [33]. After application, the column was washed with the same buffer to the zero optical density of the eluant. IgG or IgM were eluted initially with a NaCl concentration gradient (0.2-2.0 M) and then with 3 M NaCl (ABsalt), and, finally, with 40 mM glycine-HCl buffer (pH 2.6) (ABacid). All AB fractions were dialyzed against 50 mM Tris-HCl (pH 7.5) buffer and then concentrated. After every stage of the purification, the protein concentration was measured by the Bradford method with BSA used for the calibration.
Testing proteolytic activity of the ABs. The reaction mixture (10-20 µl) for analyzing hydrolysis of oligopeptides contained 50 mM Tris-HCl (pH 7.5), 30 mM NaCl, 0.5-2.0 mM OP, and 0.01-0.03 mg/ml IgGmix or IgMmix separated on IN-Sepharose (or 0.2 mg/ml hd-IgGmix or hd-IgMmix). The mixtures were incubated for 1-24 h at 37°C. As substrate, we used a nonspecific X-OP25 (X-AQGTLSKIFKLGGRDSRSGSPMARR) corresponding to one of the immunogenic sequences of human MBP that was found to include a number of specific cleavage sites of this protein by IgG-abzymes [27, 29, 31-34]. On the N-terminus of this OP, there was a fluorescent residue 6-O-(carboxymethyl)fluorescein ethyl ester (X). Oligopeptides, corresponding to two immunogenic sequences of HIV IN (in-OP1 and in-OP2) that contained a number of specific cleavage sites by IN-abzymes from HIV-infected patients, were used as a control [19, 20].
Products of hydrolysis of X-OP25 (0.5-2 µl of the reaction mixture) were analyzed by thin-layer chromatography (TLC) on plates with Kieselgel 60 F254 using an acetic acid–n-butanol–H2O solvent system (1 : 4 : 5 v/v). Then the plates were dried and photographed. The relative intensity of fluorescence spots after TLC was compared with that of control oligopeptides incubated without ABs. The intensity of X-OP25 cleavage was assessed by a decrease (in %) of the initial X-OP25 fluorescence and accumulation of new fluorescent products. The total fluorescence intensity of non-hydrolyzed OP and of all cleavage products was taken as 100%.
In some experiments, products of X-OP25 specific cleavage were initially separated by reversed-phase chromatography (RPC) on a column (4.6 × 250 mm) with Nucleosil C-18 sorbent using 0.05% trifluoroacetic acid and acetonitrile gradient (0-80%). The relative quantity of the cleavage products was calculated based on their fluorescence. The fluorescence was excited at 320 nm, and emission was determined at 490 nm. Fractions of different peaks were collected, evaporated to minimal volume, and the hydrolysis products were analyzed by TLC (see above) and MALDI mass spectrometry (see below).
Analysis of AB-dependent hydrolysis of oligopeptides by MALDI mass spectrometry. In all cases, products of X-OP25 hydrolysis were analyzed by MALDI-TOF mass spectrometry with a Reflex III system (Bruker, Germany) equipped by a nitrogen laser (337 nm) (VSL-337 ND; Laser Science, USA) with impulse duration of 3 ns. A saturated solution of cyano-4-hydrocinnamic acid in mixture of 0.1% acetonitrile and trifluoroacetic acid (1 : 2 v/v) was used as a matrix. To 1 µl of reaction mixture containing hydrolyzed OP before or after the separation of hydrolysis products by RPC or TLC, 1 µl of 0.2% trifluoroacetic acid and 1 µl of the matrix were added, then 1 µl of the final mixture was applied onto a target for MALDI mass spectrometry and the target was dried in air. The MALDI spectra were calibrated using protein and oligopeptide standard kits (I and II; Bruker Daltonics, Germany) as internal and external standards.
Analysis of proteolytic activity type of the abzymes. In accordance with [17, 18], for analyzing the proteolytic activity of IgGmix and IgMmix, preparations of these ABs (0.5 mg/ml) were preincubated for 30 min at 25°C with one of the specific inhibitors of classical proteases: iodoacetamide (4 mM), pepstatin A (50 µM), AEBSF (0.15 mM), or EDTA (50 mM). Then aliquots (2 µl) of these mixtures were added into the standard reaction mixture (20 µl) that contained 0.3 mg/ml IN, 50 mM Tris-HCl (pH 7.5), and 30 mM NaCl as described in [17, 18]. Then the reaction products were analyzed by SDS-PAGE in 12% polyacrylamide gel. The efficiencies of MBP hydrolysis were assessed from diminution of the protein substrate in the initial band of IN as compared to the control (incubation of IN without ABs) using the Gel-Pro Analyzer computer program, version 3.1. Then the decrease in the activity of ABs incubated with the above-mentioned specific inhibitors was calculated (in %) for comparison with ABs incubated without them.
Analysis of homology. The homology (% of identical amino acid residues) in the sequences of oligopeptides, MBP, and HIV integrase was assessed using the lalign program (http://www.ch.embnet.org/software/LALIGN_form.html). This program revealed in the analyzed sequences not only identical amino acid residues, but also residues with similar physicochemical features. Amino acid residues, identical and similar in features, were searched for based on an algorithm described in work [36].
Statistical analysis. The results are presented as mean values ± mean deviation of at least three independent measurements. The error of the presented values was no more than 7-12%.
RESULTS
Characteristics of abzymes. Electrophoretically homogenous preparations of IgG and IgM were obtained from blood of healthy donors and HIV-infected patients by sequential affinity chromatography of blood plasma proteins on protein A-Sepharose as described in [17, 18]. IgM was separated from IgA and IgG by high performance gel filtration of the total AB fraction as described in [17, 18]. Then the proteolytic activities of the IgG and IgM preparations were tested. The specific hydrolysis of IN only by abzymes from blood of HIV-infected patients that were used in the present work was described earlier [33].
In the present work, averaged characteristics of the antibodies were analyzed using the same mixtures of equal quantities of homogenous preparations of IgG (IgGmix) and IgM (IgMmix) from the blood of seven HIV-infected patients and also of seven healthy donors (hd-IgGmix and hd-IgMmix) that were described in [33]. Then IgGmix and IgMmix possessing affinity for IN were separated by affinity chromatography on Sepharose with immobilized IN as described in [33]. For subsequent studies, we used IgGmix and IgMmix fractions eluted from IN-Sepharose with 3 M NaCl (ABsalt) and acidic buffer with pH 2.6 (ABacid). To exclude the possible presence of traces of canonic proteases, purified preparations of IgGmix and IgMmix eluted with NaCl solution and acidic buffer were separated by SDS-PAGE under non-reducing and reducing conditions. Then the proteolytic activity was evaluated in fractions corresponding to extracts of small pieces of the gel (2-3 mm) as done in works [17-20]. Under the non-reducing conditions, the IN-hydrolyzing activity was detected only in the gel zones that corresponded to positions of intact IgGmix (150 kDa), and under the reducing conditions such activity was detected only in the zone of isolated light chains of IgGmix and IgMmix (intact IgM with molecular weight of 970 kDa did not enter the gel). Thus, these data (together with the absence of activity in the hd-IgGmix and hd-IgMmix preparations from healthy donors) directly proved that neither one of the IgGmix and IgMmix preparations from blood of HIV-infected patients purified on IN-Sepharose contained admixtures of canonic proteases. Moreover, it was shown similarly to data of works [17-20, 33] that, as discriminated from canonic proteases, IgGmix and IgMmix preparations purified on IN-Sepharose could specifically hydrolyze only globular integrase, but not other globular proteins studied – bovine serum albumin, lactoferrin, and casein of human milk, egg lysozyme, p66 reverse transcriptase of HIV-1, and MBP.
AB-dependent hydrolysis of oligopeptides. It was shown earlier that IgGmix and IgMmix from blood of HIV-infected patients hydrolyzed intact IN, its 20-mer OPs, short nonspecific tri- and tetrapeptides, and 21-mer OP (X-OP21) that corresponded to MBP [17-20, 33, 34]. Initially, it was shown that anti-IN ABs hydrolyzed X-OP25 corresponding to human MBP, whereas ABs from healthy donors did not cleave this OP (Fig. 1b, lane K3). To analyze the X-OP25 hydrolysis by anti-IN abzymes in detail, IgG(salt)mix, IgM(salt)mix, IgG(acid)mix, and IgM(acid)mix eluted from IN-Sepharose, respectively, with 3 M NaCl and acidic buffer were used.
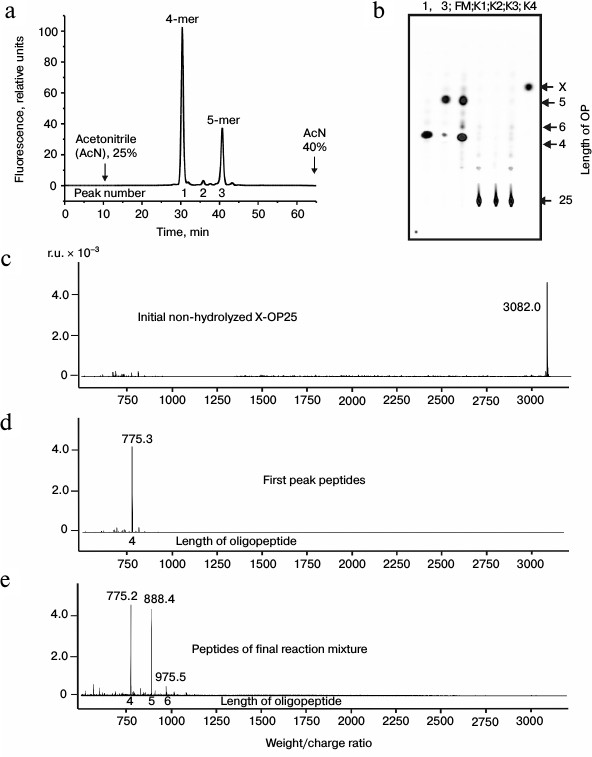
Fig. 1. RPC profile of products of the X-OP25 extensive hydrolysis (24 h) by anti-IN abzymes of the IgG(acid)mix fraction (a) and analysis of the X-OP25 hydrolysis products corresponding to RPC peaks (a) by TLC (b) and mass spectrometry (c-e). b) Numbers of lanes in TLC plate correspond to numbers of RPC peaks (a). Lanes: FM) final reaction mixture; K1, K2) X-OP25 incubated in the absence of ABs and before incubation, respectively; K3) X-OP25 incubated with IgG from healthy donors; K4) free fluorescent label X. c-e) MALDI spectra of: c) initial X-OP25 (m/z = 3082.0); d) products of peak 1 (a); e) final reaction mixture before its separation into fractions.
MALDI mass spectrometry analysis of X-OP25 hydrolysis. Figure 1 shows that virtually full hydrolysis of the specific X-OP25 by anti-IN abzymes of IgG(acid)mix during 24 h results in two major, one minor, and several poorly fluorescing OPs. Amino acid sequences of the hydrolysis products could not be definitely identified by TLC alone (Fig. 1b) because their mobility on the plate depended on many factors including the number of amino acid residues, relative hydrophobicity, the nature of the terminal amino acid residues, etc. To identify the major sites of IgG-dependent proteolysis of X-OP25, the hydrolysis products were analyzed using three methods: RPC (Fig. 1a), TLC (Fig. 1b), and mass spectrometry (Fig. 1, c-e). The two major products were 4- and 5-mer OPs, and a minor product was a 6-mer OP (Fig. 1). Note, that only tetramers and pentamers were produced immediately after the beginning of X-OP25 hydrolysis and remained as the major products up to the full cleavage of the OP. It should be noted that according to data of RPC (very small peaks in Fig. 1a) and TLC (very weak spots in Fig. 1b), the final reaction mixture (FM) contained several X-OPs in very small amounts (≤0.5%). Such trace products of hydrolysis were neglected for further analysis of the results.
Standard MALDI-TOF spectrometry analysis is a semiquantitative approach and mainly gives information about molecular weights of analyzed compounds. In some cases, to identify additionally products of X-OP25 cleavage by mass spectrometry, compounds were analyzed that corresponded to different spots obtained by TLC. The relative contents of X-OPs with different length in the final reaction mixture (FM) were assessed taking into account the ratio of relative fluorescence of the corresponding spots after the TLC (Fig. 1b): 4-mer OP > 5-mer OP ≥ 6-mer OP > all poorly visible unidentified products (Table 1).
Table 1. Results of analysis of molecular
weights and relative amounts of fluorescing OPs produced after
incubation of X-OP25 with anti-IN abzymes of IgG(acid)mix
from the data of RPC, TLC, and MALDI mass spectrometry (Fig. 1)
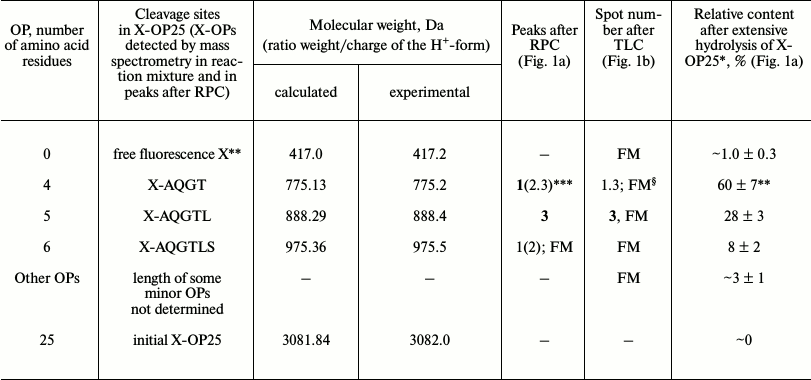
* The total amount of hydrolysis products is taken as 100%; each value
is calculated as the mean from data of three TLC experiments.
** The initial X-OP25 contained the fluorescent label X.
*** Numbers of peaks containing the major part of the product are given
in bold, and numbers of peaks containing the same OP as the minor
product are in parentheses.
§ The same products of X-OP25 hydrolysis in peaks after
RPC (Fig. 1a) were revealed not only by MALDI mass
spectrometry but also by TLC (Fig. 1b). FM
indicates the presence of a fluorescent peak of this product in the
combined final reaction mixture analyzed by TLC (Fig. 1b) and peaks of RPC (Fig. 1a).
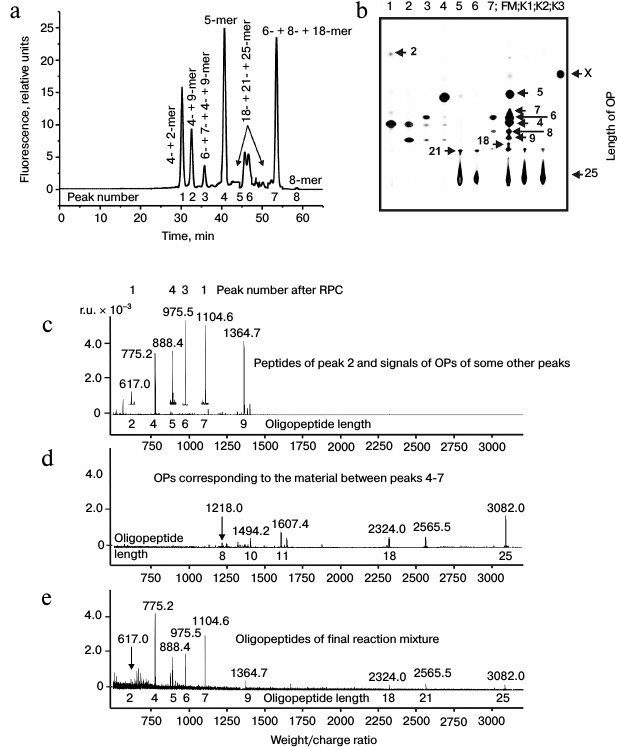
It was reasonable to suppose that abzymes eluted from IN-Sepharose by NaCl solution and acidic buffer may possess different specificity during X-OP25 hydrolysis. Therefore, products of X-OP25 hydrolysis by IgG(salt)mix preparations were analyzed additionally. In the RPC profile, seven major and seven very small peaks of fluorescent products of the X-OP25 hydrolysis were revealed (Fig. 2a).
Fig. 2. RPC profile of products of extensive hydrolysis (24 h) of X-OP25 by anti-IN abzymes of the IgG(salt)mix fraction (a) and analysis of X-OP25 hydrolysis products corresponding to RPC peaks (a) by TLC (b) and mass spectrometry (c-e). b) Numbers of lanes on TLC plate correspond to numbers of the RPC peaks (a). Lanes: FM) final reaction mixture; K1, K2) X-OP25 incubated in the absence of ABs and before the incubation, respectively; K3) free fluorescent label X. c-e) MALDI spectra of: c) peak 2 (a) products; d) OPs eluted in the two major and some minor peaks between peaks 4 and 7 (a); e) peptides of the final reaction mixture before its separation into fractions.
Products of all peaks resulting from RPC were analyzed using TLC (Fig. 2b) and mass spectrometry (e.g. Figs. 2c and 2d). The TLC revealed that some peaks after the RPC contained a number of hydrolysis products. This indicated that some individual products of the X-OP25 hydrolysis could be eluted from the sorbent by different concentrations of acetonitrile. The reaction mixture contained Tris-HCl and trifluoroacetic acid, which could interact with positively and negatively charged amino acid residues of the X-OP25 hydrolysis products. Moreover, some products of OPs could produce interpeptide complexes. The generation of numerous products of X-OP25 hydrolysis led to elution of the same products by different concentrations of acetonitrile. Products of X-OP25 hydrolysis were identified using three different approaches: RPC, TLC, and MALDI mass spectrometry. Relative contents of X-OP with different length in the final reaction mixture were evaluated taking into account the ratio of relative fluorescence of the corresponding spots after TLC (Fig. 2b): 5-mer OP > 4-mer OP > 6-mer OP ≥ 7-mer OP ≈ 8-mer OP ≈ 9-mer OP ≥ 21-mer OP ≥ 18-mer OP ≈ X ≥ 2-mer OP (Table 2).
Table 2. Molecular weights and relative
quantities of fluorescent OPs produced after incubation of X-OP25 with
anti-IN abzymes of IgG(salt)mix from the data of RPC, TLC,
and MALDI mass spectrometry (Fig. 2)
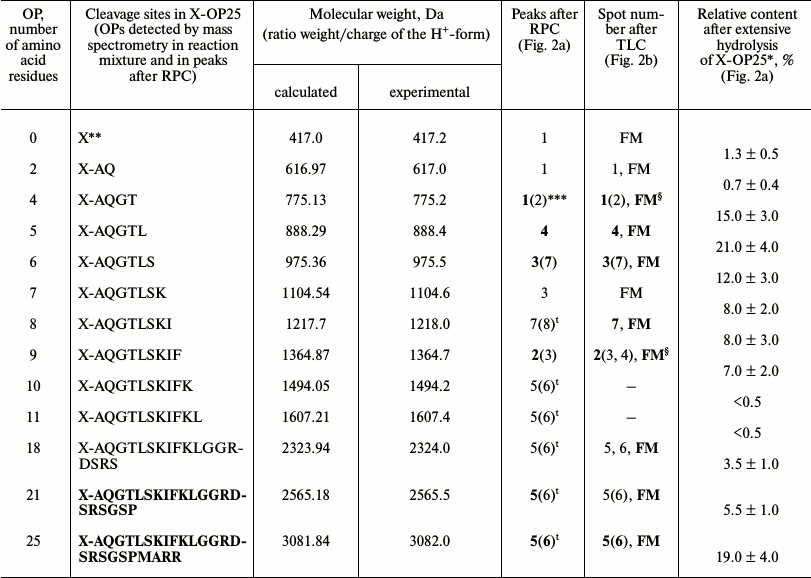
* Total amount of hydrolysis product is taken as 100%; each value is
calculated as the mean of data of three TLC experiments.
** The initial X-OP25 contained the fluorescent label X.
*** Numbers of peaks containing the major part of the product are given
in bold, and numbers of peaks containing the same OP as the minor
product are in parentheses.
§ The same products of X-OP25 hydrolysis in peaks after
RPC (Fig. 2a) were revealed not only by MALDI mass
spectrometry but also by TLC (Fig. 2b). FM
indicates the presence of fluorescing peak of this product in the
combined final reaction mixture analyzed by TLC (Fig. 2b) and peaks of RPC (Fig. 2a).
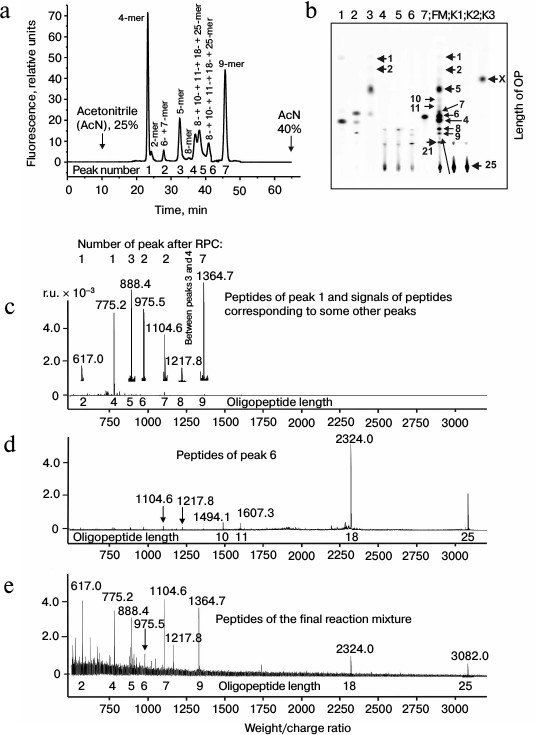
The immune system produced initially IgM and later IgG [1]. It might be that IgM and IgG would have different substrate specificities. At first we analyzed products of X-OP25 hydrolysis by the IgM(acid)mix preparation eluted from IN-Sepharose by 3 M NaCl (Fig. 3 and Table 3) as described above for IgG preparations. The relative contents of the X-OP hydrolysis products were evaluated: 4-mer OP ≥ 5-mer OP ≥ 6-mer OP ≥ 8-mer OP ≥ 9-mer OP ≈ 18-mer OP ≥ 7-mer OP ≥ 12-mer OP ≈ 21-mer OP ≈ 11-mer OP ≈ 10-mer OP ≈ 2-mer OP ≈ X (Table 3).
Fig. 3. RPC profile of products of the X-OP25 extensive hydrolysis by anti-IN abzymes of the IgM(acid)mix fraction (a) and analysis of X-OP25 hydrolysis products corresponding to RPC peaks (a) by TLC (b) and mass spectrometry (c-e). b) Numbers of lanes in the TLC plate correspond to numbers of the RPC peaks (a). Lanes: FM) final reaction mixture; K1, K2) X-OP25 incubated in the absence of ABs and before the incubation, respectively; K3) X-OP25 incubated with IgG from healthy donor. c-e) MALDI mass spectrometry spectra of: c, d) products of peaks 4 and 6 (a); e) peptides of final reaction mixture before its separation into fractions.
Table 3. Results of analysis of molecular
weights and relative amounts of fluorescing OPs produced after
incubation of X-OP25 with anti-IN abzymes of IgM(acid)mix
from the data of RPC, TLC, and MALDI mass spectrometry (Fig. 3)
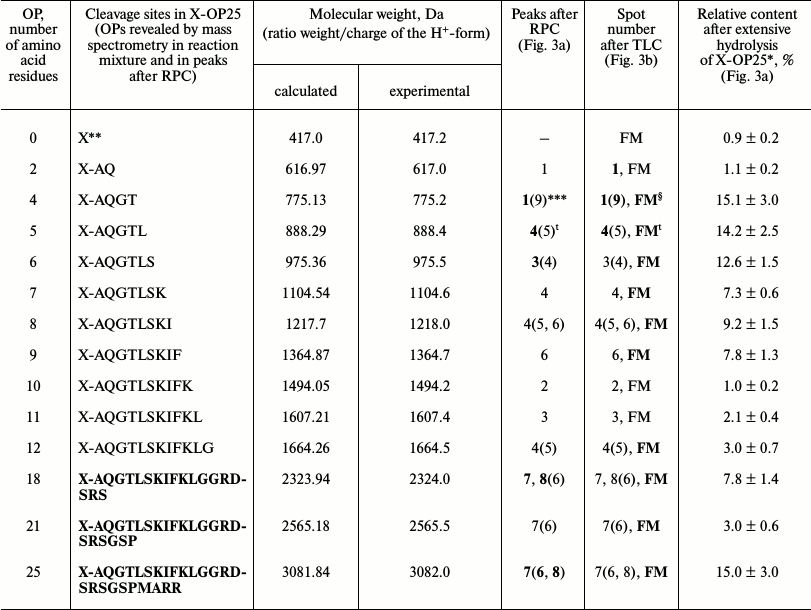
* The total amount of hydrolysis product is taken as 100%; each value is
calculated as the mean from data of three TLC experiments.
** The initial X-OP25 contained the fluorescent label X.
*** Numbers of peaks containing the major part of the product are
printed in bold, and numbers of peaks containing the same OPs as the
minor product are in parentheses.
§ The same products of X-OP25 hydrolysis in peaks after
RPC (Fig. 3a) were revealed not only by MALDI mass
spectrometry, but also by TLC (Fig. 3b). FM
indicates the presence of a fluorescent peak of this product in the
combined final reaction mixture analyzed by TLC (Fig. 3b) and in peaks of RPC (Fig. 3a).
ŧ All major and minor peaks between the fourth and
seventh including the fifth and sixth peaks (Fig. 3a).
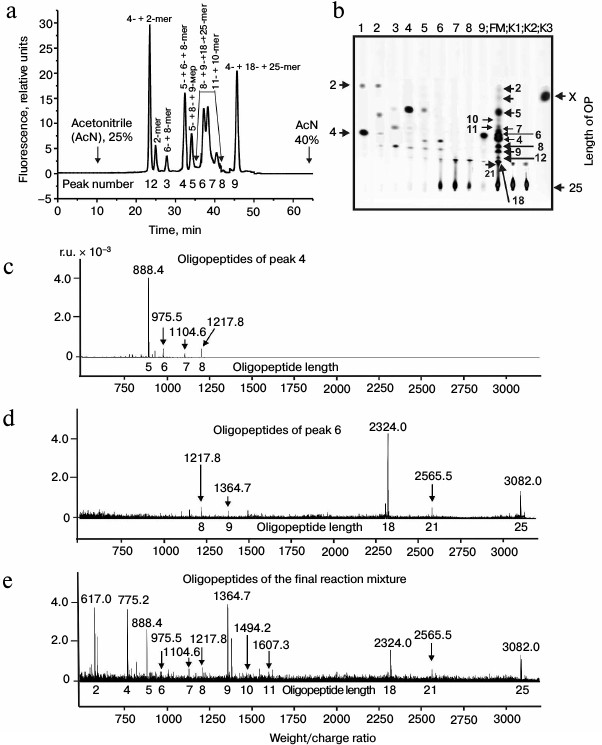
Then products of X-OP25 hydrolysis by IgM(salt)mix preparation were assessed (Fig. 4 and Table 4). The relative contents of the X-OP25 hydrolysis products decreased in the order: 5-mer OP ≈ 4-mer OP ≥ 6-mer OP ≥ 7-mer OP ≥ 8-mer OP ≥ 21-mer OP ≥ 9-mer OP ≥ 10-mer OP ≥ 18-mer OP ≈ 1-mer OP ≈ 11-mer OP ≈ 2-mer OP ≈ X (Table 4).
Fig. 4. RPC profile of products of X-OP25 extensive hydrolysis by anti-IN abzymes of the IgM(salt)mix fraction (a) and analysis of the X-OP25 hydrolysis products corresponding to RPC peaks (a) by TLC (b) and mass spectrometry (c-e). b) Numbers of lanes in the TLC plate correspond to numbers of the RPC peaks (a). Lanes: FM) final reaction mixture; K1, K2) X-OP25 incubated in the absence of ABs and before the incubation, respectively; K3) free fluorescent label X. c-e) MALDI mass spectrometry spectra of: c) peak 1 (a) products are presented in relative units (RU); d) OP peak 6 (a); e) peptides of the final reaction mixture before its separation into fractions. In total, the cleavage sites of X-OP25 and X-OP21 by ABs against MBP and IN are grouped into two clusters (Fig. 5) highly homologous with in-OP1 and in-OP2 (Fig. 7).
Table 4. Results of analysis of molecular
weights and relative amounts of fluorescent OPs produced after
incubation of X-OP25 with anti-IN abzymes of IgM(salt)mix
from the data of RPC, TLC, and MALDI mass spectrometry (Fig. 4)
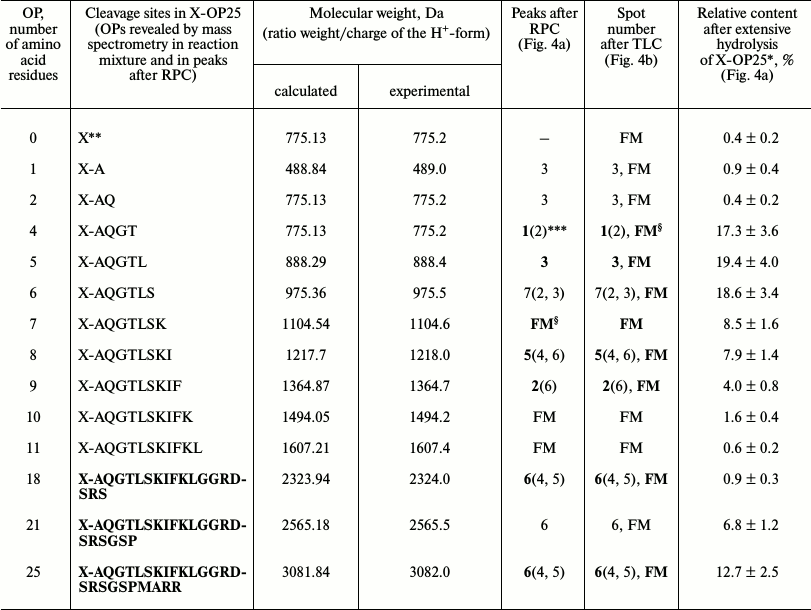
* The total amount of hydrolysis product is taken as 100%; each value is
calculated as the mean of data of three TLC experiments.
** The initial X-OP25 contained the fluorescent label X.
*** Numbers of peaks containing the major part of the product are
printed bold, and numbers of peaks containing the same OPs as the minor
product are in parentheses.
§ The same products of X-OP25 hydrolysis in the peaks
after RPC (Fig. 4a) were revealed not only by MALDI
mass spectrometry, but also by TLC (Fig. 4b). FM
indicates the presence of a fluorescent peak of this product in the
combined final reaction mixture analyzed by TLC (Fig. 4b) and in peaks of RPC (Fig. 4a).
These data confirmed that specific fractions of IgG and IgM with different affinity for IN could contain abzymes with different efficiency in hydrolysis of oligopeptides at their different cleavage sites.
DISCUSSION
Recently, it was shown that anti-MBP IgG and IgM from blood of patients with MS [24-26] and SLE [29, 30] effectively hydrolyzed only globular MBP, whereas anti-IN IgG and IgM from blood of HIV-infected patients effectively hydrolyzed only intact globular IN [17-20]. These abzymes did not cleave other studied globular proteins. IgG and IgM preparations from blood of healthy donors did not hydrolyze MBP [24-26, 29, 30] or IN [17-20]. Sites of MBP cleavage by MS and SLE abzymes [27, 29-32] similarly to sites of IN hydrolysis by antibodies of HIV-infected patients [19] determined by mass spectrometry were localized, respectively, in four and seven known immunodominant sequences of MBP and IN.
Note that for identification of MBP hydrolysis sites, only large peptides were initially used that were produced as a result of AB-dependent hydrolysis of intact globular MBP within a short time of incubation (Fig. 5c) [27]. Six major sites of cleavage were detected in four immunogenic sequences of MBP. However, we found that long-term incubation (48-72 h) of MBP with ABs from blood of patients with MS and SLE, especially with abzymes possessing high proteolytic activity, resulted in production of short and very short protein fragments [29, 30]. This situation was observed for anti-IN abzymes from blood of HIV-infected patients on hydrolysis of viral integrase [19]. That seemed to indicate that the total pool of different antibodies could contain subfractions of ABs against one of the proteins, e.g. MBP or IN, and were unable to hydrolyze specific proteins – antigens and their fragments with different efficiency, at least, at some sites.
Fig. 5. a-c) Cleavage sites of proteins and oligopeptides by trypsin and abzymes. For all sequences, major, moderate, and weak cleavage sites are shown, respectively, by thick arrows, rhombs, and short arrows. a) Sites of hydrolysis of oligopeptide X-OP25 of MBP by two IgG preparations and by two IgM preparations of abzymes from blood of HIV-infected patients identified by combination of RPC, TLC, and mass spectrometry. Sites of the OP cleavage are clustered in two closely spaced fragments of the sequences. b) Four trypsin cleavage sites of the OP25 sequence within the globular intact MBP and one site of MBP hydrolysis corresponding to the OP25 sequence at the initial moment of MBP incubation with anti-MBP abzymes from blood of MS and SLE patients; sites of X-OP25 cleavage by anti-MBP abzymes of MS and SLE patients, and also all sites of the X-OP25 cleavage by anti-IN IgG and IgM abzymes at HIV-infection. c) Sites of the OP21 cleavage by trypsin, one site of the MBP hydrolysis corresponding to the OP21 sequence at the initial moment of MBP incubation with anti-MBP abzymes from blood of MS and SLE patients; all sites of the X-OP21 hydrolysis by IgG and IgM anti-MBP antibodies [30], and all sites of the X-OP21 hydrolysis by IgG and IgM anti-IN antibodies from blood of HIV-infected patients [33]. For analyzing the cleavage sites of OP21 and OP25, IgGmix and IgMmix preparations described in work [33] were used.
It was shown in some works that nonspecific short OPs could be substrates of abzymes that on the level of globular proteins specifically hydrolyzed only globular protein antigens. Anti-thyroglobulin IgG with a proteolytic activity efficiently hydrolyzed not only thyroglobulin but also Pro-Phe-Arg-methylcoumarin amide (MCA) by the Arg–MCA bond with a significantly lower affinity and efficiency that thyroglobulin [37]. IgG preparations from blood of patients with rheumatoid arthritis also could hydrolyze Pro-Phe-Arg-MCA [38]. Bence-Jones proteins cleaved a number of different MCA-peptides [39].
It was shown earlier that anti-IN abzymes of HIV-infected patients efficiently (during 0.5-3 h of incubation) hydrolyzed not only specific OPs corresponding to immunogenic sequences of the viral IN but also nonspecific tri- and tetrapeptides [19, 20, 34]. Moreover, anti-IN abzymes efficiently hydrolyzed 20-25-mer OPs corresponding to immunogenic sequences of RT of HIV [34] and one 21-mer OP corresponding to one of the immunodominant sequences of human MBP [33]. It should be emphasized that anti-MBP abzymes from blood of MS and SLE patients were unable to hydrolyze efficiently nonspecific tri- and tetrapeptides as well as 20-25-mer OPs corresponding to RT and IN of the immunodeficiency virus; noticeable hydrolysis could be observed only after incubation for 4-6 days [30, 31]. In the present work, we tried to analyze possible reasons of significant differences in the hydrolysis of globular proteins and relatively short and long specific and nonspecific oligopeptides by specific antibodies against different proteins.
The efficient hydrolysis of X-OP25 by anti-IN abzymes could be, in particular, due to a high homology between the sequences efficiently cleaved by anti-IN and anti-MBP abzymes. Therefore, it was interesting to analyze possible differences in X-OP25 cleavage sites by anti-MBP and anti-IN abzymes as well as possible homology of sequences of oligopeptides cleaved by anti-MBP and anti-IN abzymes between themselves and with full sequences of MBP and IN.
Figure 5a presents summarized data on hydrolysis of X-OP25 (by cleavage sites) by four preparations of anti-IN abzymes. Extensive hydrolysis of this OP by IgG(acid)mix for 24 h generates two major and one minor OPs: 4-mer OP > 5-mer OP > 6-mer OP (Table 1). In the initial period of the hydrolysis, only the 4- and 5-mer OPs were observed. The same was also observed for IgG(salt)mix (Fig. 2 and Table 2). However, in the last case there were additional cleavage products: 7-, 8-, 9-, 18-, and 21-mer OPs. In the case of IgM(acid)mix and IgM(salt)mix preparations, the number of X-OP25 cleavage sites was higher than for the IgG(acid)mix and IgG(salt)mix preparations. IgM(acid)mix hydrolyzed X-OP25 with additional production of 11-, 12-, 18-, and 21-mer X-OPs. In the case of IgM(salt)mix, no 11- and 12-mer OPs were produced but a 10-mer OP was generated (Fig. 5a and Tables 3 and 4). In general, all anti-IN abzymes hydrolyzed X-OP25 by nine clustered sites starting from the 4th (Thr–Leu) and to the 12th (Gly–Gly) amino acid residue of OP25 (Fig. 5b, lower line). Two more cleavage sites of the OP corresponded to positions 18 and 21 of amino acid residues (the bonds Ser–Gly and Ser–Pro, respectively). It should be noted that only the major products of extensive hydrolysis of X-OP25 were observed also upon incubation for a short time (1-3 h).
Patterns of the oligopeptide X-OP25 hydrolysis by trypsin (the cleavage sites were calculated theoretically; Fig. 5b, first line), by anti-MBP antibodies from blood of MS patients, and by anti-IN abzymes of HIV-infected patients, as well as patterns of globular MBP hydrolysis by anti-MBP antibodies from MS and SLE patients, were compared (Fig. 5c). One of the cleavage sites of X-OP25 by anti-IN antibodies coincided with one of the five possible cleavage sites of the OP by trypsin (Fig. 5b, first and fourth lines).
For abzymes of MS patients, the major site of the AB-dependent cleavage of the OP25 sequence was identified earlier within intact MBP with the corresponding generation of a 10-mer OP (Fig. 5b, second line) [27]. Anti-MBP antibodies did not hydrolyze X-OP25 at the trypsinolysis sites or with production of the 10-mer OP (Fig. 5b, third line). The anti-IN abzymes did not cleave X-OP25 at three of five trypsin-dependent sites but hydrolyzed with low efficiency the bonds Lys–Ile and Lys–Leu with production of the 7- and 10-mer OPs (Fig. 5b).
Some cleavage sites in the first cluster TLSKIFKLGG of the oligopeptide X-OP25 occurred to be the same for both anti-MBP abzymes (Fig. 5b, third line) and anti-IN antibodies (Fig. 5b, fourth line). Moreover, in both cases the cleavage of the TLS sequence resulted in production of 4- and 5-mer OPs as the main products of hydrolysis. However, as discriminated from anti-MBP antibodies, anti-IN abzymes did not have cleavage sites in the first cluster of X-OP25 corresponding to production of 1-, 2-, and 3-mer OPs. In the second cluster, which corresponded to production of 18- and 20-mer products in the case of anti-IN abzymes (Fig. 5b, fourth line), these products were absent on the OP cleavage by anti-MBP abzymes (third line).
Figure 5c presents the earlier published data on the X-OP21 hydrolysis by trypsin (line 1), by anti-MBP antibodies from MS and SLE patients (line 2), and also by anti-IN IgGmix and IgMmix abzymes from HIV-infected patients (line 3) that were used earlier [33] and in the present work. Moreover, in Fig. 5c (line 4) data are presented on hydrolysis of X-OP21 by anti-MBP abzymes from blood of SLE patients [31]. One of two possible cleavage sites of X-OP21 by trypsin coincided with the cleavage site of the amino acid sequence of OP21 within the intact MBP at the early stages of hydrolysis of this protein by anti-MBP antibodies from blood of MS and SLE patients (compare lines 1 and 2). Although the cleavage sites of X-OP25 by anti-MBP antibodies and anti-IN abzymes were very similar but nevertheless different (Fig. 5b), in the case of X-OP21 they virtually coincided except for one site (Fig. 5b, compare lines 3 and 4).
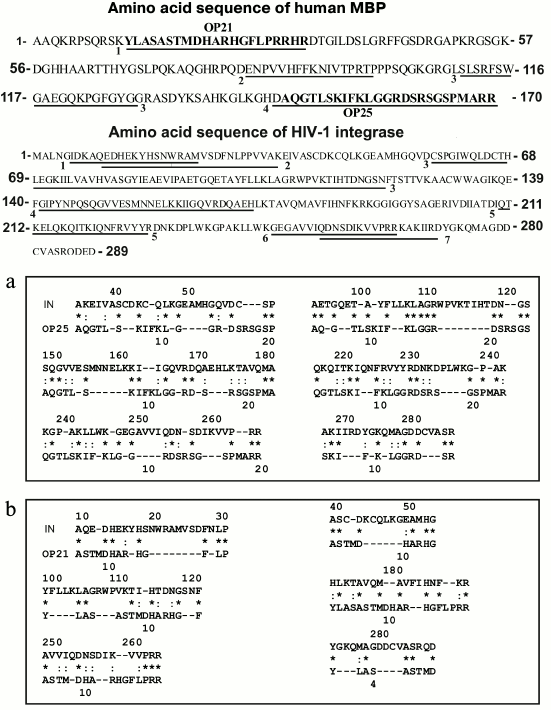
According to the literature, IN of HIV contains seven immunogenic amino acid sequences (IAS) that included the residues 5-22 (IAS1), 14-35 (IAS2) [40], 58-141 (IAS3), 141-172 (IAS4), 248-264 (IAS5) [41], 208-228 (IAS6), and 251-271 (IAS7) [42] (Fig. 6). In the present work, we analyzed for the first time homologies of OP25 and OP21 sequences in comparison with that of IN (Fig. 6).
Fig. 6. Full amino acid sequences of human MBP and HIV-1 integrase. Fragments of four and seven known immunodominant sequences of MBP and IN, respectively, are underlined and positions are shown of sequences of the OP25 and OP21 peptides within the full sequence of MBP (denominations are presented on the figure). a) Data on homology of oligopeptides OP25 (a) and OP21 (b) with different fragments of HIV integrase sequence; the homology data are presented for six of 10 fragments of the protein sequences. Identical amino acid residues are shown by asterisks (*) and amino acid residues with similar physicochemical features are shown by colons (:).
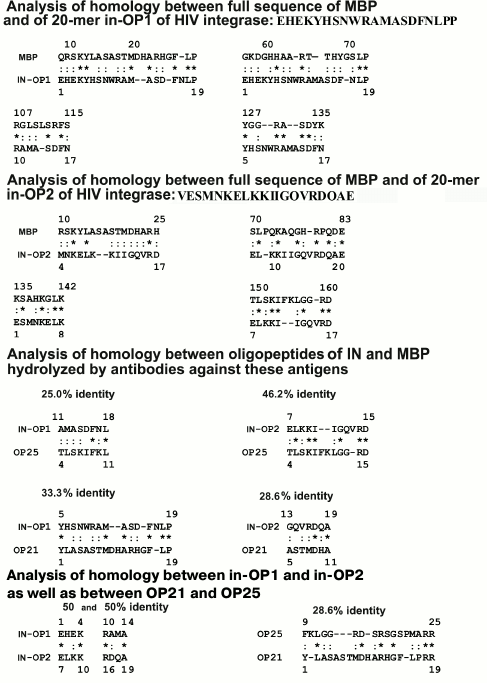
Both oligopeptides (OP25 and OP21) were 28-40% homologous to 10 fragments of the full sequence of IN. Figure 6 presents data only for six of 10 fragments of IN homologous to OP25 and OP21; some amino acid residues were fully coincident (indicated by asterisks), whereas others manifested good correlation in structure and physicochemical features (indicated by colons). Note that OP25 and OP21 themselves had noticeable homology — 28.6% (Fig. 7).
Fig. 7. Data on homology of oligopeptides in-OP1 and in-OP2 with different fragments of the full sequence of human MBP (data are presented on homology for four of seven fragments of the protein sequences), between oligopeptides of IN (in-OP1 and in-OP2) and MBP (OP21 and OP25) hydrolyzed by abzymes against these proteins, as well as between themselves of in-OP1 – in-OP2 and OP21 – OP25. Identical amino acids are indicated by asterisks (*) and those with similar physicochemical features by colon (:).
As shown in work [34], anti-IN abzymes effectively cleaved oligopeptides corresponding to cleavage sites of viral RT by ABs against this protein from blood of HIV-infected patients. Thus, it might be that partial homology and structural similarity of amino acids of sequences of different OPs and IN-sequences including antibody-dependent cleavage sites can be sufficient for them to cleave effectively nonspecific OPs by anti-IN abzymes.
According to the literature, MBP contains four immunogenic amino acid sequences that include residues 11-32 (IAS1), 83-99 (IAS2), 111-129 (IAS3), and 145-170 (IAS4) [27] (Fig. 6). It is interesting that 20-mer oligopeptides in-OP1 and in-OP2 corresponding to immunogenic sequences of IN containing sites of AB-dependent cleavage are also partially homologous (20-50%) to some fragments of MBP. Figure 7 shows only four of seven differently homologous fragments of MBP and in-OP. Moreover, two 4-mer fragments of in-OP1 and in-OP2 are 50% homologous (Fig. 7).
In total, sites of cleavage of X-OP25 and X-OP21 by ABs against MBP and IN are grouped in two clusters (Fig. 5) that are highly homologous to in-OP1 and in-OP2 (Fig. 7); the full identity of amino acid residues varies from 25 to 46.2% (Fig. 7). A similar partial homology (18-50% identity of amino acid residues) was also observed for some fragments of the MBP sequence and OPs corresponding to the cleaved sequences of HIV integrase (in-OP1 and in-OP2; Fig. 7). However, from our standpoint, the question about factors important for cleavage of foreign OPs by abzymes can be much more complicated. In addition to the complete identity of amino acid residues of the sequences, a spatial structure of oligopeptides that depends not only on a full coincidence of some amino acids but also on the presence in these sequences of other amino acids with similar physicochemical features can also play an important role. On taking into account these two factors, likeness of studied sequences will be much higher. Thus, for the case of 18% identity (indicated by the asterisk in Fig. 7), considering amino acids with similar physicochemical features (indicated by the colon in Fig. 7), the identity of common physicochemical parameters reaches 71% and for OPs with 46-50% of identical amino acids the identity of physicochemical features of these sequences reaches 75-88%.
Nevertheless, in the case of abzymes against other proteins (not IN), partial homology and structural similarity of the sequences can be insufficient for effective hydrolysis of foreign OPs. Thus, anti-MBP abzymes do not cleave IN and RT peptides [31, 33, 34]. Therefore, we suppose that in the presence of some ABs (of the anti-IN abzyme type) a partial homology and a general similarity in the structure of the sequences can play an important role for cleavage of “foreign” peptides and be of no importance in other cases.
It is interesting why abzymes are very specific to globular proteins but many of them can efficiently cleave nonspecific tri- and tetrapeptides [34, 37-39]. Catalytic centers of proteolytic abzymes are usually located in the variable part of the light chain, whereas the heavy chain is responsible for a specific recognition of the antigen and an increase in the antigen affinity for AB. Thus, using SDS-electrophoresis (conditions for dissociation in the presence of dithiothreitol) and a gel containing a polymerized substrate, the active center localization on the light chains was shown for DNA- and RNA-hydrolyzing ABs from blood of patients with different autoimmune [43-45], bacterial, and viral diseases [46, 47], for human milk [48], and blood of autoimmune mice [49]. Using SDS electrophoresis under conditions for dissociation with subsequent analysis of activity of eluates of many gel fragments, localization of the active center on the light chains was shown for antibodies hydrolyzing MBP [29], integrase of HIV-1 [17, 18], oligosaccharides [50], and ATP [51]. Moreover, monoclonal light chains were shown to be active in hydrolyzing DNA [52, 53], a vasoactive neuropeptide [54], prothrombin [55], β-amyloid [56], a chemokine receptor CCR-5 [57], gp120 protein of HIV-1 [58], a peptide of hepatitis C [58], and proteins of Helicobacter pylori [60]. Intact globular proteins usually interact with both chains of immunoglobulins and thus provide for specific recognition of proteins and their cleavage [1-6]. The affinity of short OPs, including 20-25-mer oligopeptides [27, 34, 39, 61, 62] and also short oligonucleotides [52, 53], for intact abzymes and separate light chains of antibodies is approximately 100-1000 times lower than for the corresponding globular proteins and elongated nucleic acids. Consequently, tri- and tetrapeptides and also the longer OPs (and short oligonucleotides) can interact with the light chain independently of the heavy chain. Therefore, depending on the amino acid sequence, hydrolysis of OPs can be less specific or fully nonspecific. Moreover, in the case of globular proteins hydrolysis sites can be determined by their specific interaction with the heavy chains. Active centers of the light chains can hydrolyze globular proteins by the bonds that occur nearer to these centers due to specific interactions of globular proteins with the heavy chains. This is confirmed by data on differences in the cleavage sites of globular proteins and oligopeptides. Figure 5 shows that the number of cleavage sites of OPs corresponding to hydrolyzed sequences of globular proteins is much higher (compare line 2 with lines 3 and 4 (b) and line 2 with line 3 (c)). Note, that in the case of oligopeptides, hydrolysis by the cleavage sites of globular proteins can even fail to occur (Fig. 5b; compare line 2 with line 3), or the major cleavage site in the case of MBP hydrolysis by anti-MBP antibodies (Fig. 5b, line 2) can be minor on the cleavage of OP (Fig. 5b, line 4). This suggests that significant differences should exist in the interaction of antibodies with the same sequences on the level of globular proteins and oligopeptides. It should be emphasized that abzymes cleave OPs not at all peptide bonds. However, clustered positions of the great number of sites of oligopeptide hydrolysis (Fig. 5) can suggest production of some alternative complexes with the light chains possessing preferred oligopeptide sequences. It is interesting that on the AB-dependent hydrolysis of OPs, there is no pronounced specificity that is characteristic for trypsin and chymotrypsin; abzymes are more like elastase, which cleaves proteins at very different amino acid sequences.
In the presence of the above-described mutual homology between the cleaved sequences of IN and MBP, anti-IN abzymes effectively cleave OPs corresponding to MBP, whereas anti-MBP antibodies are virtually inactive with respect to IN oligopeptides. However, the affinities of the same short OPs for anti-IN and for anti-MBP abzymes are comparable [20, 31-34]. Thus, it might be that similarly to the case of classical enzymes [63, 64], when affinities of different ligands for active centers are similar, the efficiency of the mutual conformational adaptation stage of the ligand and abzyme can be essential in catalysis by antibodies. It seems that upon binding with the light chain only those short OPs capable of changing their conformation up to optimal for catalysis can be efficiently cleaved.
It was said above that 40 cleavage sites of IN were revealed in seven known immunodominant sequences of IN [19]. Note that each individual preparation of ABs from HIV-infected patients had a specific ratio of hydrolysis sites within different immunodominant sequences, whereas some preparations of antibodies hydrolyzed IN only in a limited number of sites [19]. Figure 8 exemplifies the difference in the cleavage sites of IN for three individual preparations of anti-IN abzymes. These preparations were analyzed earlier in work [19] and used in the present work for obtaining IgGmix. One can see that IgG1, IgG6, and IgG7 cleave IN at 35, 28, and 6 sites, some of which are inherent only in some of these preparations of abzymes. A similar situation can also occur for ABs against other proteins, and it seems that specific features of abzymes against the same protein can depend on the disease. Thus, polyclonal anti-MBP abzymes from blood of MS patients were mainly serine proteases because the activity of 12 preparations was more effectively suppressed after their incubation with PMSF (on average, by 84.5 ± 10.5%) but not with EDTA (on average, by 15.8 ± 3.5%) [29]. However, a pool of 12 preparations of ABs from blood of SLE patients mainly contained abzymes with metal-dependent protease activity; EDTA suppressed their activity, on average, by 76.3 ± 11.7% and PMSF only by 20.9 ± 14.0% [29]. Therefore, it was not a surprise when four preparations of IgGmix and IgMmix from HIV-infected patients with different affinities for MBP demonstrated noticeable differences in the hydrolysis of X-OP25.
Fig. 8. Comparison of cleavage sites of HIV integrase (indicated by arrows) under the influence of anti-IN abzymes from blood of three HIV-infected patients [19].
The studied proteolytic abzymes were mainly serine proteases, and their activity significantly decreased on incubation with specific inhibitors of serine proteases, PMSF and/or AEBSF. Such abzymes also included IgG hydrolyzing a vasoactive intestinal peptide, thyroglobulin, prothrombin, and protein factor VIII from blood of autoimmune patients [38, 55, 65, 66], casein-hydrolyzing ABs from human milk and from blood of HIV-infected patients [67], and IgM hydrolyzing protein gp120 of HIV from blood of HIV-infected patients [68]. Moreover, small fractions of IgG and IgM from blood of MS and SLE patients possessed not only a serine-like activity, but also a metal-dependent protease activity [24, 25, 29-31].
Anti-IN ABs from blood of HIV-infected patients are specific abzymes. Specific inhibitors of serine proteases inhibited preparations of IN-hydrolyzing IgG and IgM only from 20% of HIV-infected proteins, and the activity was low or completely absent [17, 18]. Thus, AEBSF inhibited the IgG activity in two of 10 patients by 42 and 49%. The combined pools of IgG and IgM in five patients of 10 (50%) contained ABs with metal-dependent activity; an addition of EDTA decreased the activity of the corresponding five IgG preparations by 0-98%. The activity of two of 10 preparations (20%) of IgG was inhibited by specific inhibitors of acidic proteases by 0-59%. Iodoacetamide, a specific inhibitor of thiol proteases, usually insignificantly influenced the activity of the above-described proteolytic abzymes, i.e. the inhibition varied from ≤3 to 7% ([17, 18] and references in these works). Therefore, it was quite unexpected that inhibition of the IN-hydrolyzing activity in 100% of abzymes preparations from HIV-infected patients varied from 12 to 98% (the mean value for 10 preparations of IgG was 65.7 ± 20.6%). Thus, on average, for 10 studied preparations of IgG the trypsin-like proteolytic activity was not the major one in the case of anti-IN abzymes and decreased in the order: thiol > metalloprotease > acidic ≥ serine-like [18]. A similar situation was observed for anti-IN IgM antibodies [17].
The action of some specific inhibitors of proteases on IgG and IgM preparations was analyzed in detail in parallel experiments. Thus, the activity of one IgM preparation was significantly inhibited by four specific inhibitors: 46 ± 5% by AEBSF, 88 ± 9% by pepstatin A, 64 ± 4% by EDTA, and 91 ± 10% by iodoacetamide. In general, as differentiated from the known proteolytically active abzymes, the activity of 50-60% of IgG and IgM preparations from blood of HIV-infected patients was suppressed by inhibitors of serine, acidic, metal-dependent, and thiol proteases [17, 18]. Thus, this suggests that the immune system of some HIV-infected patients could induce generation of anti-IN abzymes with complex structure of the active center, which included amino acid residues typical for four different types of proteases.
In parallel experiments, we evaluated the relative decrease in the IN-hydrolyzing activity of IgGmix and IgMmix used in this work after their incubation with AEBSF, EDTA, pepstatin A, and iodoacetamide. IgGmix lost its activity after incubation with any of these inhibitors, and the loss in the activity was as follows: the serine-like activity was decreased by 9 ± 0.7%, the acidic protease activity by 12 ± 0.7%, the metalloprotease activity by 35 ± 2%, and the thiol-dependent activity by 69 ± 4%. The relative percent of inhibition of this IgGmix preparation by the four inhibitors was in a good agreement with the average values of above-mentioned parameters for the earlier studied 10 individual preparations of IgG. Similar values of the inhibition by the four specific inhibitors were obtained in the case of IgMmix: AEBSF (6 ± 1%), pepstatin A (20 ± 2%), EDTA (44 ± 3%), and iodoacetamide (80 ± 4%).
Thus, the trypsin-like proteolytic activity was not the main activity of anti-IN abzymes, and the majority of individual preparations did not have any such activity [17, 18]. Most likely, anti-IN abzymes can hydrolyze X-OP25 at additional sites in comparison with anti-MBP antibodies due to their activities similar to those of acidic, metal-dependent, and especially of thiol proteases; different monoclonal ABs constituting pools of each HIV-infected patient can possess different substrate specificity.
In total, the efficient cleavage of specific IN and its OPs, X-OP25 and X-OP21 oligopeptides of MBP as well as nonspecific di-, tri-, and tetrapeptides by anti-IN abzymes from HIV-infected patients can be due to a combination of a number of factors – partial homology of amino acid sequences and structural similarity of OPs with some fragments of IN, interaction with these OPs of only the light chain of abzymes at significantly lower affinity compared to globular protein, and, probably, a lower specificity of anti-IN abzymes, which can be due to manifestation of four different proteolytic activities.
This work was supported by the Russian Academy of Sciences Presidium program “Molecular and Cell Biology”, by the Russian Foundation for Basic Research (projects Nos. 12-04-00315, 13-04-00205, 13-04-00208, 14-04-00735), and by the foundation of the Siberian Division of the Russian Academy of Sciences.
REFERENCES
1.Keinan, E. (ed.) (2005) Catalytic
Antibodies, Wiley-VCH Verlag GmbH and Co. KgaA, Weinheim,
Germany.
2.Nevinsky, G. A., and Buneva, V. N. (2002) Human
catalytic RNA- and DNA-hydrolyzing antibodies, J. Immunol.
Methods, 269, 235-249.
3.Nevinsky, G. A., and Buneva, V. N. (2005) Natural
catalytic antibodies – abzymes, in Catalytic
Antibodies (Keinan, E., ed.) VCH-Wiley, Weinheim, Germany, pp.
503-567.
4.Nevinsky, G. A. (2010) Natural catalytic antibodies
in norm and in autoimmune diseases, in Autoimmune Diseases:
Symptoms, Diagnosis and Treatment (Brenner, K. J., ed.) Nova
Science Publishers, Inc., USA.
5.Nevinsky, G. A. (2011) Natural catalytic antibodies
in norm and in HIV-infected patients, in Understanding HIV/AIDS
Management and Care – Pandemic Approaches the 21st
Century (Kasenga, F. H., ed.) InTech, Rijeka, Croatia, pp.
151-192.
6.Nevinsky, G. A., and Buneva, V. N. (2012)
Autoantibodies and natural catalytic antibodies in health, multiple
sclerosis, and some other diseases, Adv. Neuroimmune Biol.,
3, 157-182.
7.Fauci, A. S., Braunwald, E., Kasper, D. L., Hauser,
S. L., Longo, D. L., and Jameson, J. L. (2008) Harrison’s
Principles of Internal Medicine, 7th Edn., McGraw-Hill
Professional, New York.
8.Katz, A. R., and Skalka, A. M. (1994) HIV-1
integrase: structural organization, conformational changes, and
catalysis, Ann. Rev. Biochem., 63, 133-173.
9.Litvak, S. (1996) Retroviral reverse
transcriptases, in Molecular Biology Intelligency Unit Series
(Landes, R., ed.) Chapman and Hall/Springer Verlag, Heidelberg.
10.Skalka, A. M., and Goff, S. P. (1993) Reverse
Transcriptase, Cold Spring Harbor Laboratory Press, New York.
11.Asante-Appiah, E., and Skalka, A. M. (1999) HIV-1
integrase: structural organization, conformational changes, and
catalysis, Adv. Virus Res., 52, 351-369.
12.Verkoczy, L., and Diaz, M. (2014) Autoreactivity
in HIV-1 broadly neutralizing antibodies: implications for their
function and induction by vaccination, Curr. Opin. HIV AIDS,
9, 224-234.
13.Zandman-Goddard, G., and Shoenfeld, Y. (2002) HIV
and autoimmunity, Autoimmun. Rev., 1, 329-337.
14.Gololobov, G. V., Mikhalap, S. V., Starov, A. V.,
Kolesnikov, A. F., and Gabibov, A. G. (1994) DNA–protein
complexes. Natural targets for DNA-hydrolyzing antibodies, Appl.
Biochem. Biotechnol., 47, 305-314.
15.Odintsova, E. S., Kharitonova, M. A.,
Baranovskii, A. G., Sizyakina, L. P., Buneva, V. N., and Nevinsky, G.
A. (2006) DNA-hydrolyzing IgG antibodies from the blood of patients
with acquired immune deficiency syndrome, Mol. Biol., 40,
857-864.
16.Odintsova, E. S., Kharitonova, M. A.,
Baranovskii, A. G., Sizyakina, L. P., Buneva, V. N., and Nevinsky, G.
A. (2006) Proteolytic activity of IgG antibodies from blood of acquired
immunodeficiency syndrome patients, Biochemistry (Moscow),
71, 251-261.
17.Baranova, S. V., Buneva, V. N., Kharitonova, M.
A., Sizyakina, L. P., Calmels, C., Parissi, V., Andreola, M. L.,
Buneva, V. N., Zakharova, O. D., and Nevinsky, G. A. (2010) HIV-1
integrase-hydrolyzing IgM antibodies from sera of HIV-infected
patients, Int. Immunol., 22, 671-680.
18.Baranova, S. V., Buneva, V. N., Kharitonova, M.
A., Sizyakina, L. P., Calmels, C., Andreola, M. L., Parissi, V., and
Nevinsky, G. A. (2009) HIV-1 integrase-hydrolyzing antibodies from sera
of HIV-infected patients, Biochimie, 91,
1081-1086.
19.Odintsova, E. S., Baranova, S. V., Dmitrenok, P.
S., Rasskazov, V. A., Calmels, C., Parissi, V., Andreola, M. L.,
Buneva, V. N., Zakharova, O. D., and Nevinsky, G. A. (2011) Antibodies
to HIV integrase catalyze site-specific degradation of their antigen,
Int. Immunol., 23, 601-612.
20.Odintsova, E. S., Dmitrenok, P. S., Buneva, V.
N., and Nevinsky, G. A. (2013) Specific anti-integrase abzymes from
HIV-infected patients: a comparison of the cleavage sites of intact
globular HIV integrase and two 20-mer oligopeptides corresponding to
its antigenic determinants, J. Mol. Recognit., 26,
121-135.
21.O’Connor, K. C., Bar-Or, A., and Hafler, D.
A. (2001) Neuroimmunology of multiple sclerosis, J. Clin.
Immunol., 21, 81-92.
22.Archelos, J. J., Storch, M. K., and Hartung, H.
P. (2000) The role of B cells and autoantibodies in multiple sclerosis,
Ann. Neurol., 47, 694-706.
23.Hemmer, B., Archelos, J. J., and Hartung, H. P.
(2002) New concepts in the immunopathogenesis of multiple sclerosis,
Nat. Rev. Neurosci., 3, 291-301.
24.Polosukhina, D. I., Kanyshkova, T. G., Doronin,
B. M., Tyshkevich, O. B., Buneva, V. N., Boiko, A. N., Gusev, E. I.,
Favorova, O. O., and Nevinsky, G. A. (2004) Hydrolysis of myelin basic
protein by polyclonal catalytic IgGs from the sera of patients with
multiple sclerosis, J. Cell. Mol. Med., 8, 359-368.
25.Polosukhina, D. I., Buneva, V. N., Doronin, B.
M., Tyshkevich, O. B., Boiko, A. N., Gusev, E. I., Favorova, O. O., and
Nevinsky, G. A. (2005) Hydrolysis of myelin basic protein by IgM and
IgA antibodies from the sera of patients with multiple sclerosis,
Med. Sci. Monit., 11, BR266-BR272.
26.Polosukhina, D. I., Kanyshkova, T. G., Doronin,
B. M., Tyshkevich, O. B., Buneva, V. N., Boiko, A. N., Gusev, E. I.,
Nevinsky, G. A., and Favorova, O. O. (2006) Metal-dependent hydrolysis
of myelin basic protein by IgGs from the sera of patients with multiple
sclerosis, Immunol. Lett., 103, 75-81.
27.Ponomarenko, N. A., Durova, O. M., Vorobiev, I.
I., Belogurov, A. A., Kurkova, I. N., Petrenko, A. G., Telegin, G. B.,
Suchkov, S. V., Kiselev, S. L., Lagarkova, M. A., Govorun, V. M.,
Serebryakova, M. V., Avalle, B., Tornatore, P., Karavanov, A., Morse,
H. C. 3rd, Thomas, D., Friboulet, A., and Gabibov, A. G. (2006)
Autoantibodies to myelin basic protein catalyze site-specific
degradation of their antigen, Proc. Natl. Acad. Sci. USA,
103, 281-286.
28.Hhachn, B. Ch. (1996) Systemic lupus
erythematosus, in Internal Diseases (Braunvald, E. E.,
Isselbakher, K. D., Petersdorf, R. G., Wilson, D. D., Martin. D. B.,
and Fauchi, A. S., eds.) [Russian translation], Meditsina, Moscow.
29.Bezuglova, A. M., Konenkova, L. P., Doronin, B.
M., Buneva, V. N., and Nevinsky, G. A. (2011) Affinity and catalytic
heterogeneity and metal-dependence of polyclonal myelin basic
protein-hydrolyzing IgGs from sera of patients with systemic lupus
erythematosus, J. Mol. Recognit., 24, 960-974.
30.Bezuglova, A. M., Dmitrenok, P. S., Konenkova, L.
P., Buneva, V. N., and Nevinsky, G. A. (2012) Multiple sites of the
cleavage of 17- and 19-mer encephalitogenic oligopeptides corresponding
to human myelin basic protein (MBP) by specific anti-MBP antibodies
from patients with systemic lupus erythematosus, Peptides,
37, 69-78.
31.Timofeeva, A. M., Dmitrenok, P. S., Konenkova, L.
P., Buneva, V. N., and Nevinsky G. A. (2013) Multiple sites of the
cleavage of 21- and 25-mer encephalitogenic oligopeptides corresponding
to human myelin basic protein (MBP) by specific anti-MBP antibodies
from patients with systemic lupus erythematosus, PLoS One,
8, e51600.
32.Bezuglova, A. M., Konenkova, L. P., Buneva, V.
N., and Nevinsky, G. A. (2012) IgGs containing light chains of the
λ- and κ-type and of all subclasses (IgG1-IgG4) from the
sera of patients with systemic lupus erythematosus hydrolyze myelin
basic protein, Int. Immunol., 12, 759-770.
33.Odintsova, E. S., Dmitrenok, P. S., Timofeeva, A.
M., Buneva, V. N., and Nevinsky, G. A. (2014) Why specific
anti-integrase antibodies from HIV-infected patients can efficiently
hydrolyze 21-mer oligopeptide corresponding to antigenic determinant of
human myelin basic protein, J. Mol. Recognit., 27,
32-45.
34.Odintsova, E. S., Baranova, S. V., Dmitrenok, P.
S., Calmels, C., Parissi, V., Andreola, M. L., Buneva, V. N., and
Nevinsky, G. A. (2012) Anti-integrase abzymes from the sera of
HIV-infected patients specifically hydrolyze integrase but
nonspecifically cleave short oligopeptides, J. Mol. Recognit.,
25, 193-207.
35.Caumont, A., Jamieson, G., de Soultrait, V.,
Parissi, V., Fournier, M., Zakharova, O. D., Bayandin, R., Litvak, S.,
Tarrago-Litvak, L., and Nevinsky, G. A. (1999) High affinity
interaction of HIV-1 integrase with specific and nonspecific
single-stranded short oligonucleotides, FEBS Lett., 455,
154-158.
36.Huang, X., and Miller, W. (1991) Local alignment
of two-base encoded DNA sequence, Adv. Appl. Math., 12,
337-357.
37.Paul, S., Li, L., Kalaga, R., O’Dell, J.,
Dannenbring, R. E., Swindells, S., Hinrichs, S., Cauturegli, P., and
Rose, N. R. (1997) Characterization of thyroglobulin-directed and
polyreactive catalytic antibodies in autoimmune disease, J.
Immunol., 159, 1530-1536.
38.Kalaga, R., Li, L., O’Dell, J. R., and
Paul, S. (1995) Unexpected presence of polyreactive catalytic
antibodies in IgG from unimmunized donors and decreased levels in
rheumatoid arthritis, J. Immunol., 155, 2695-2702.
39.Paul, S., Li, L., Kalaga, R., Wilkins-Stevens,
P., Stevens, F. J., and Solomon, A. (1995) Natural catalytic
antibodies: peptide-hydrolyzing activities of Bence–Jones
proteins and VL fragment, J. Biol. Chem., 270,
15257-15261.
40.Yi, J., Arthur, J. W., Dunbrack, R. L., and
Skalka, A. M. (2000) An inhibitory monoclonal antibody binds at the
turn of the helix-turn-helix motif in the N-terminal domain of HIV-1
integrase, J. Biol. Chem., 275, 38739-38748.
41.Bizub-Bender, D., Kulkosky, J., and Skalka, A. M.
(1994) Monoclonal antibodies against HIV type 1 integrase: clues to
molecular structure, AIDS Res. Hum. Retroviruses, 10,
1105-1115.
42.Nilsen, B. M., Haugan, I. R., Berg, K., Olsen,
L., Brown, P. O., and Helland, D. E. (1996) Monoclonal antibodies
against human immunodeficiency virus type 1 integrase: epitope mapping
and differential effects on integrase activities in vitro, J.
Virol., 70, 1580-1587.
43.Baranovskii, A. G., Buneva, V. N., Doronin, B.
M., and Nevinsky, G. A. (2008) Immunoglobulins from blood of patients
with multiple sclerosis are catalytically heterogeneous nucleases,
Ros. Immunol. Zh., 2, 405-419.
44.Andrievskaya, O. A., Buneva, V. N., Baranovskii,
A. G., Gal’vita, A. V., Benzo, E. S., Naumov, V. A., and
Nevinsky, G. A. (2002) Catalytic diversity of polyclonal
RNA-hydrolyzing IgG antibodies from the sera of patients with systemic
lupus erythematosus, Immunol. Lett., 81,
191-198.
45.Galvita, A. V., Baranovskii, A. G., Kuznetsova,
I. A., Vinshu, N. V., Galenok, V. A., Buneva, V. N., and Nevinsky, G.
A. (2007) Peculiarities of DNA hydrolysis by antibodies from blood of
patients with pancreatic diabetes, Rus. J. Immunol., 1,
116-131.
46.Parkhomenko, T. A., Buneva, V. N., Tyshkevich, O.
B., Generalov, I. I., Doronin, B. M., and Nevinsky, G. A. (2010)
DNA-hydrolyzing activity of IgG antibodies from the sera of patients
with tick-borne encephalitis, Biochimie, 92, 545-554.
47.Parkhomenko, T. A., Odintsova, E. S., Buneva, V.
N., Kunder, E. V., Zhyltsov, I. V., Senkovich, S. A., Generalov, I. I.,
and Nevinsky, G. A. (2009) DNA-hydrolyzing activity of IgG antibodies
from the sera of patients with diseases caused by different bacterial
infections, J. Cell. Mol. Med., 13, 2875-2887.
48.Kanyshkova, T. G., Semenov, D. V., Buneva, V. N.,
and Nevinsky, G. A. (1999) Human milk lactoferrin binds two molecules
of DNA with different affinities, FEBS Lett., 451,
235-237.
49.Kuznetsova, I. A., Orlovskaya, I. A., Buneva, V.
N., and Nevinsky, G. A. (2007) Activation of DNA-hydrolyzing antibodies
from the sera of autoimmune-prone MRL-lpr/lpr mice by different metal
ions, Biochim. Biophys. Acta, 1774, 884-896.
50.Andryushkova, A. A., Kuznetsova, I. A.,
Orlovskaya, I. A., Buneva, V. N., and Nevinsky, G. A. (2006) Antibodies
with amylase activity from the sera of autoimmune-prone MRL/MpJ-lpr
mice, FEBS Lett., 580, 5089-5095.
51.Andryushkova, A. A., Kuznetsova, I. A.,
Orlovskaya, I. A., Buneva, V. N., and Nevinsky, G. A. (2009)
Nucleotide-hydrolyzing antibodies from the sera of autoimmune-prone
MRL-lpr/lpr mice, Int. Immunol., 21, 935-945.
52.Botvinovskaya, A. V., Kostrikina, I. A., Buneva,
V. N., and Nevinsky, G. A. (2013) Systemic lupus erythematosus:
molecular cloning of several recombinant DNase monoclonal kappa light
chains with different catalytic properties, J. Mol. Recognit.,
26, 450-460.
53.Kostrikina, I. A., Buneva, V. N., and Nevinsky,
G. A. (2014) Systemic lupus erythematosus: molecular cloning of
fourteen recombinant DNase monoclonal kappa light chains with different
catalytic properties, Biochim. Biophys. Acta, 1840,
1725-1737.
54.Sun, M., Gao, Q. S., Kirnarskiy, L., Rees, A.,
and Paul, S. (1997) Cleavage specificity of a proteolytic antibody
light chain and effects of the heavy chain variable domain, J. Mol.
Biol., 271, 374-385.
55.Thiagarajan, P., Dannenbring, R., Matsuura, K.,
Tramontano, A., Gololobov, G., and Paul, S. (2000) Monoclonal antibody
light chain with prothrombinase activity, Biochemistry,
39, 6459-6465.
56.Rangan, S. K., Liu, R., Brune, D., Plaque, S.,
Paul, S., and Sierks, M. R. (2003) Degradation of beta-amyloid by
proteolytic antibody light chains, Biochemistry,
42, 14328-14334.
57.Mitsuda, Y., Hifimi, E., Tsuruhata, R., Fujinami,
H., Yamamoto, N., and Uda, T. (2004) Catalytic antibody light chain
capable of cleaving a chemokine receptor CCR-5 peptide with a high
reaction rate constant, Biotechnol. Bioeng., 86,
217-225.
58.Nidhiyama, Y., Karle, S., Planque, S., Taguchi,
H., and Paul, S. (2007) Antibodies to the superantigenic site of HIV-1
gp120, hydrolytic and binding activities of the light chain subunit,
Mol. Immunol., 44, 2707-2718.
59.Taguchi, H., Keck, Z., Foung, S. K., Paul, S.,
and Nishiyama, Y. (2004) Antibody light chain-catalyzed hydrolysis of a
hepatitis C virus peptide, Bioorg. Med. Chem. Lett., 14,
4529-4532.
60.Hifumi, E., Morihara, F., Hatiuchi, K., Okuda,
T., Nishisono, A., and Uda, T. (2008) Catalytic features and
eradication ability of antibody light-chain UA15-L against
Helicobacter pylori, J. Biol. Chem., 283,
899-907.
61.Li, L., Paul, S., Tyutyulkova, S., Kazatchkine,
M. D., and Kavery, S. (1995) Catalytic activity of anti-thyroglobulin
antibodies, J. Immunol., 154, 3328-3332.
62.Gao, Q. S., Sun, M., Tyutyulkova, S., Webster,
D., Rees, A., Tramontano, A., Massey, R. J., and Paul, S. (1994)
Molecular cloning of a proteolytic antibody light chain, J. Biol.
Chem., 269, 32389-32393.
63.Fersht, A. (1985) Enzyme Structure and
Mechanism, 2nd Edn., W. H. Freeman, Co., N. Y.
64.Nevinsky, G. A. (2003) in Protein Structures:
Kaleidoscope of Structural Properties and Functions (Uversky, V.
N., ed.) Research Signpost, Kerala, pp. 133-222.
65.Paul, S., Volle, D. J., Beach, C. M., Johnson, D.
R., Powell, M. J., and Massey, R. J. (1989) Catalytic hydrolysis of
vasoactive intestinal peptide by human autoantibody, Science,
244, 1158-1162.
66.Lacroix-Desmazes, S., Moreau, A., Sooryanarayana,
Bonnemain, C., Stieltjes, N., Pashov, A., Sultan, Y., Hoebeke, J.,
Kazatchkine, M. D., and Kaveri, S. V. (1999) Catalytic activity of
antibodies against factor VIII in patients with hemophilia A, Nat.
Med., 5, 1044-1047.
67.Odintsova, E. S., Buneva, V. N., and Nevinsky, G.
A. (2005) Casein-hydrolyzing activity of sIgA antibodies from human
milk, J. Mol. Recognit., 18, 413-421.
68.Paul, S., Karle, S., Planque, S., Taguchi, H.,
Salas, M., Nishiyama, Y., Handy, B., Hunter, R., Edmundson, A., and
Hanson, C. (2004) Naturally occurring proteolytic antibodies: selective
immunoglobulin M-catalyzed hydrolysis of HIV gp120, J. Biol.
Chem., 279, 39611-39619.