Reorganization of Low-Molecular-Weight Fraction of Plasma Proteins in the Annual Cycle of Cyprinidae
A. M. Andreeva1*, N. E. Lamas2,3, M. V. Serebryakova4, I. P. Ryabtseva1, and V. V. Bolshakov1
1Papanin Institute for Biology of Inland Waters, Russian Academy of Sciences, 152742 Borok, Russia; fax: (485) 472-4042; E-mail: aam@ibiw.yaroslavl.ru; molbiol@ibiw.yaroslavl.ru; victorb@ibiw.yaroslavl.ru2Zhirmunsky Institute of Marine Biology, Far-Eastern Branch of the Russian Academy of Sciences, ul. Palchevskogo 17, 690041 Vladivostok, Russia; E-mail: ninalamash@yandex.ru
3Far-Earstern Federal University, ul. Sukhanova 8, 690922 Vladivostok, Russia
4Lomonosov Moscow State University, Leninsky Gory 1, 119991 Moscow, Russia; E-mail: mserebr@mail.ru
* To whom correspondence should be addressed.
Received August 21, 2014; Revision received September 26, 2014
Reorganization of the low-molecular-weight fraction of cyprinid plasma was analyzed using various electrophoretic techniques (disc electrophoresis, electrophoresis in polyacrylamide concentration gradient, in polyacrylamide with urea, and in SDS-polyacrylamide). The study revealed coordinated changes in the low-molecular-weight protein fractions with seasonal dynamics and related reproductive rhythms of fishes. We used cultured species of the Cyprinidae family with sequenced genomes for the detection of these interrelations in fresh-water and anadromous cyprinid species. The common features of organization of fish low-molecular-weight plasma protein fractions made it possible to make reliable identification of their proteins. MALDI mass-spectrometry analysis revealed the presence of the same proteins (hemopexin, apolipoproteins, and serpins) in the low-molecular-weight plasma fraction in wild species and cultured species with sequenced genomes (carp, zebrafish). It is found that the proteins of the first two classes are organized as complexes made of protein oligomers. Stoichiometry of these complexes changes in concordance with the seasonal and reproductive rhythms.
KEY WORDS: fish, plasma proteins, mass spectra, MALDIDOI: 10.1134/S0006297915020078
Blood plasma is one of the most complicated objects for studies using methods of proteomics [1]. Along with the “true plasma proteins”, extracellular proteins tagged after the synthesis of amino acids with signaling peptide, plasma also contains some amount of tissue proteins entering the bloodstream due to the destruction of cells [2]. In addition, the proteins of some viruses, bacteria, and yeast have been found in plasma [3]. The technique of 2D-electrophoresis can be used to differentiate the plasma into tens of proteins [4, 5], and, using MALDI and SELDI mass spectrometry, more than a thousand polypeptides were identified in these proteins [6, 7]. However, with such large number of proteins in the plasma, the whole variety of its “true proteins” can be grouped in the four main electrophoretic fractions defined by Tiselius [8] as albumins and α-, β-, and γ-globulins.
Similarly to humans, about one thousand various products of separate genes were found in the fish plasma proteome [9]. The studies of fish plasma proteome are interesting because these organisms belong to the lower vertebrates. The main protein fractions of plasma typical for all vertebrates appeared and formed in lower taxa in particular [10-13]. At the same time, it is interesting that the fish plasma proteome is very changeable. Dynamics are also a characteristic of the human plasma proteome: the contents of certain proteins exhibit large variability [14], which is especially true for serum albumin. Its concentration in plasma varies by more than 10 orders of magnitude [1]. In fish, as in poikilotherms, the dynamics of plasma proteome should be manifested not only in varying concentrations of certain proteins, but also in other features reflecting its presumable concordance with seasonal and consequent reproductive rhythms.
The main goal of the present work was to study the patterns of changes in the composition of the most variable protein fraction, the low-molecular-weight proteins, during the annual cycle of cyprinid fishes.
MATERIALS AND METHODS
Study objects. The following fishes of order Cypriniformes, family Cyprinidae were studied: 1) “wild species” – bream Abramis brama, roach Rutilus rutilus, blue bream Abramis ballerus, bleak Alburnus alburnus, sabrefish Pelecus cultratus, silver bream Blicca bjoerkna, crucian carp Carassius auratus (Volga river basin, Rybinsk reservoir); redfins of the Tribolodon genus – Pacific redfin T. brandtii and large-scaled redfin T. hakonensis (Vostok Bay, Sea of Japan; Razdolnaya river); 2) cultivated species – common carp Cyprinus carpio (reared in experimental ponds of the Institute for Biology of Inland Waters, Russian Academy of Sciences) and zebrafish Danio rerio (kept in aquaria).
Sampling of fish blood serum and plasma. Blood was sampled from caudal blood vessels immediately after fish were caught. To obtain serum, the blood was settled at 4°C, to obtain plasma. The blood was collected to test tubes containing 1% solution (w/v) of heparinoid. Precipitated erythrocytes were removed by centrifugation at room temperature for 10 min at 14,000g.
Determination of maturity stage of gonads. The stages of gonad maturity were determined following the routine common in fish biology and farming practice [15].
Electrophoretic methods. Low-molecular-weight fraction (LMW-fraction) was assessed by the number of proteins, their molecular weight (MW), electrophoretic mobility (m) of proteins, and their localization in 7.5% polyacrylamide gel on disc electrophoresis.
The pattern of organization (monomer/oligomer) of proteins was determined by position, number, and MW of proteins on nondenaturing 2D-electrophoresis within 5-40% concentrations of polyacrylamide gel (2D-PAGE) and on denaturing PAGE in 11% polyacrylamide gel with 8 M urea [16] as well as on 12.5% SDS-PAGE under reducing conditions [17]. Disc electrophoresis in polyacrylamide gel in the first direction was performed according to [18, 19] using a 3% concentrating (stacking) gel (pH 6.9) and 4.9% separating gel (pH 8.9). For the unidimensional disc electrophoresis, we used 3% concentrating (stacking) and 7.5% separating gel.
Following the electrophoresis under nondenaturing conditions and in polyacrylamide gel with urea, the gels were fixed in 10% trichloroacetic acid, washed, and stained with Coomassie R-250 (0.01% solution in ethanol–acetic acid–water, 10 : 1 : 30 v/v). Following the SDS-PAGE, the gels were fixed in 70% isopropyl alcohol and then stained with Coomassie R-250 (0.04% solution in isopropanol–ethanol–acetic acid–water, 2 : 1 : 1 : 6 v/v) according to protocol [17].
If in the polyacrylamide gel gradient and in polyacrylamide with urea the protein was present as one “spot”, it was considered a monomer; if there were several “spots” in the urea-containing polyacrylamide, the protein was considered as oligomer stabilized by hydrogen bonds. If the number of “spots” in the polyacrylamide with urea was less than in SDS-polyacrylamide under reducing conditions, we considered that the protein consists of several polypeptide chains linked by S–S bonds.
As MW markers, we used polymers of human serum albumin (HSA; 67, 134, 201, 268, 335 kDa) and of ovalbumin (OA; 45, 90, 135 kDa) and the PageRulerTM Prestained Protein Ladder Plus (11, 17, 28, 36, 55, 72, 95, 130, 250 kDa) (Fermentas, Lithuania). For densitometry and calculations of relative contents of proteins and their molecular weights, we used ONE-Dscan, v. 1.31 (Scananalytic Inc., USA) software.
MALDI mass spectrometry. All procedures for preparation of the samples of analyzed proteins for mass spectrometry followed a protocol described elsewhere [20]. The mass spectra (ms) of trypsin-digested proteins were obtained using a MALDI-TOF/TOF mass-spectrometer (UltrafleXtreme Bruker Daltonics, Germany) equipped with Nd laser in reflecto-mode. Monoisotopic [MH+] ions were measured in the 700-4500 m/z range with a tolerance of 50 ppm. Fragment ion spectra were obtained in Lift mode. The accuracy of fragment ion mass peak measurements was within 1 Da. To analyze mass spectra, FlexAnalysis 3.3 software (Bruker Daltonics, Germany) was used.
The proteins were identified using the MASCOT search software (“peptide fingerprint” option; www.matrixscience.com). The search was carried out in databases NCBI and/or EST vertebrates. Candidate proteins were considered as reliably identified when score > 83 (p < 0.05). For the search of candidate proteins in combined ms + (ms-ms) data, Biotools 3.0 (Bruker Daltonics) was used.
RESULTS
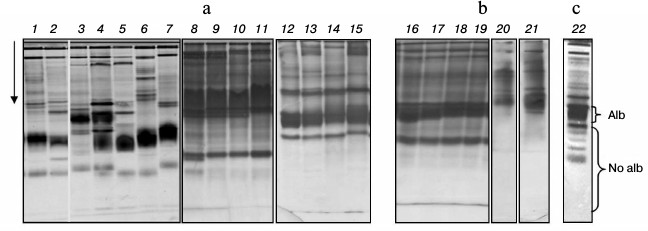
Composition of low-molecular-weight protein fraction in plasma of cyprinid fish on electrophoretic gel. Disc electrophoresis showed LMW-fraction of studied species plasma to be situated in the m 0.37-0.87 range. It is considered low-molecular-weight traditionally, although proteins within this fraction possess molecular masses of over 40-50 kDa. This fraction contains two subfractions in all species. One dominant protein making up to 50% and over of the whole protein of the fraction is found in the first. The second subfraction consists of several proteins, from two to 15 (Fig. 1, a and b).
Fig. 1. Plasma disc electrophoresis: a) bream (1, 2), blue bream (3, 4), bleak (5), sabrefish (6), silver bream (7), goldfish (8-11), redfin (12-15); b) cultured carp (16-19), zebrafish (20, 21); c) subfractions Alb and No alb of LMW-fraction of bream (22).
We have previously shown that subfractions differ in their ability to bind albumin-specific Evans Blue dye: the first subfraction bound the dye during electrophoresis, while the second did not [20, 21]. For this reason, we have designated the first subfraction as albumin-like proteins (Alb) and the second as non-albumin proteins (No alb) (Fig. 1c).
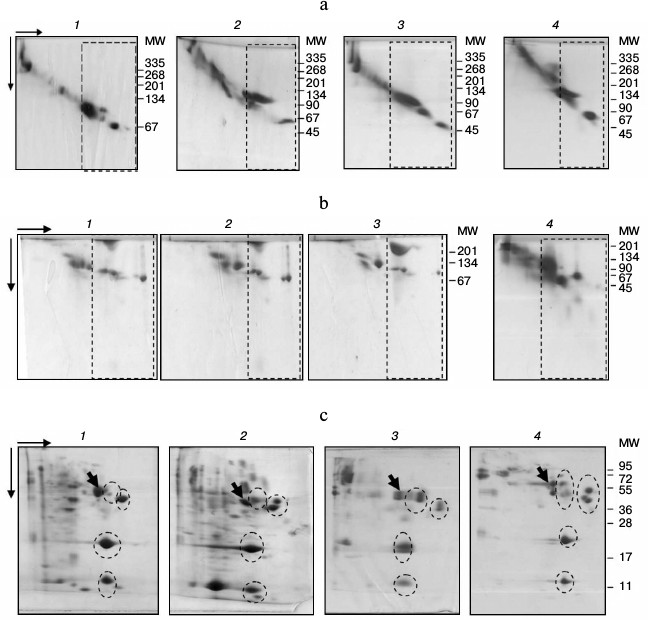
2D-PAGE showed the subfractions to be organized similarly in all studied species (Fig. 2). Thus, under nondenaturing conditions, the fraction contained one dominant protein with MW in the 100-120 kDa range in the Alb-subfraction and 2-15 proteins with MW ranging from 45 to 100 kDa within No alb-subfraction (Fig. 2a). The fraction structure under denaturing conditions was similar (Fig. 2, b and c). In SDS-PAGE, four groups of proteins with MW around 14, 25, 50, and 60 kDa were present within the fraction of all species studied (Fig. 2c). Therefore, protein composition of plasma LMW-fraction in different studied fish species using electrophoretic methods is characterized by significant similarity under both nondenaturing and denaturing conditions.
Fig. 2. 2D-PAGE of fish plasma: a) in polyacrylamide gel gradient: bream (1), roach (2), redfin (3), carp (4); b) in polyacrylamide with urea: blue bream (1), bream (2), roach (3), redfin (4); c) in SDS-polyacrylamide: roach (1), goldfish (2), redfin (3), carp (4). LMW-fractions are framed with rectangles (a, b), and groups of proteins with ovals (c). Arrows point to transferrins.
This fact allows utilizing results of protein identification in cultivated species with sequenced genome (carp, zebrafish) for future identification of proteins in wild species. Prior to that, we determine fraction boundaries on the plasma proteomic map, identifying globulins adjacent to the fraction with carp – a species with partly sequenced genome as an example.
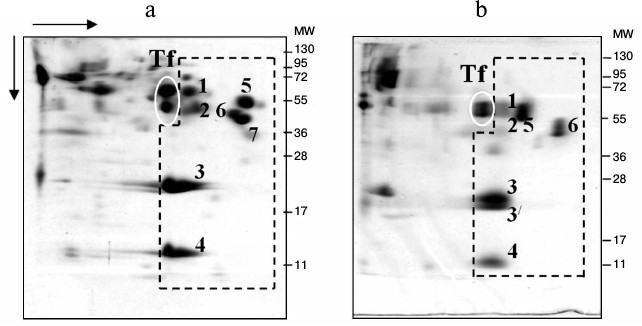
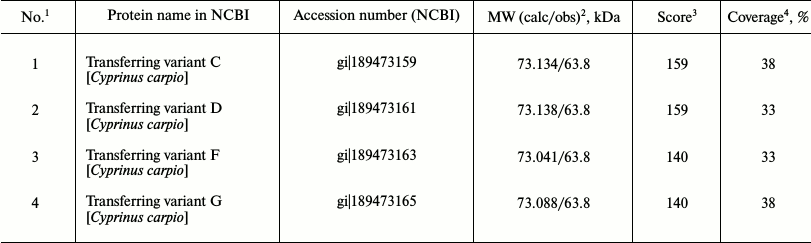
Identification of globulins adjacent to low-molecular-weight fraction proteins on the plasma proteomic map in carp and Trilobodon redfins. We previously assumed that proteins adjacent to the LMW-fraction of fish plasma on the 2D-electrophoregram from the globulin side are transferrins [22] because they were stained by Muller reagent [23]. To identify them precisely, we cut two gel fragments (containing proteins) from the track of Muller-positive proteins (Fig. 3a), carried out their trypsinolysis, and acquired ms of their proteomic map. The mass spectra of these proteins matched, evidencing their homology. All potential candidates were transferrins of carp exclusively (Table 1).
Fig. 3. SDS-PAGE of carp (a) and redfin plasma proteins (b). Proteins (1-7) of low-molecular-weight fraction are denoted by dotted line, and transferrins by ovals.
Table 1. Candidate proteins for carp plasma
globulin with MW ~ 63 kDa
1 Number of the candidate protein.
2 Calculated and experimental (observed) value of molecular
weight.
3 Probabilistic coefficient of significance.
4 Amino acid sequence overlap.
Then we attempted to identify possible transferrins in a wild species, redfin T. brandtii, following the same routine. Similarly to carp, redfin was found to contain homologous 62-kDa proteins in the zone adjacent to the LMW-fraction (Fig. 3b). We could not identify them using ms. But the search using ms-ms revealed the same candidate proteins as in the case with carp (Table 1) with the same Accession numbers and score values. Overlap area of amino acid sequences with candidates was naturally smaller than in carp, being 13, 13, 11, and 11%, correspondingly. Nevertheless, high score values (159, 159, 140, and 140, correspondingly) and affiliation of the candidate and the experimental fish species to one family suggest that the identification is quite reliable.
Therefore, the LMW-fraction bordering the globulins is adjacent to transferrins on the proteomic map of cyprinid fish plasma. Due to simple staining in the gel, these proteins (transferrins) can be used as a marker to determine localization of the LMW-fraction on the fish plasma proteomic map.
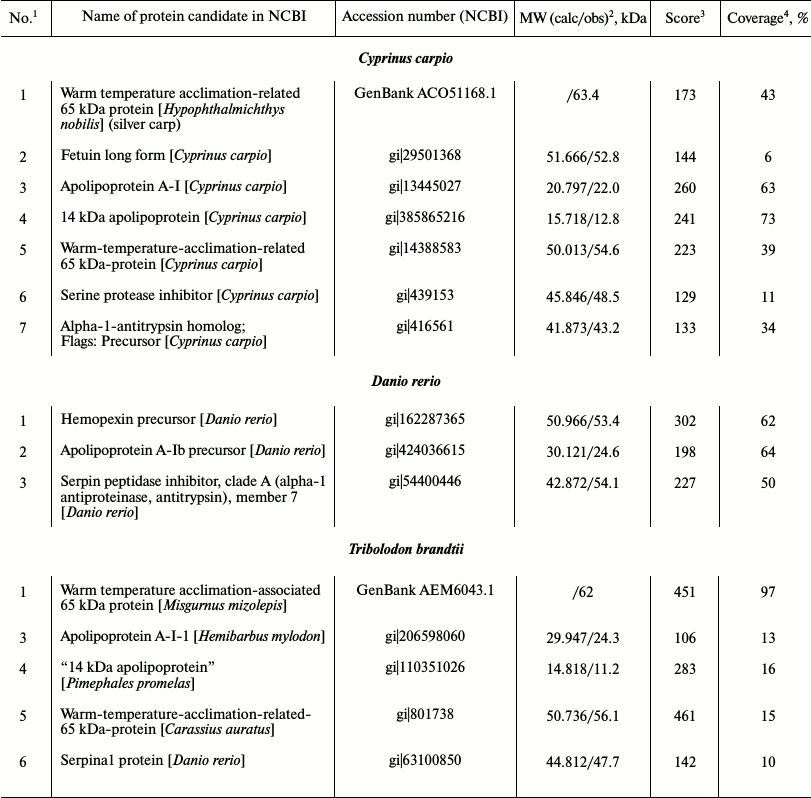
Identification of proteins from low-molecular-weight fraction of plasma using carp proteins as an example. Seven proteins from the fraction were taken for identification. Proteins 1, 3, 4, and 5 were identified using ms, proteins 2 and 6 using ms-ms. One candidate with the highest score value for each protein is presented in Table 2.
Table 2. Identification of carp, zebrafish,
and redfin proteins
Note: See notes 1-4 for Table 1.
Homologs of protein 1 (Fig. 3a) with experimental value of MW close to 64 kDa were only found in the unannotated database EST, so its calculated molecular weight is unclear. Candidate sequences for this protein had score > 83 and were represented by cDNA libraries constructed on the basis of mRNA populations of carp liver (CF662379). We were able to find a homolog of this particular protein, “warm temperature acclimation-related 65-kDa protein”, in silver carp by inserting amino acid sequence reconstructed on the basis of mRNA into Protein BLAST and searching among all vertebrate proteins. The protein contains in its structure hemopexin-like repeats, and it binds heme and transports it to the liver (GenBank ACO51168.1). A candidate for protein 2 (MW around 53 kDa) is fetuin (Fig. 3a). Similarly to albumin, it participates in the transport of metabolites in blood. On the other side, it has a cystatin-like domain in its structure characteristic for the cysteine proteinase inhibitors family [24]. Proteins 3 and 4 (Fig. 3a) are identified as nonhomologous A-I and “14 kDa” apolipoproteins.
We identified three proteins within the “non-albumin proteins” subfraction; they are designated as 5, 6, and 7 on Fig. 3a and in Table 2. Protein 5 possesses hemopexin features [25], while proteins 6 and 7 are inhibitors of serine proteinases.
The results have shown coincidence of candidates for proteins 1 and 5. In both cases, the same carp protein is being considered, probably “warm-temperature-acclimation-related-65 kDa”. However, cDNA libraries cover different regions of the protein in different candidates – N-terminal for protein 1 (GenBank ACO51168.1) and the whole amino acid sequence (1-439) for protein 5 (gi|14388583).
We obtained similar results during identification of Danio rerio proteins (Table 2).
Identification of low-molecular-weight fraction proteins in wild fish species. We were unable to identify LMW-fraction proteins of roach blood (gi|291530602, gi|339326474), and only one protein-candidate “unnamed protein product” [Tetraodon nigroviridis] with low score value of 59 (gi|47220181) was found for albumin-like protein of bream (gi|47220181). However, it was possible to identify proteins of redfin T. brandtii (Table 2) using ms-ms. The protein-candidates were identical to those of carp.
The search for protein 1 candidates (Fig. 3b) with MW around 62 kDa using ms was fruitless. Search using ms-ms revealed homologs in the EST database, and so the calculated MW of the protein was not determined. Candidate sequences with high significance values (score 245) are represented by cDNA libraries constructed based on roach mRNA populations (EG541453). Inserting the amino acid sequence reconstructed on the basis of mRNA into Protein BLAST and searching among all vertebrate proteins, we found a protein 1 homolog “warm-temperature-acclimation-associated 65-kDa protein” in mud loach Misgurnus mizolepis from the order Cypriniformes. Just like the carp’s corresponding protein candidate, it contains hemopexin-like repeats, binds heme, and transports it to liver [26]. Similar to carp’s corresponding protein, redfin’s proteins 1 and 5 contain hemopexin-like repeats, bind heme, and transport it to liver [27]. Therefore, the same protein of spawning redfin is present on the electrophoregram in different regions and has different motility (m) and MW. It should be noted, however, that at the end of the intensive feeding period, “warm-temperature-acclimation-associated 65-kDa protein” was found only in the lane of the dominant protein.
Just like in carp, redfin proteins 3 and 4 (Fig. 3b) are nonhomologous apolipoproteins, and protein 6 is an inhibitor of serine proteinases (Table 2).
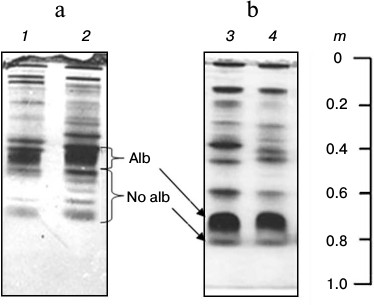
Main changes in composition of LMW-fraction plasma proteins and their alternation in fish yearly cycle. Analysis of m and MW values of fish plasma LMW-fraction proteins during a year allowed us to determine the main changes observed in the fraction. They are identified as two discontinuous types designated “basal” and “plastic”. The types are sharply different in heterogeneity, m, and MW values of proteins in the fraction (Fig. 4).
Fig. 4. “Basal” (a) and “plastic” (b) organization of bream plasma low-molecular-weight fraction (1-4) in 7.5% polyacrylamide gel. Alb and No alb, subfractions of plasma LMW-fraction.
The LMW-fraction of basal type is characteristic for: 1) maximum heterogeneity of “Non-albumin proteins” subfraction; 2) low values of electrophoretic motility m of the dominating protein from “Albumins” subfraction, and 3) low values of m of the most motile protein from “Non-albumin proteins” subfraction.
The LMW-fraction of plastic type is characteristic for: 1) minimal heterogeneity of “Non-albumin proteins” subfraction; 2) high values of electrophoretic motility m of dominating protein from “Albumins” subfraction, and 3) maximum values of m of the most motile protein from “Non-albumin proteins” subfraction (Fig. 4).
The mass of the non-denatured dominant protein of basal type was always higher (around 120 kDa) than in the plastic type protein (around 100 kDa).
Such structure of the LMW-fraction was typical for all studied fish species, while absolute values of m and the degree of fraction heterogeneity were species-specific. Thus, disc electrophoresis showed that the dominant protein of basal type had motility m 0.60-0.62 (bleak, sabrefish, and silver bream), 0.55-0.57 (bream), 0.48-0.54 (Pacific redfin, zebrafish, carp), 0.45-0.46 (blue bream), and 0.39-0.42 (goldfish). Subfraction “Non-albumin proteins” was represented by several (up to 10 or more) proteins with either one protein clearly seen on the electrophoregram and multiple minor ones (Pacific redfin) or several clearly seen (usually up to six) proteins (bream, goldfish) and complete absence of minor ones. In the LMW-fractions of plastic type, the dominant protein had higher values of m (mmax = 0.70) and non-albumin proteins were represented by a minimal number of proteins (or a single protein) with mmax = 0.83 (Fig. 4).
The basal type of LMW-fraction is characteristic for adult fish caught in summer-autumn and winter, while plastic type is characteristic for fish caught in spring (April, May) in the pre-spawning period. Gonads of fish with basal type of plasma LMW-fraction were at stage I-III of maturity, and plastic type were at stage IV.
Reorganization of plasma LMW-fractions during fish yearly cycle. We found both monomeric as well as oligomeric proteins among plasma LMW-fraction proteins. The latter were found when comparing 2D-electrophoregrams under nondenaturing and denaturing conditions (Fig. 2).
In all studied fish, the dominant protein in the polyacrylamide gel concentration gradient was usually represented by one “spot”, and under denaturing conditions there were 3-12 proteins. It is necessary to add that number of subunits within dominant protein in the “Albumins” subfraction was comparable in both PAGE with urea and SDS (Fig. 2). This is why we assumed that the dominant protein from the “Albumins” subfraction is a complex consisting of several proteins stabilized by hydrogen bonds. This assumption is further confirmed and illustrated by results of mass spectrometry of proteins from Pacific redfin T. brandtii caught in the pre-spawning period.
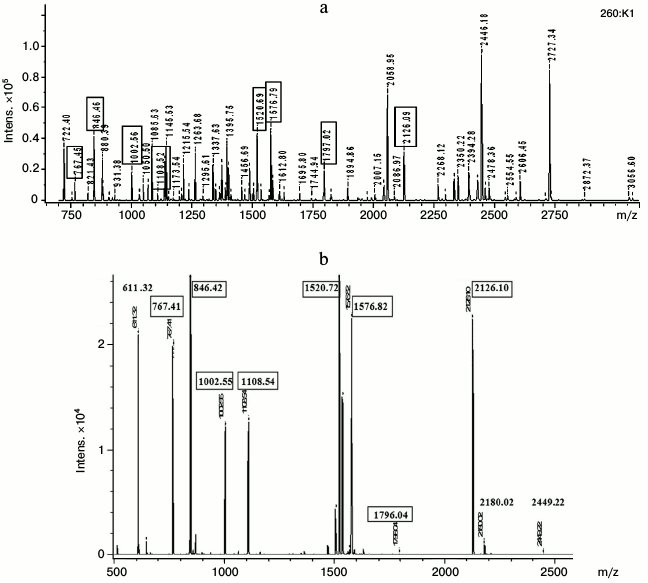
To perform mass spectrometry, we took: 1) several fragments of gel from the region of non-denatured dominant protein “Albumins”, and 2) 10 fragments of 2D-SDS-gel containing subunits from the lane of the dominant protein. The results showed coincidence of mass spectra of all samples from the region of the non-denatured dominant protein. These ms contained a large number of signals, suggesting the presence of several polypeptide chains in the samples (Fig. 5a).
Fig. 5. Mass spectra of non-denatured protein complex (a) and its component, 14-kDa apolipoprotein (b). Matching m/z values of peptides from the protein complex and from the apolipoprotein are denoted by boxes.
Of 10 subunits taken for mass spectrometry, only four had pronounced signals in mass spectra of non-denatured dominant protein, and one of them is presented in the figure (Fig. 5b).
The most intensive signals belong to subunits with MW around 23 kDa (subunits 3 and 3′ on Fig. 3a), 12 kDa (Fig. 5b), and 60 kDa, which (as shown above) are nonhomologous A-I and 14-kDa apolipoproteins and “warm temperature acclimation-associated 65-kDa protein”. The results indicate that the native dominant protein is organized as a complex consisting of oligomers of different composition but with approximately the same MW values, around 100 kDa.
It is hard to evaluate complex stoichiometry from the data of mass spectrometry, but the intensity of protein spots on the electrophoregram suggests apolipoproteins and “warm temperature acclimation-associated 65-kDa protein” as the main components of the complex. Different combinations of proteins and lipids bound with them giving a value of around 100 kDa in sum are possible.
Decrease in non-denatured protein mass from 120 to 100 kDa in Pacific redfin in the pre-spawning period is not accompanied by a change in its subunit composition. This fact indicates that the dominant protein (organized as a protein complex) changes its structural organization prior to spawning. The MW of the non-denatured dominant protein is around 120 kDa for most of the yearly cycle. This corresponds to the presence of the following elements in its composition: dimers of “warm temperature acclimation-associated 65-kDa protein” and oligomers of each of the apolipoproteins or complexes of apolipoproteins associated with one another or complexes of the listed proteins with lipids. The complex MW decreases to approximately 100 kDa in the pre-spawning period. Admittedly, this corresponds to the presence of an oligomer made of three monomers – “warm temperature acclimation-associated 65-kDa protein” and two apolipoproteins in the complex.
DISCUSSION
An important result of our work that helps to discover patterns of fish plasma LMW-fraction protein reorganization was finding a similar structure of a fraction consisting of two subfractions. Plasma LMW-fractions of other teleosts (families Percidae, Esocidae, and Clupeidae) have the same structure [28].
Characteristic groups of proteins with similar molecular weights were found in all studied species fraction composition. Using MALDI, they were found to belong to the same functional classes – apolipoproteins, proteinase inhibitors, and hemopexins. Data of other researchers who identified the same proteins within plasma proteome of cyprinids [29], salmonids [30], brown-marbled groupers [31], and freshwater eels [32] support our findings.
Satisfactory results on the identification of plasma LMW-fraction proteins in wild species (genus Tribolodon) were obtained by utilizing MALDI-TOF-TOF. Using MALDI-TOF and then searching for candidates in NCBI databases among proteins of all organisms did not yield any results. However, the search by fragmentation spectra revealed the same candidates for redfin proteins as in the case with corresponding proteins of carp Cyprinus carpio and zebrafish Danio rerio. Values of sequence coverage (10-97%) and score (142-461) suggest satisfactory identification of the redfin proteins. The range of these parameters was 11-73% and 129-260 in carp and 50-64% and 198-302 in zebrafish, correspondingly.
The main result of our work is finding seasonal rhythm of protein molecular mass and motility fluctuations as well as fraction heterogeneity. During most of the year – summer, autumn, and winter – hemopexins and apolipoproteins formed 120-kDa protein complexes stabilized by hydrogen bonds. They are probably constituted of associated protein oligomers: hemopexin dimers, tetramers of apolipoprotein A-I, and associates of 14-kDa apolipoprotein of higher order (up to 10 protein chains). Other combinations of proteins within the complex yielding the same seeming value of its mass are also probable. It is equally probable that complexes may contain significant amounts of lipids bound by proteins. In spring, complex mass decreased to 100 kDa. Due to the fact that its composition remained the same, we assumed that it consists of oligomers organized by hemopexin and apolipoprotein monomers.
If we try to determine what peculiarities of plasma proteins and microenvironments facilitate protein aggregation and considering the fact that practically all fish plasma proteins (including albumin-like ones the low-molecular-weight fraction) are glycoproteids [11, 33], we can explain fish protein affinity for association also by the process of glycosylation, which facilitates association [34]. Normally, mammal blood does not contain such protein complexes. However, it is known that glycosylation of serum albumin provokes its aggregation during pathologies (diabetes) [35].
In addition to the conformity of plasma LMW-fraction protein fluctuations with seasonal dynamics, we found a connection between protein fluctuations and fish reproductive biological rhythms. By matching the organization of plasma LMW-fraction with gonad maturity stages, we found that the basal type complex (120 kDa) observed in fish for most of the year was only present in fish at gonad maturity stages I-III, while plastic type complex (100 kDa) observed in spring prior to spawning was present in fish at gonad maturity stage IV. Therefore, prior to spawning the dominant protein of the LMW-fraction organized in oligomers associated into the complex has been reorganized. During this reorganization, we registered the appearance of protein with hemopexin features not only in the zone of motility of the complex, but also in the zone of free protein motility. The mass of the latter was lower than that of the protein contained within the complex. We relate it to possible plastic function of the protein and its utilization (by means of proteolytic degradation) as a source of amino acids during biosynthetic processes linked with gonad maturation.
Dynamic reformation of plasma LMW-fraction takes place in spring, in the period of activation of metabolic processes, and are probably controlled by sex hormones. Experiments with rainbow trout hepatocytes showed that albumin-synthesizing activity evaluated by the level of mRNA transcribed from the albumin gene was clearly dependent on estradiol dose and time of its action [36]. Plasma proteins are among the most likely suppliers of amino acids for biosynthetic processes [37]. However, in Tribolodon redfins, in both immature fry and adult spawners, reorganization of plasma low-molecular-weight fractions in spring follows the same (plastic) type. This indicates that reformations of LMW-fraction in spring reflect the intensity of metabolic processes rather than reproductive biorhythms.
Reorganization of the LMW-fraction taking place according to the described scenario was also observed during adaptation of fish to elevated salinity. Thus, single cases of immature bream and roach manifest the same reorganization of fraction as seen in adult fish prior to spawning at salinity of 20‰. In the majority of experiments, reorganization led to decrease in complex concentration in plasma and increase in low-molecular-weight protein concentration in the muscle fluid [38], so it is safe to assume that single cases of fraction reorganization in plasma reflect the initial stage of transcapillary rearrangement of plasma protein in conditions of high salinity. Therefore, reorganization of plasma LMW-fraction during plastic and water metabolism adaptations take place according to a single algorithm.
In conclusion, we would like to note that our research was complicated by the absence of sequences of wild fish species proteins in question in the DB Proteins NCBI, which made it impossible to identify bream and roach proteins. Nevertheless, the tasks of studying wild fish proteins are being solved thanks to the presence of cyprinid species popular in aquaculture and commonly used by specialists in genome sequencing. Presently, proteins of cultivated fish species including those from plasma are present in DB Proteins: 4344 for carp, 3496 for goldfish, and 77,560 protein sequences for zebrafish. The work on the accumulation of data on genomes and proteins of wild and cultivated fish species continues [39, 40].
Authors are grateful to V. A. Brykov from the Institute of Marine Biology, Far-Eastern Branch of the Russian Academy of Sciences, for help with material collection.
This study was supported by the Russian Foundation for Basic Research (project No. 13-04-00427-a). The MALDI MS facility became available to us in the framework of the Moscow State University Development Program (PNG 5.13).
REFERENCES
1.Anderson, N. L., and Anderson, N. G. (2002) The
human plasma proteome: history, character, and diagnostic prospects,
Mol. Cell. Proteom., 1, 845-867.
2.Liotta, L. A., and Petricoin, E. F. (2006) Serum
peptidome for cancer detection: spinning biologic trash into diagnostic
gold, J. Clin. Invest., 116, 26-30.
3.Anderson, N. L., Polanski, M., Pieper, R.,
Gatlin, T., Tirumalai, R. S., Conrads, T. P., Veenstra, T. D., Adkins,
J. N., Pounds, J. G., Fagan, R., and Lobley, A. (2004) The human plasma
proteome: a nonredundant list developed by combination of four separate
sources, Mol. Cell. Proteom., 3, 311-326.
4.Bouwman, F. G., Roos, B., Rubio-Aliaga, I.,
Crosley, L. K., Duthie, S. J., Mayer, C., Horgan, G., Polley, A. C.,
Heim, C., Coort, S., Evelo, C. T., Mulholland, F., Johnson, I. T.,
Elliott, R. M., Daniel, H., and Mariman, E. (2011) 2D-electrophoresis
and multiplex immunoassay proteomic analysis of different body fluids
and cellular components reveal known and novel markers for extended
fasting, BMC Med. Genom., 4, 1-12.
5.Babaei, F., Ramalingam, R., Tavendale, A., Liang,
Y., Yan, L. S., Ajuh, P., Cheng, S. H., and Lam, Y. W. (2013) Novel
blood collection method allows plasma proteome analysis from single
zebrafish, J. Proteome Res., 12, 1580-1590.
6.Zhang, Z., Bast, R. C., Yu, Y., Li, J., Sokoll, L.
J., Rai, A. J., Rosenzweig, J. M., Cameron, B., Wang, Y. Y., Meng,
X. Y., Berchuck, A., Van Haaften-Day, C., Hacker, N. F., de Bruijn, H.
W., van der Zee, A. G., Jacobs, I. J., Fung, E. T., and Chan, D. W.
(2004) Three biomarkers identified from serum proteomic analysis for
the detection of early stage ovarian cancer, Cancer Res.,
64, 5882-5890.
7.Luczak, M., Formanowicz, D., Pawliczak, E.,
Wanic-Kossowska, M., Wykretowicz, A., and Figlerowicz, M. (2011)
Chronic kidney disease-related atherosclerosis – proteomic
studies of blood plasma, Proteome Sci., 9, 1-12.
8.Tiselius, A. (1937) Electrophoresis of serum
globulin: electrophoretic analysis of normal and immune sera,
Biochem. J., 31, 1464-1472.
9.Lucitt, M. B., Price, T. S., Pizarro, A., Wu, W.,
Yocum, A. K., Seiler, C., Pack, M. A., Blair, I. A., FitzGerald, G. A.,
and Grosser, T. (2008) Analysis of the zebrafish proteome during
embryonic development, Mol. Cell Proteom., 7,
981-994.
10.Danis, M. H., Filosa, M. F., and Youson, J. H.
(2000) An albumin-like protein in the serum of non-parasitic brook
lamprey (Lampetra appendix) is restricted to pre-adult phases of
the life cycle in contrast to the parasitic species Petromyzon
marinus, Comp. Biochem. Physiol. B Biochem. Mol. Biol.,
127, 251-260.
11.Metcalf, V. J., George, P. M., and Brennan, S. O.
(2007) Lungfish albumin is more similar to tetrapod than to teleost
albumins: purification and characterization of albumin from the
Australian lungfish Neoceratodus forsteri, Comp. Biochem.
Physiol. B Biochem. Mol. Biol., 147, 428-437.
12.Fanali, G., Ascenzi, P., Bernardi, G., and
Fasano, M. J. (2012) Sequence analysis of serum albumins reveals
the molecular evolution of ligand recognition properties, Biomol.
Struct. Dyn., 29, 691-701.
13.Xue, Z., Pang, Y., Liu, X., Zheng, Z., Xiao, R.,
Jin, M., Han, Y., Su, P., Lv, L., Wang, J., and Li, Q. (2013) First
evidence of protein G-binding protein in the most primitive vertebrate:
serum lectin from lamprey (Lampetra japonica), Dev. Comp.
Immunol., 41, 618-630.
14.Shevchenko, V. E., Kovalev, S. V., Yurchenko, V.
A., Matveev, V. B., and Zaridze, D. G. (2011) Human blood plasma
proteome mapping in norm and in renal light cell carcinoma,
Onkourologiya, 3, 65-69.
15.Sakun, O. F., and Butskaya, N. A. (1968)
Determination of Maturity Stages and Study of Fish Sex Products
[in Russian], PINRO, Murmansk, p. 46.
16.Creighton, T. E. (1979) Electrophoretic analysis
of the unfolding of proteins by urea, J. Mol. Biol., 129,
235-264.
17.Laemmli, U. K. (1970) Cleavage of structural
proteins during the assembly of the head of bacteriophage,
Nature, 227, 680-685.
18.Ornstein, L. (1964) Disk-electrophoresis. I.
Background and theory, Ann. N. Y. Acad. Sci., 121,
321-349.
19.Davis, B. J. (1964) Disk-electrophoresis. II.
Method and application to human serum proteins, Ann. N. Y. Acad.
Sci., 121, 404-427.
20.Andreeva, A. M. (2013) Identification of some
proteins of blood and tissue liquid in fishes with undecoded genome,
Zh. Evol. Biokhim. Fiziol., 49, 394-402.
21.Andreeva, A. M. (2011) Mechanisms of the
plurality of Scorpaena porcus L. serum albumin, Open J.
Marine Sci., 1, 31-35.
22.Andreeva, A. M., Serebryakova, M. V., Lamash, N.
E., Fedorov, R. A., and Ryabtseva, I. P. (2014) Organization of
low-molecular-weight fraction of plasma proteins in Far-Eastern redfins
of Tribolodon genus and other Cyprinidae species, Biol.
Morya, in press.
23.Palmour, R. M., and Sutton, H. E. (1971)
Vertebrate transferrins molecular-weight, clinical composition and iron
binding studies, Biochemistry, 10, 4026-4032.
24.Tsai, P. L., Chen, C. H., Huang, C. J., Chou, C.
M., and Chang, G. D. (2004) Purification and cloning of an endogenous
protein inhibitor of carp nephrosin, an astacin metalloproteinase,
J. Biol. Chem., 279, 11146-11155.
25.Kinoshita, S., Itoi, S., and Watabe, S. (2001)
cDNA cloning and characterization of the
warm-temperature-acclimation-associated protein Wap65 from carp,
Cyprinus carpio, Fish Physiol. Biochem., 24,
125-134.
26.Cho, Y. S., Kim, B. S., Kim, D. S., and Nam, Y.
K. (2012) Modulation of warm-temperature-acclimation-associated 65
kDa-protein genes (Wap65-1 and Wap65-2) in mud loach (Misgurnus
mizolepis, Cypriniformes) liver in response to different
stimulatory treatments, Fish Shellfish Immunol., 32,
662-669.
27.Kikuchi, K., Yamashita, M., Watabe, S., and Aida,
K. (1995) The warm temperature acclimation-related 65-kDa protein,
Wap65, in goldfish and its gene expression, J. Biol. Chem.,
270, 17087-17092.
28.Andreeva, A. M. (2012) Structural and
Functional Organization of Fish Blood Proteins, Nova Science
Publisher, N. Y., p. 188.
29.Dietrich, M. A., Arnold, G. J., Nynca, J.,
Frohlich, T., Otte, K., and Ciereszko, A. (2014) Characterization of
carp seminal plasma proteome in relation to blood plasma, J.
Proteom., 98, 218-232.
30.Braceland, M., Bickerdike, R., Tinsley, J.,
Cockerill, D., Mcloughlin, M. F., Graham, D. A., Burchmore, R. J.,
Weir, W., Wallace, C., and Eckersall, P. D. (2013) The serum proteome
of Atlantic salmon, Salmo salar, during pancreas disease (PD)
following infection with salmonid alphavirus subtype 3 (SAV3), J.
Proteom., 94, 423-436.
31.Low, C. F., Shamsudin, M. N., Chee, H. Y.,
Aliyu-Paiko, M., and Idrus, E. S. (2014) Putative apolipoprotein A-I,
natural killer cell enhancement factor, and lysozyme γ are
involved in the early immune response of brown-marbled grouper,
Epinephelus fuscoguttatus, Forskal, to Vibrio
alginolyticus, J. Fish Dis., 37, 693-701.
32.Metcalf, V. J., Brennan, S. O., Chambers, G., and
George, P. M. (1999) High density lipoprotein (HDL), and not
albumin, is the major palmitate binding protein in New Zealand
long-finned (Anguilla dieffenbachii) and short-finned eel
(Anguilla australis schmidtii) plasma, Biochim. Biophys.
Acta, 1429, 467-475.
33.Metcalf, V. J., Brennan, S. O., Chambers, G. K.,
and George, P. M. (1998) The albumin of the brown trout (Salmo
trutta) is a glycoprotein, Biochim. Biophys. Acta,
1386, 90-96.
34.Aleksandrov, V. Ya. (1985) Cell Reactivity and
Proteins [in Russian], Nauka, Leningrad.
35.Mohd. Wajid Ali Khan, Zafar Rasheed, Wahid Ali
Khan, and Rashid Ali (2007) Biochemical, biophysical, and thermodynamic
analysis of in vitro glycated human serum albumin,
Biochemistry (Moscow), 72, 146-152.
36.Flouriot, G., Ducouret, B., Byrnes, L., and
Valotaire, Y. (1998) Transcriptional regulation of expression of the
rainbow trout albumin gene by estrogen, J. Mol. Endocrinol.,
20, 355-362.
37.Novikov, G. G. (2000) Growth and Development
in Early Ontogenesis of Teleost Fishes [in Russian], Editorial
URSS, Moscow.
38.Andreeva, A. M. (2010) Role of structural
organization of blood plasma proteins in stabilization of aqueous
metabolism in teleost fishes (Teleostei), Voprosy Ikhtiol.,
50, 570-576.
39.Papakostas, S., Vasemägi, A., Himberg, M.,
and Primmer, C. R. (2014) Proteome variance differences within
populations of European whitefish (Coregonus lavaretus)
originating from contrasting salinity environments, J. Proteom.,
105, 144-150.
40.Armengaud, J., Trapp, J., Pible, O., Geffard, O.,
Chaumot, A., and Hartmann, E. M. (2014) Non-model organisms, a
species endangered by proteogenomics, J. Proteom., 105,
5-18.