Y-Box Binding Protein 1 (YB-1) Promotes Detection of DNA Bulky Lesions by XPC-HR23B Factor
E. E. Fomina1, P. E. Pestryakov1, E. A. Maltseva1, I. O. Petruseva1, D. A. Kretov2, L. P. Ovchinnikov2, and O. I. Lavrik1,3,4*
1Institute of Chemical Biology and Fundamental Medicine, Siberian Division of the Russian Academy of Sciences, 630090 Novosibirsk, Russia; fax: +7 (383) 363-5153; E-mail: lavrik@niboch.nsc.ru2Institute for Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia
3Novosibirsk State University, 630090 Novosibirsk, Russia
4Altai State University, 656049 Barnaul, Russia
* To whom correspondence should be addressed.
Received August 27, 2014; Revision received October 10, 2014
The nucleotide excision repair system (NER) is one of the main mechanisms protecting cellular DNA from lesions caused by such significant environmental factors as UV radiation, the influence of polycyclic aromatic hydrocarbons, and medical treatment by several antitumor drugs, e.g. cisplatin. One of the major NER components is XPC-HR23B, the key factor during the damage recognition step of repair. Binding of XPC-HR23B to DNA that contains different bulky lesions impairing the structure of DNA is the basis for the wide substrate specificity of this DNA repair pathway. The multifunctional protein YB-1 among other protein factors has high affinity towards damaged DNA. Involvement of YB-1 in the cellular response to genotoxic stress and its ability to interact with damaged DNA harboring lesions of various origins pinpoint its putative involvement as a modulatory factor in DNA damage recognition and verification steps of NER. In the present work, we assayed functional interactions of protein factors XPC-HR23B and YB-1 upon binding to DNA structures mimicking damaged DNA containing single bulky lesions, as substrates of NER, and bulky lesions combined with abasic sites as an example of clustered lesions. The results indicate that YB-1 and XPC-HR23B stimulate each other in binding to DNA containing a bulky or clustered lesion, which suggests the involvement of YB-1 in the regulation of DNA repair by the NER mechanism.
KEY WORDS: nucleotide excision repair, bulky DNA lesions, clustered DNA lesions, DNA–protein complexes, Y-box binding protein 1 (YB-1), XPC-HR23BDOI: 10.1134/S000629791502008X
Abbreviations: AP site, apurinic/apyrimidinic site; BER, base excision repair; HR23B, human protein homologous to yeast repair factor Rad23; NER, nucleotide excision repair; XPC, Xeroderma pigmentosum complementation group C factor; YB-1, Y-box-binding protein 1.
The DNA molecules in living human cells are constantly exposed to
exogenous and endogenous factors causing damage. Base excision repair
(BER) and nucleotide excision repair (NER) belong to the major systems
that effectively repair DNA damage thus preventing the emergence of
mutations [1, 2]. The main task
of BER is to repair damage arising in the course of redox reactions
involving heterocyclic bases of the DNA molecule, such as spontaneous
depurination, alkylation, oxidation, or reduction of heterocyclic
bases. The bulky lesions arising under the action of chemical
carcinogens, UV irradiation, and even certain drugs are substrates of
the second system, NER. An interesting feature of the NER system,
namely, its broadest substrate specificity, consists in the ability to
repair damage of the various chemical nature. This property is related
to a principle of damage detection, the key factor of which is the
protein complex XPC-HR23B that (together with additional protein
factors XPA and RPA) recognizes local disturbance of the
double-stranded DNA near the damage [3, 4]. Experiments on cells of human fibroblasts confirm
that XPC-HR23B is a major component of the detection-verification NER
ensemble initiating the assembly of a preincision complex [4]. Despite the key role of XPC-HR23B in the NER
system, it has previously been shown to physically interact with DNA
damages, eliminated by the base excision repair (BER), to which
apurinic/apyrimidinic sites (AP sites) should belong [5]. Such observations might warrant inclusion of the
XPC-HR23B factor into the list of proteins that mediate interaction of
recognition components of NER and BER. Another protein involved in
direct interaction with damaged DNA is the multifunctional
Y-box-binding protein YB-1 [6]. Under normal
conditions, YB-1 is localized predominantly in the cytoplasm, but under
conditions of genotoxic stress caused by a wide variety of factors such
as UV irradiation or treatment with mitomycin C, cisplatin, large
amounts of YB-1 transfer into the nucleus [7-9]. Furthermore, YB-1 has a higher affinity to damaged
DNA containing AP sites or other distortions of the double-stranded DNA
and single-stranded DNA rather than to a double-stranded one [10-12]. Finally, it is known
that YB-1 physically and functionally interacts with key proteins and
enzymes of BER such as APE1, DNA polymerase β, and
DNA glycosylases NEIL2 and NTH1 [7, 13-16]. Based on YB-1
intracellular localization and the network of its functional
interactions, YB-1 can be considered as a modulating component of the
repair complex, providing a functional link of the BER system with
other pathways of DNA integrity maintenance and repair. The available
literature indicate that YB-1 is involved in the repair of DNA bulky
lesions such as cyclobutane pyrimidine dimers, cisplatin adducts, and
products of interaction of DNA with mitomycin C [8, 17]. Due to such wide
specificity of YB-1 and XPC-HR23B towards the damaged DNA spectrum,
these proteins can be regarded as participants of repair of DNA cluster
damages consisting of individual lesions of different nature and
structure. In this work, we analyzed the possibility of interaction of
YB-1 and XPC-HR23B with DNA structures containing both damages such as
a BER substrate/intermediate – AP site – and a fluorescein
residue modeling a DNA bulky lesion.
MATERIALS AND METHODS
Proteins and oligonucleotides. Recombinant YB-1 and XPC-HR23B were isolated and purified according to published protocols [18, 19]. Recombinant APE1 and uracil-DNA glycosylase from E. coli (Ung) were generous gifts from Dr. S. N. Khodyreva (Institute of Chemical Biology and Fundamental Medicine, Siberian Division of the Russian Academy of Sciences, Novosibirsk). The T4 phage polynucleotide kinase was from Biosan (Russia). The sequences of the oligonucleotides used in this work – ODN1, ODN2 (Bioset, Russia), and ODN3 (a generous gift from Dr. V. N. Silnikov, Institute of Chemical Biology and Fundamental Medicine) are given in Table 1.
Table 1. Designations and sequences of
oligonucleotides

Note: U, 2′-deoxyuridine residue; F,
fluorescein-5(6)-carboxyamidocaproyl-[5-(3-aminoallyl)-2′-deoxyuridine]
residue.
To visualize DNA in gel retardation assay experiments, the ODN1 chain was 5′-radioactively labeled using T4 phage polynucleotide kinase and [γ-32P]ATP (Institute of Chemical Biology and Fundamental Medicine). The unreacted [γ-32P]ATP was removed from the mixture by gel filtration using a MicroSpin G-25 column (GE Healthcare Life Sciences, Sweden) according to the manufacturer’s protocol. Oligonucleotide duplexes were prepared by hybridization of ODN1 oligonucleotide with a complementary ODN2 or ODN3 chain in molar ratio 1 : 1.2. AP sites in DNA substrates were prepared in situ immediately before the reaction by incubating uracil-containing DNA substrates with Ung (0.15 U/pmol DNA) for 20 min at 37°C (symbolic designations of the resulting duplexes and their schematic images are given in Table 2).
Table 2. Designations and structure of
double-stranded oligonucleotides
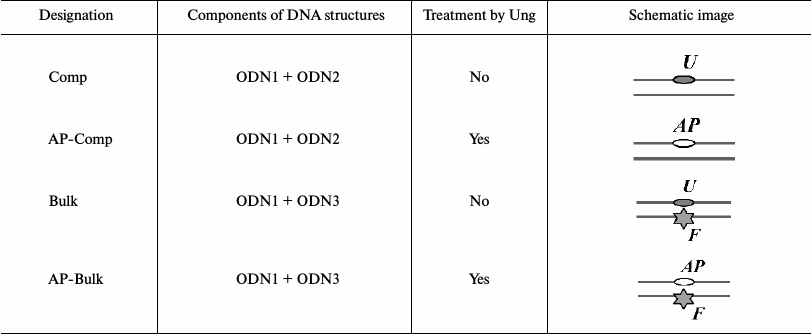
Note: U, 2′-deoxyuridine residue; F,
fluorescein-5(6)-carboxyamidocaproyl-[5-(3-aminoallyl)-2′-deoxyuridine]
residue; AP, AP site.
In fluorescence titration experiments, a fluorescein residue in DNA structures (Bulk, AP-Bulk) (wavelengths of excitation and fluorophore emission are 495 and 520 nm, respectively) was used as the reporter group. Anisotropy and fluorescence intensity were measured in plates on a POLARstar Optima apparatus (BMG Labtech, Germany). The values obtained for each reaction mixture at one scanning step corresponded to the calculated mean value from 50 measurements. To improve the reliability of the values obtained at one scanning step, the kinetic scan mode was used by 10 times scanning of the entire plate containing a series of reaction samples with subsequent averaging of data for each experimental sample in the series.
Determination of active protein concentration in preparations of YB-1 and XPC-HR23B. A protein able to bind DNA is hereafter termed as “active protein”, and “total protein concentration” corresponds to protein concentration in the preparation determined by the Bradford method. To determine the concentration of active protein in YB-1 and XPC-HR23B preparations, a titration of DNA by the analyzed protein preparation under stoichiometric binding conditions (i.e. putative concentration of the active protein and DNA concentration were at least an order of magnitude higher than the putative dissociation constant of the protein–DNA complex) was conducted according to a procedure described earlier [20]. In the case of YB-1, reaction mixtures (150 µl) contained standard buffer components (buffer RB: 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 0.6 g/liter BSA), the analyzed protein at variable concentration, and fluorescently labeled Bulk DNA at 197 nM concentration. Incubation (10 min) and measurements of fluorescence intensity and anisotropy of reaction mixtures were carried out at 25°C. The XPC-HR23B preparation was analyzed similarly except that the Bulk DNA duplex concentration was 10–8 M.
Protein binding to the fluorescein-containing DNA was monitored by an increase in the fluorescence anisotropy in a series of reaction mixtures containing a fixed amount of DNA and protein at various total concentration. The fraction of DNA complexed with the active protein, Fb, for the individual reaction mixture was calculated by formula (1) with a correction for fluorophore quenching during binding [21]:
Fb = (A – Af)/((Qb/Qf)(Ab – A) + A – Af), (1)
where Fb is fraction of DNA complexed with active protein; A is observed fluorescence anisotropy; Af and Ab are fluorescence anisotropy of free DNA and DNA totally bound to protein, respectively; Qf and Qb are specific fluorescence intensities of free DNA and DNA totally bound to protein, respectively. Measurement of the Af, Qf, Ab, and Qb parameters for completely free and totally bound DNA was determined in a series of five independent experiments and averaged.
The decrease in fluorescence intensity upon binding of XPC-HR23B with DNA appeared to be less than 10%, which allows assuming Qf = Qb. Thus, for calculations in this case, formula (1) becomes:
Fb = (A – Af)/(Ab – Af). (2)
Under conditions of stoichiometric titration, the dependence of Fb on the analyzed protein total concentration in the mixture is linear, reaching a plateau (at Fb = 1) at the point of “equivalence” corresponding to the total saturation of DNA present in the sample by bound protein and defined as the point of intersection of tangents to the linear sections of the plot. Equimolar content of DNA and the active protein in the mixture at the point of “equivalence” allows determination of the concentration of active protein in the preparation. In further experiments studying YB-1 and XPC-HR23B interaction with different DNA structures, the term “protein concentration” means the concentration of the active protein in YB-1 and XPC-HR23B preparations determined by this procedure.
Determination of parameters of YB-1 and XPC-HR23B interaction with fluorescent DNA. To determine YB-1 and XPC-HR23B binding constants with fluorescently labeled DNA, a DNA titration by the analyzed protein preparation under equilibrium binding conditions (i.e. concentration of protein and DNA was chosen to be less than the putative dissociation constant of the protein–DNA complex) was carried out according to the procedure described earlier [20]. The reaction mixtures (150 µl) contained buffer RB, analyzed protein (from 60 pM to 375 nM of YB-1, from 20 pM to 125 nM of XPC-HR23B), and fluorescently labeled Bulk or AP-Bulk DNA at concentration of 1.5 or 0.5 nM (in experiments with YB-1 or XPC-HR23B, respectively). Incubation (10 min) and measurements of fluorescence intensity and anisotropy of reaction mixtures were carried out at 25°C.
Protein binding of the fluorescein-containing DNA was monitored by an increase in the fluorescence anisotropy following the increase in protein concentration in the mixture. For each experimental point, the fraction of DNA complexed with protein (Fb) and the equilibrium concentration of protein–DNA complex ([DNA–protein] = Fb × B0, where B0 is total DNA concentration in the mixture) were calculated by formulas (1) or (2). Binding isotherms were analyzed within the framework of a model assuming the existence of a single DNA–protein binding site [21]:
[DNA–protein] = 1/2(A0 + B0 + Kd + ((A0 + B0 + Kd)2 – 4A0B0)1/2), (3)
where A0 and B0 are total concentrations of the active protein and DNA, and Kd is dissociation constant of DNA–protein complex. Approximation of the titration data with the equation of the theoretical curve (formula (3)) with Kd determination was performed by nonlinear regression using the Origin Pro 8.0 program (OriginLab, USA).
Study of YB-1 and XPC-HR23B complexing with DNA by gel retardation assay. Reaction mixtures (5 µl) containing 25 mM sodium phosphate, pH 7.5, 100 mM NaCl, 2.5 mM MgCl2, 0.5 mM EDTA, 0.1 g/liter BSA, 8 nM radioactively labeled DNA, and indicated concentrations of analyzed proteins were incubated at 37°C for 5 min and then subjected to electrophoresis in nondenaturing conditions at voltage of 10 V/cm in 7% polyacrylamide gel equilibrated with 50 mM Tris-borate buffer (50 mM Tris, 50 mM H3BO3, 1 mM EDTA, pH 8.3). The position of radioactively labeled DNA and protein–DNA complexes was determined by autoradiography using FX phosphorimager (BioRad, USA).
RESULTS AND DISCUSSION
XPC-HR23B factor is a key protein of DNA damage recognition of the NER pathway. It is believed that XPC-HR23B in most cases is responsible for direct search and interaction with DNA carrying local defects in the structure of the double-stranded DNA usually induced by bulky DNA lesions. Among proteins interacting with the NER specific substrates, other components of the system should be noted, e.g. the factor DDB1-DDB2, specifically interacting with DNA damaged by UV irradiation, and proteins of the damage verification stage, XPA and RPA, which were considered for a long time as candidates for the role of direct DNA damage sensors [22]. However, it is known that some other proteins that traditionally do not belong to the repair system are capable of high-affinity binding to damaged DNA. One of the most interesting representatives of this group is the multifunctional protein YB-1 [6]. Increased affinity of the protein to DNA treated with cisplatin and UV irradiation, noted in experiments in vitro [7, 10, 12], as well as participation of YB-1 in cell response to treatment with these damaging agents in vivo [7, 23, 24], attract attention to YB-1 as an additional factor modulating NER pathway activity at the stage of damage detection. In this work, the functional interaction of YB-1 with XPC-HR23B was investigated using DNA substrates bearing a nucleotide residue modified by fluorescein. Due to such structure, oligonucleotides can be used as model substrate for the NER system because the uracil residue in DNA strand modified by introducing the fluorescein group is recognized as a bulky lesion [25, 26]. Additionally, we used the DNA structure carrying a single AP site or AP site in combination with bulky lesion located in the opposite strand. In the latter case, two lesions (of different chemical nature) were in the oligonucleotide structure, whose repair in the cell usually occurs by two different systems – NER and BER.
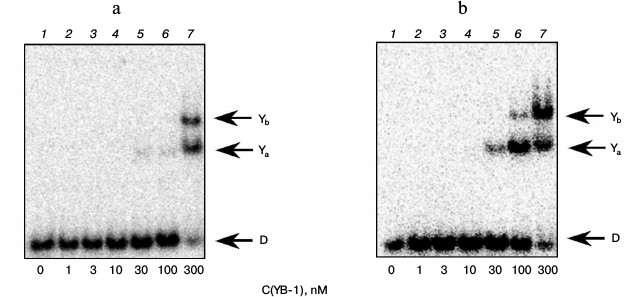
The gel retardation experiments showed that under the conditions used two types of complexes of YB-1 with DNA structures that did not contain bulky lesion are formed (Fig. 1, bands Ya and Yb). These data can be interpreted to mean that the complex with a higher electrophoretic mobility corresponds to the binding of one molecule of YB-1 with DNA, and a second complex to the binding of two protein molecules. The results indicate that YB-1 has a higher affinity for DNA containing an AP site (Fig. 1b) compared with the intact substrate (Fig. 1a), which is consistent with the literature indicating the interaction of YB-1 with DNA containing an AP site [11]. Moreover, as we showed earlier, interaction of the protein molecule amino group and the AP site aldehyde form leads to the formation of a Schiff base upon the formation of such complex [5]. Apparently, additional stabilization of the formed complex of YB-1 with AP site-containing DNA compared to intact substrate can be explained by the formation of a covalent bond in this case [5].
Fig. 1. Formation of complexes of YB-1 protein with DNA structures Comp (a) and AP-Comp (b). Autoradiographs of the reaction products separated in 7% nondenaturing polyacrylamide gel are presented. Ya and Yb, YB-1 complexes with DNA; D, free DNA. Reaction mixtures (5 µl) contained the standard components of the reaction buffer, 8 nM radiolabeled DNA Comp (a) or AP-Comp (b), and YB-1 at the concentrations indicated below the figure.
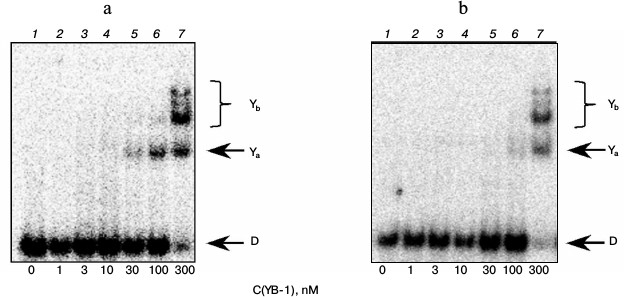
In experiments with model DNA carrying a bulky or cluster lesion, we also noted a rather high affinity of YB-1 to such structures (Fig. 2). Furthermore, we observed the appearance of additional protein–DNA complexes with lower electrophoretic mobility (compare Figs. 1 and 2, bands Yb). Under the experimental conditions, we observed fluorescence quenching of a residue simulating a DNA bulky lesion when DNA interacted with the protein, suggesting a direct interaction of the YB-1 protein globule with the fluorescein. Apparently, additional complexes are different in the conformation of the protein or DNA from complexes detectable in oligonucleotides containing no bulky lesion, and therefore they might have different electrophoretic mobilities. On the other hand, the observed phenomenon might be due to the difference in the stoichiometry of the observed complexes at the expense of binding of the third and fourth protein molecules with a model DNA. In this case, as evidenced by the results, the formation of such complexes is specific exactly for the DNA containing the analog of a bulky or clustered lesion.
Fig. 2. Formation of complexes of YB-1 protein with DNA structures Bulk (a) and AP-Bulk (b). Autoradiographs of the reaction products separated in 7% nondenaturing polyacrylamide gel are presented. Ya and Yb, YB-1 complexes with DNA; D, free DNA. Reaction mixtures (5 µl) contained the standard components of the reaction buffer, 8 nM radiolabeled DNA Bulk (a) or AP-Bulk (b), and YB-1 at concentrations indicated below the figure.
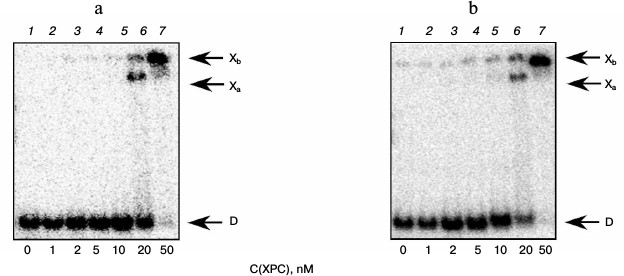
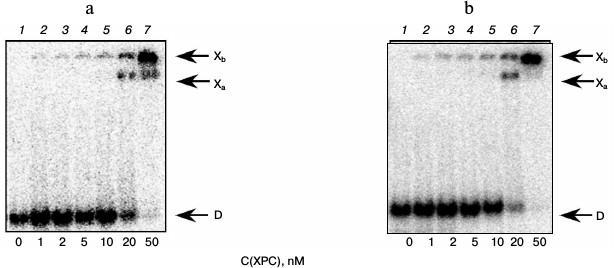
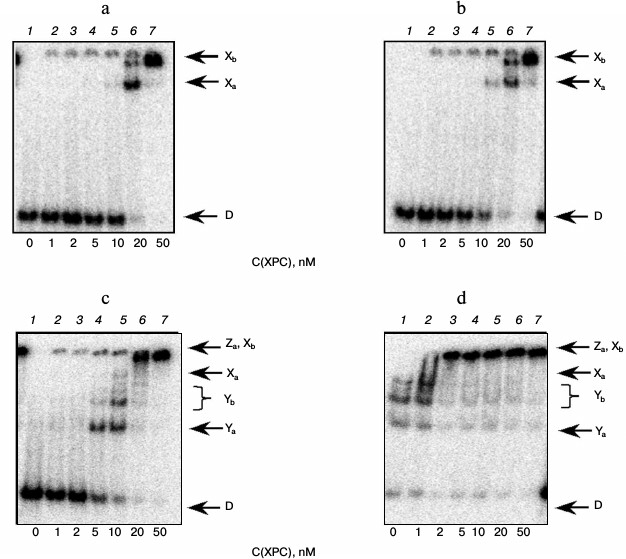
Under the conditions used, XPC-HR23B factor similarly to YB-1 effectively bound to all model DNA used in this work (Figs. 3 and 4). During the experiments, at least two types of protein–DNA complexes were formed by XPC-HR23B with all used oligonucleotides that can be distinguished by both stoichiometry of protein binding with DNA or differences in their conformation in the forming complex (Figs. 3 and 4, bands Xa and Xb). Investigation of interactions of individual proteins with the DNA structures suggests higher DNA-binding activity of XPC-HR23B compared to YB-1. This follows from the comparison of the concentration of one or the other protein required for binding of the same amount of DNA under the same reaction conditions (compare Figs. 1 and 3 as well as Figs. 2 and 4). This assumption was tested for the fluorescein-containing DNA structures by using the fluorescence titration method. The dissociation constants of the complexes calculated from isotherms of the proteins binding to DNA substrates are presented in Table 3. As can be seen, the dissociation constants of complexes with DNA comprising a single bulky lesion for both proteins are very low, and this is quite sufficient for effective interaction with such DNA at the damage recognition stage. Determination of dissociation constants of protein complexes with DNA mimicking a DNA cluster lesion succeeded only in case of YB-1. High instrumental imprecision in the specified range did not allow reliable quantification of the interaction of XPC-HR23B with “cluster-damaged” DNA. However, preliminary analysis of the binding isotherms showed that the expected value of dissociation constant for such complexes is even lower than for DNA containing a single “bulky” adduct. In contrast to XPC-HR23B, the affinity of YB-1 to AP-Bulk DNA was lower than for Bulk DNA. Thus, despite the low values of dissociation constants for complexes of the analyzed proteins with damaged DNA, differences in the interaction should be noted. In accordance with the established point of view, at the stage of damage detection by NER system, XPC-HR23B acts as sensor of areas of DNA double helix significantly destabilized by the presence of lesions [3]. A similar effect was observed previously in the case when a noncanonical pair of nucleotides was placed in the model DNA near the bulky lesion analog [25]. The presence of an AP site apparently also contributes to extra disturbances in DNA structure, which is reflected in a decrease in the dissociation constant of corresponding complexes. On the other hand, the interaction of YB-1 with the reviewed structures and individual lesions in the cluster is apparently of a different character. This is reflected in a relative decrease in the protein affinity to the cluster damage. Thus, in terms of absolute values of the binding constants with the fluorescein-containing DNA, and in terms of the universality of interaction with duplex-destabilized regions of DNA, XPC-HR23B has an advantage over the YB-1, which is quite consistent with its key role in the detection of lesions specifically processed by the NER pathway.
Fig. 3. Formation of complexes of XPC-HR23B protein with DNA structures Comp (a) and AP-Comp (b). Autoradiographs of the reaction products separated in 7% nondenaturing polyacrylamide gel are presented. Xa and Xb, XPC-HR23B complexes with DNA; D, free DNA. Reaction mixtures (5 µl) contained the standard components of the reaction buffer, 8 nM radiolabeled DNA Comp (a) or AP-Comp (b), and XPC-HR23B at concentrations indicated below the figure.
Fig. 4. Formation of complexes of XPC-HR23B protein with DNA structures Bulk (a) and AP-Bulk (b). Autoradiographs of the reaction products separated in 7% nondenaturing polyacrylamide gel are presented. Xa and Xb, XPC complexes with DNA; D, free DNA. Reaction mixtures (5 µl) contained the standard components of the reaction buffer, 8 nM radiolabeled DNA Bulk (a) or AP-Bulk (b), and XPC-HR23B at concentrations indicated below the figure.
Table 3. Dissociation constants of XPC and
YB-1 complexes with DNA mimicking a bulky lesion
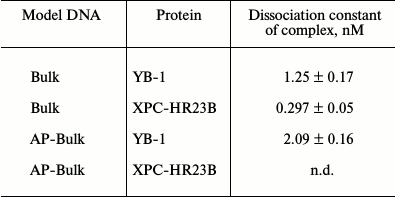
Note: n.d., not determined.
To determine the functional interactions between YB-1 and XPC-HR23B factors, we carried out experiments in which the binding of XPC-HR23B with Bulk DNA containing the bulky lesion was performed in the absence (Fig. 5a) or presence of 10 nM (Fig. 5b), 100 nM (Fig. 5c), or 300 nM YB-1 (Fig. 5d). Analysis of the results shows that under conditions when the presence of 10 nM YB-1 in the reaction mixture does not directly lead to the formation of the YB-1 complexes with DNA (Fig. 5b, lane 1), the protein, however, stimulates formation of XPC-HR23B–Bulk complexes (bands Xa and Xb, for comparison see Figs. 5a and 5b, lanes 5 and 6). Higher YB-1 concentrations (100 and 300 nM) resulted in the formation of complexes of YB-1 with DNA with different electrophoretic mobility (Figs. 5c and 5d, complexes Ya and Yb), which are not formed under the same conditions without XPC-HR23B (for comparison, see Fig. 5c, lanes 1 and 5, 6), indicating the stimulation of DNA-binding activity of YB-1 by protein XPC-HR23B. Under these conditions it can be assumed also the formation of a ternary complex XPC-HR23B–YB-1–DNA (band Za), which is indistinguishable from the complexes XPC-HR23B–DNA (Xb) by its electrophoretic mobility under the used conditions of reaction product separation. In the presence of 100 and 300 nM YB-1, bands of the indicated electrophoretic mobility appear under substantially less XPC-HR23B concentration in the reaction mixture than in the absence of YB-1 (for comparison, see Fig. 5a (lane 7), Fig. 5c (lane 6), and Fig. 5d (lane 3)). Similar results were obtained for the DNA structure containing a clustered lesion of AP-Bulk (data not shown).
Fig. 5. Binding XPC-HR23B to the DNA structure Bulk in the absence (a) and presence of different concentrations of YB-1 (b-d). Autoradiographs of the reaction products separated in 7% nondenaturing polyacrylamide gel are presented. Xa and Xb, XPC-HR23B complexes with DNA; Ya and Yb, YB-1 complexes with DNA; D, free DNA. Reaction mixtures (5 µl) contained the standard components of the reaction buffer, 8 nM radiolabeled DNA Bulk, YB-1 in concentrations of 0 (a), 10 (b), 100 (c), and 300 nM (d), and XPC-HR23B at concentrations indicated below the figure.
Thus, the data show that the YB-1 and XPC-HR23B proteins have high affinity to DNA containing an analog of a “bulky” DNA lesion, and, moreover, they mutually stimulate binding to DNA having a clustered or bulky lesion. The nature of such effect may be a physical interaction between the YB-1 and XPC-HR23B protein factors, leading to a formation of protein–protein complex that is more effective in recognition of the DNA lesion. Another explanation is mutual influence mediated by protein–nucleic acid interactions. The values of the dissociation constants of the YB-1 and XPC-HR23B complexes with damaged DNA that fall into the nanomolar and subnanomolar range suggest this. During such interactions, one protein individually binds DNA and changes its own conformation or DNA ligand conformation with the latter being more preferable for stable binding of another protein. In a number of cases, the first protein is then displaced from the ternary complexes in a competition for a ligand DNA. The described interactions lead to token transfer of DNA ligand from protein to protein in the repair process, similar to transfer of damaged DNA from the stage of primary damage recognition to the stage of its verification in the NER system [3, 27]. The revealed ability of the multifunctional YB-1 protein to influence the key factor of the initial stages of NER, XPC-HR23B, along with the ability of YB-1 to interact directly with damaged DNA and components of the protein ensembles [6, 12, 28] involved in other repair pathways, suggests a putative modulating role of YB-1 in regulation of interaction between several repair processes. These undoubtedly include the processes of base excision repair, repair of noncanonical base pairs, and nonhomologous end joining [7, 15, 16], which suggests the crucial role of YB-1 in systems related to the conservation of DNA integrity and realization of genetic information.
The authors thank Kaoru Sugasawa, Biosignal Research Center, Kobe University, Kobe, Japan for providing recombinant DNA encoding XPC and HR23B subunits.
This work was supported by the Russian Science Foundation (grant 14-24-00038) and the Russian Foundation for Basic Research (grant 14-04-00268).
REFERENCES
1.Lindahl, T., and Wood, R. D. (1999) Quality control
by DNA repair, Science, 286, 1897-1905.
2.Scharer, O. D. (2003) Chemistry and biology of DNA
repair, Angew. Chem. Int. Ed. Engl., 42, 2946-2974.
3.Sugasawa, K., Okamoto, T., Shimizu, Y., Masutani,
C., Iwai, S., and Hanaoka, F. (2001) A multistep damage recognition
mechanism for global genomic nucleotide excision repair, Genes
Dev., 15, 507-521.
4.Volker, M., Mone, M. J., Karmakar, P., Van Hoffen,
A., Schul, W., Vermeulen, W., Hoeijmakers, J. H. J., Van Driel, R., Van
Zeeland, A. A., and Mullenders, L. H. F. (2001) Sequential assembly of
the nucleotide excision repair factors in vivo, Mol.
Cell, 8, 213-224.
5.Pestryakov, P., Zharkov, D. O., Grin, I., Fomina,
E. E., Kim, E. R., Hamon, L., Eliseeva, I. A., Petruseva, I. O., Curmi,
P. A., Ovchinnikov, L. P., and Lavrik, O. I. (2012) Effect of the
multifunctional proteins RPA, YB-1, and XPC repair factor on AP site
cleavage by DNA glycosylase NEIL1, J. Mol. Recognit., 25,
224-233.
6.Eliseeva, I. A., Kim, E. R., Guryanov, S. G.,
Ovchinnikov, L. P., and Lyabin, D. N. (2011) Y-box-binding protein 1
(YB-1) and its functions, Biochemistry (Moscow), 76,
1402-1433.
7.Das, S., Chattopadhyay, R., Bhakat, K. K., Boldogh,
I., Kohno, K., Prasad, R., Wilson, S. H., and Hazra, T. K. (2007)
Stimulation of NEIL2-mediated oxidized base excision repair via YB-1
interaction during oxidative stress, J. Biol. Chem., 282,
28474-28484.
8.Cohen, S. B., Ma, W., Valova, V. A., Algie, M.,
Harfoot, R., Woolley, A. G., Robinson, P. J., and Braithwaite, A. W.
(2010) Genotoxic stress-induced nuclear localization of oncoprotein
YB-1 in the absence of proteolytic processing, Oncogene,
29, 403-410.
9.Koike, K., Uchiumi, T., Ohga, T., Toh, S., Wada,
M., Kohno, K., and Kuwano, M. (1997) Nuclear translocation of the Y-box
binding protein by ultraviolet irradiation, FEBS Lett.,
417, 390-394.
10.Gaudreault, I., Guay, D., and Lebel, M. (2004)
YB-1 promotes strand separation in vitro of duplex DNA
containing either mispaired bases or cisplatin modifications, exhibits
endonucleolytic activities and binds several DNA repair proteins,
Nucleic Acids Res., 32, 316-327.
11.Hasegawa, S. L., Doetsch, P. W., Hamilton, K. K.,
Martin, A. M., Okenquist, S. A., Lenz, J., and Boss, J. M. (1991) DNA
binding properties of YB-1 and dbpA: binding to doublestranded,
single-stranded, and abasic site containing DNAs, Nucleic Acids
Res., 19, 4915-4920.
12.Ise, T., Nagatani, G., Imamura, T., Kato, K.,
Takano, H., Nomoto, M., Izumi, H., Ohmori, H., Okamoto, T., Ohga, T.,
Uchiumi, T., Kuwano, M., and Kohno, K. (1999) Transcription factor
Y-box binding protein 1 binds preferentially to cisplatin-modified DNA
and interacts with proliferating cell nuclear antigen, Cancer
Res., 59, 342-346.
13.Marenstein, D. R., Ocampo, M. T., Chan, M. K.,
Altamirano, A., Basu, A. K., Boorstein, R. J., Cunningham, R. P., and
Teebor, G. W. (2001) Stimulation of human endonuclease III by Y
box-binding protein 1 (DNA-binding protein B). Interaction between a
base excision repair enzyme and a transcription factor, J. Biol.
Chem., 276, 21242-21249.
14.Chattopadhyay, R., Das, S., Maiti, A. K.,
Boldogh, I., Xie, J., Hazra, T. K., Kohno, K., Mitra, S., and Bhakat,
K. K. (2008) Regulatory role of human AP-endonuclease (APE1/Ref-1) in
YB-1-mediated activation of the multidrug resistance gene MDR1, Mol.
Cell Biol., 28, 7066-7080.
15.Prasad, R., Williams, J. G., Hou, E. W., and
Wilson, S. H. (2012) Pol β associated complex and base excision
repair factors in mouse fibroblasts, Nucleic Acids Res.,
40, 11571-11582.
16.Kim, E. R., Selyutina, A. A., Buldakov, I. A.,
Evdokimova, V., Ovchinnikov, L. P., and Sorokin, A. V. (2013) The
proteolytic YB-1 fragment interacts with DNA repair machinery and
enhances survival during DNA damaging stress, Cell Cycle,
12, 3791-3803.
17.Ohga, T., Koike, K., Ono, M., Makino, Y.,
Itagaki, Y., Tanimoto, M., Kuwano, M., and Kohno, K. (1996) Role of the
human Y box-binding protein YB-1 in cellular sensitivity to the
DNA-damaging agents cisplatin, mitomycin C, and ultraviolet light,
Cancer Res., 56, 4224-4228.
18.Selivanova, O. M., Guryanov, S. G., Enin, G. A.,
Skabkin, M. A., Ovchinnikov, L. P., and Serdyuk, I. N. (2010) YB-1 is
capable of forming extended nanofibrils, Biochemistry (Moscow),
75, 115-120.
19.Nishi, R., Okuda, Y., Watanabe, E., Mori, T.,
Iwai, S., Masutani, C., Sugasawa, K., and Hanaoka, F. (2005) Centrin 2
stimulates nucleotide excision repair by interacting with xeroderma
pigmentosum group C protein, Mol. Cell Biol., 25,
5664-5674.
20.Kim, C., Paulus, B. F., and Wold, M. S. (1994)
Interactions of human replication protein A with oligonucleotides,
Biochemistry, 33, 14197-14206.
21.Lundblad, J. R., Laurance, M., and Goodman, R. H.
(1996) Fluorescence polarization analysis of protein–DNA and
protein–protein interactions, Mol. Endocrinol., 10,
607-612.
22.Rechkunova, N. I., Krasikova, Y. S., and Lavrik,
O. I. (2011) Nucleotide excision repair: DNA damage recognition and
preincision complex assembly, Biochemistry (Moscow), 76,
24-35.
23.Ohga, T. (1998) Direct involvement of the Y-box
binding protein YB-1 in genotoxic stress-induced activation of the
human multidrug resistance 1 gene, J. Biol. Chem., 273,
5997-6000.
24.Yahata, H., Kobayashi, H., Kamura, T., Amada, S.,
Hirakawa, T., Kohno, K., Kuwano, M., and Nakano, H. (2002) Increased
nuclear localization of transcription factor YB-1 in acquired
cisplatin-resistant ovarian cancer, J. Cancer Res. Clin. Oncol.,
128, 621-626.
25.Krasikova, Y. S., Rechkunova, N. I., Maltseva, E.
A., Petruseva, I. O., Silnikov, V. N., Zatsepin, T. S., Oretskaya, T.
S., Scharer, O. D., and Lavrik, O. I. (2008) Interaction of nucleotide
excision repair factors XPC-HR23B, XPA, and RPA with damaged DNA,
Biochemistry (Moscow), 73, 886-896.
26.Krasikova, Y. S., Rechkunova, N. I., Maltseva, E.
A., Petruseva, I. O., and Lavrik, O. I. (2010) Localization of
xeroderma pigmentosum group A protein and replication protein A on
damaged DNA in nucleotide excision repair, Nucleic Acids Res.,
38, 8083-8094.
27.Maria Berra, C., de Oliveira, C. S., Machado
Garcia, C. C., Reily Rocha, C. R., Koch Lerner, L., de Andrade Lima, L.
C., da Silva Baptista, M., and Martins Menck, C. F. (2013) Nucleotide
excision repair activity on DNA damage induced by photoactivated
methylene blue, Free Radic. Biol. Med., 61C, 343-356.
28.De Souza-Pinto, N. C., Mason, P. A., Hashiguchi,
K., Weissman, L., Tian, J., Guay, D., Lebel, M., Stevnsner, T. V.,
Rasmussen, L. J., and Bohr, V. A. (2009) Novel DNA mismatch-repair
activity involving YB-1 in human mitochondria, DNA Repair
(Amsterdam), 8, 704-719.