Supramolecular Organization of Hfq-Like Proteins
V. N. Murina1*, O. M. Selivanova1, A. O. Mikhaylina1, A. S. Kazakov2, E. Yu. Nikonova1, N. V. Lekontseva1, S. V. Tishchenko1, and A. D. Nikulin1
1Institute of Protein Research, Russian Academy of Sciences, ul. Institutskaya 4, 142290 Pushchino, Moscow Region, Russia; E-mail: thyrada@rambler.ru2Institute for Biological Instrumentation, Russian Academy of Sciences, ul. Institutskaya 7, 142290 Pushchino, Moscow Region, Russia
* To whom correspondence should be addressed.
Received October 28, 2014
Bacterial Hfq proteins are structural homologs of archaeal and eukaryotic Sm/Lsm proteins, which are characterized by a 5-stranded β-sheet and an N-terminal α-helix. Previously, it was shown that archaeal Lsm proteins (SmAP) could produce long fibrils spontaneously, in contrast to the Hfq from Escherichia coli that could form similar fibrils only after special treatment. The organization of these fibrils is significantly different, but the reason for the dissimilarity has not been found. In the present work, we studied the process of fibril formation by bacterial protein Hfq from Pseudomonas aeruginosa and archaeal protein SmAP from Methanococcus jannaschii. Both proteins have high homology with E. coli Hfq. We found that Hfq from P. aeruginosa could form fibrils after substitutions in the conserved Sm2 motif only. SmAP from M. jannaschii, like other archaeal Lsm proteins, form fibrils spontaneously. Despite differences in the fibril formation conditions, the architecture of both was similar to that described for E. coli Hfq. Therefore, universal nature of fibril architecture formed by Hfq proteins is suggested.
KEY WORDS: Sm-like proteins, Hfq, quaternary structure of proteins, fibrilsDOI: 10.1134/S0006297915040070
Hfq is a posttranslational regulator of gene expression that binds small noncoding RNAs (sRNA) and promotes their interaction with mRNAs [1, 2]. It belongs to the widespread Sm/Lsm (Sm-like) protein family, members of which are found in Bacteria, Archaea, and Eukarya [3]. Eukaryotic Sm and Lsm proteins are an important part of ribonucleoprotein complexes that participate in RNA processing, including splicing, mRNA degradation, histone formation, and telomere replication [4, 5]. The function of the archaeal Lsm proteins have studied rather poorly [6]; it is known, however, they bind archaeal sRNA [7].
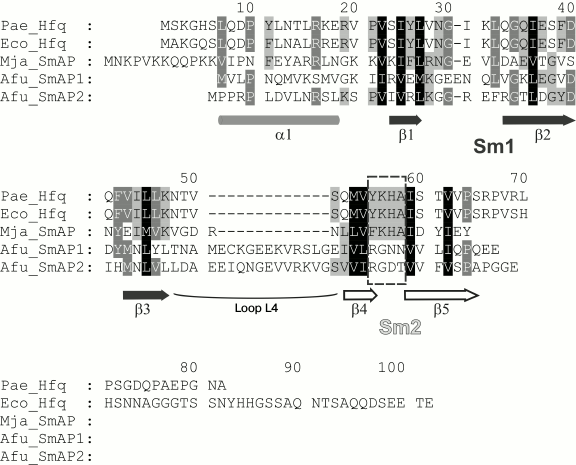
Sm/Lsm proteins have specific conserved three-dimensional structure consisting of the N-terminal α-helix, five β-strands (that is the Sm domain), and a disordered C-terminal part of variable length [8] (Fig. 1). Contacting via peripheral β4 and β5 strands belonging to the Sm domain, the proteins organize doughnut shaped ring structure from six (bacterial Hfq) or seven (archaeal SmAP, eukaryotic Sm and Lsm) monomers. This characteristic architecture of the protein quaternary structure seems to be important for the specific binding of single-stranded RNA repeating motifs by the small protein [9, 10]. The Sm domain includes two conserved motifs: Sm1 (β1, β2, and β3 strands) and Sm2 (β4 and β5 strands). They are connected by loop L4 that has length of four amino acids in bacterial Hfq and about 20 amino acids in archaeal and eukaryotic Lsm proteins [8]. It is considered that loop L4 has no influence on the quaternary structure of the protein [11]. The Sm1 and Sm2 sequences are conserved among all Lsm proteins [11, 12], but bacterial Sm2 is characterized by strictly conserved YKHA pattern, while eukaryotic Sm2 has RGxx consensus [10, 11]. It was suggested that differences in the protein sequences could affect the number of subunits in the protein particles, and the His57 of the YKHA consensus might be important for Hfq hexamer stabilization through its side-chain organized intersubunit hydrogen bonds [13]. It has been shown that the substitution of His57 for alanine, threonine, or asparagine results in a significant decrease in the thermostability of the protein, but it does not change its spatial structure [14]. Later, it was demonstrated that substitution of Tyr55 of the same consensus to alanine has also no effect on the protein structure [15]. Thus, a single substitution in the consensus YKHA leads to no change in the quaternary structure of the protein. But, will the replacement of the entire YKHA consensus affect the structural properties of the protein? To answer this question, we replaced the YKHA sequence in Hfq from Pseudomonas aeruginosa (Pae Hfq) with the RGDT sequence, which has the heptamer archaeal protein SmAP1 from Archaeoglobus fulgidus.
Fig. 1. Alignment of bacterial Hfq and archaeal SmAP protein representatives. Secondary structure elements, conserved motifs Sm1 (strands β1, β2, and β3), Sm2 (strands β4 and β5), and connecting loop L4 are marked. Conserved bacterial consensus YKHA of the Sm2 motif is indicated by the dashed rectangle.
Previously, it was found archaeal proteins SmAP from Pyrobaculum aerophilum and from Methanobacterium thermautotrophicum spontaneously form in solution non-amyloid polar fibers [16]. Their thickness of ~8 nm is comparable with the diameter of a single SmAP1 heptamer. It was suggested the protein heptamers are packed “head-to-tail” perpendicularly to the fiber axis. These fibers can polymerize into thick (50 nm) bundles of several parallel packed fibrils. Later it was found that Hfq from E. coli could also form fibers, but only after dialysis against dodecyl-β-D-maltoside and lyophilization followed by dilution of the protein in water [17]. Using electron microscopy and Fourier-transformed infrared spectroscopy, it was shown that the protein forms helical fibrils of protein hexamers arranged on a cylindrical surface. The diameter of the cylinder is 17 nm and the angle of the hexamer plane with respect to the fiber axis is 37.7° (calculated data based on helical pitch of 24 nm and interplanar spacing of 4 nm in Hfq homohexamers) [17]. Thereby, both archaeal SmAP and bacterial Hfq proteins can form fibers, but the structural organization and the conditions of their formation differ significantly. We studied the influence of the YKHA/RGDT substitution on the ability of the Pae Hfq for fibers to form and compared the results with those obtained for the SmAP protein from archaea Methanococcus jannaschii (Mja SmAP), which has the characteristic features of bacterial Hfq proteins: the Sm2 consensus is YKHA, and loop L4 between Sm1 and Sm2 motifs is short [18].
MATERIALS AND METHODS
Preparation of plasmid construct carrying gene encoding Pae Hfq with YKHA/RGDT substitution and purification of the protein. The QuikChange Site Directed Mutagenesis Kit (Stratagene, USA) was used to prepare the YKHA/RGDT substitution in the Pae Hfq protein. PCR was carried out using pET22b(+)/Hfq plasmid, and the primers contained the necessary substitution: 5′-GTCAGCCAGATGGTTCGCGGCGACACGATCTCCACCG-3′ (forward) and 5′-CGGTGGAGATCGTGTCGCCGCGAACCATCTGGCTGAC-3′ (reverse). The resulting plasmid construct was checked by sequencing. Gene expression and protein purification of wild-type Pae Hfq was performed as described earlier [14]. Pae Hfq YKHA/RGDT gene expression was made in strain E. coli BL21(DE3). The cells were grown at 42°C in LB medium to OD600 ≈ 0.8 and induced with isopropyl-d-1-thiogalactopyranoside (IPTG, 1 mM) for 2 h. Then the cells were precipitated and sonicated in 1 M NaCl, 0.25 M MgCl2, 5 mM EDTA, and 50 mM Tris-HCl buffer, pH 8.0. Cell membranes and ribosome were precipitated using stepwise centrifugation at 14,000g for 20 min and at 90,000g for 50 min, respectively. The supernatant was dialyzed in 0.1 M NaCl, 50 mM Tris-HCl, pH 8.0, loaded on heparin-Sepharose (Amersham, Sweden), and eluted in a 0.1-0.8 M NaCl gradient with 50 mM Tris-HCl, pH 8.0. The fractions were analyzed by SDS-PAGE in 15% polyacrylamide gel. Purified Pae Hfq YKHA/RGDT protein was concentrated to 12 mg/ml in 0.3 M NaCl and 50 mM Tris-HCl, pH 8.0.
Preparation of plasmid construct carrying gene encoding SmAP M. jannaschii and purification of the protein. The gene of SmAP M. jannaschii was cloned into the high-level expression vector pET11a-PL using M. jannaschii genomic DNA as a template. A DNA fragment encoding SmAP M. jannaschii flanked by a 5′-FauNDI and a 3′-EcoRI site was generated by standard PCR with primers 5′-GGAATTCCATATGAATAAGCCAGTAAAAAAACAGCAACCAAAGAAAGTC-3′ (forward) and 5′-CAATATGAATTCTTAGTATTCTATGTAGTCAATAGCATGTTTAAATACTAATAAGTTCC-3′ (reverse). The nucleotide sequences of the genes were verified by sequencing.
SmAP M. jannaschii (Mja SmAP) was overproduced in E. coli strain BL21(DE3) as a host. To avoid the potential misincorporation of lysine instead of arginine, the host strain was cotransformed with pUBS520 [19]. This plasmid carries the gene for tRNAARG AGA/AGG [20] and the kan (resistance to kanamycin) gene.
The transformed cells were cultured at 37°C in LB medium containing 100 µg/ml ampicillin and 50 µg/ml kanamycin. Protein expression was induced using 0.6 mM IPTG when the OD600 reached ~0.8 and incubated overnight at 20°C. The cells were harvested by centrifugation at 8000g for 20 min at 4°C and then resuspended in lysis buffer consisting of 50 mM Tris-HCl, pH 8.0, 1 M NaCl, 0.25 M MgCl2, and 5 mM EDTA and disrupted using an EmulsiFlex-C3 high-pressure cell disrupter (Avestin, Canada). The cell debris was removed by centrifugation at 14,000g for 30 min. The supernatant was heated at 82°C for 20 min, and denatured E. coli proteins were removed by centrifugation (14,000g, 40 min, 4°C). Ammonium sulfate was added to the supernatant to 1.5 M, and the supernatant solution was loaded onto butyl-Toyopearl 650S column previously equilibrated in 50 mM Tris-HCl buffer, pH 8.0, 1 M NaCl, 1.5 M (NH4)2SO4. The protein was eluted in a linear gradient of NaCl and (NH4)2SO4 from start buffer to 50 mM Tris-HCl, pH 8.0. Fractions containing the target protein (checked by 15% SDS-PAGE) were pooled. Mja SmAP was concentrated to 30 mg/ml and dialyzed into 0.2 M NaCl and 50 mM Tris-HCl, pH 8.0.
Analysis of protein particle size by gel filtration. For this analysis, we used an Acta Basic system (Amersham) with Superdex 200 gel-filtration column (24 ml) previously equilibrated with 100 mM NaCl, 50 mM Tris-HCl, pH 8.0. The protein sample (2 mg/ml, 100 µl) was injected into the column and eluted with velocity 0.4 ml/min. The calibration curve was modeled used BSA, ovalbumin, ribonuclease A, and aprotinin.
Size and molecular weight determination of protein particles using dynamic laser light scattering (DLS). DLS experiments were performed using a Zetasizer Nano ZS (Malvern Instruments Ltd, UK). The back-scattered light from a 4-mW He/Ne laser (wavelength 632.8 nm) was collected at the angle of 173°. Protein samples with concentration 1 mg/ml in buffer (0.1 M NaCl, 50 mM Tris-HCl, pH 7.5) were used. Temperature was maintained at 25°C. The accumulation time of the autocorrelation function was 100 s. The final autocorrelation function was the average of 10 measurements. The radius distribution versus the light scattering intensity was calculated using the following parameters: refractive index of 1.330 and the solution viscosity of 0.8882 Pa·s. The molecular weight distribution versus the light scattering intensity was calculated using the Mark–Houwink equation from the standard software for this device (Malvern Zetasizer Software; Malvern Instruments Ltd, UK):
D = K· M–a,
where D is diffusion coefficient; K and a, constants (a = 0.428 cm2/s and K = 7.67E–5); M, molecular weight in kDa.
Transmission electron microscopy. Prior to negative staining, samples of Pae Hfq and Pae Hfq YKHA/RGDT were dialyzed into 25 mM Tris-HCl (pH 8.0) buffer containing 50 mM NaCl with protein concentration of 2 mg/ml. Then they were diluted to the final concentration of 0.1 mg/ml. Samples of Mja SmAP with 20-40 mg/ml concentration stored in buffer of 0.1 M NaCl, 50 mM Tris-HCl, pH 8.0, were diluted in the same buffer to the concentration of 0.2 mg/ml.
Copper meshes (400 Mesh) coated with Formvar film (0.2%) were mounted on a sample drop (10 µl). After 5 min, the grid was negatively stained with 1% (w/v) aqueous uranyl acetate for 1-1.5 min and then examined under a transmission electron microscope (JEM-100C) at accelerating voltage 80 kV. Images were recorded on Kodak electron image film at nominal magnification of 40,000-60,000.
RESULTS AND DISCUSSION
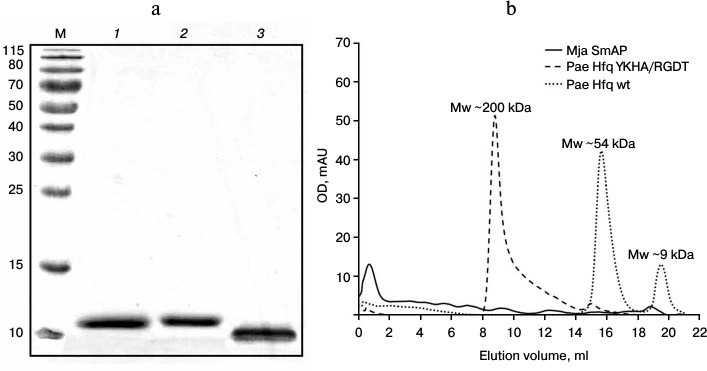
Protein preparation. Pae Hfq was purified by protocols described earlier [13]. The homogeneity was checked by SDS-PAGE, gel filtration, and DLS. Under denaturing conditions, all the studied proteins had molecular weight corresponding to the monomers (Fig. 2a): Pae Hfq, both wild type and YKHA/RGDT mutant, had molecular weight 9.5 kDa, and Mja SmAP was of 8.3 kDa. It was found that Pae Hfq YKHA/RGDT has very low solubility, and for this reason, we could not determine its crystal structure and thermostability as was done for several Pae Hfq mutant forms previously [15].
The analysis of the protein by gel filtration revealed Pae Hfq YKHA/RGDT formed multimers containing several hexamers, in contrast to the wild-type protein existing as monomers and hexamers in solution (Fig. 2b). Mja SmAP was characterized by a large peak at the beginning of the elution profile, which indicated high level of protein oligomerization.
Fig. 2. a) SDS-PAGE of the studied proteins. Lanes: M, molecular weight markers; 1) Pae Hfq wt; 2) Pae Hfq YKHA/RGDT; 3) Mja SmAP. b) Elution profiles of Pae Hfq wt, Pae Hfq YKHA/RGDT (a Superdex 200 10/30 column was used).
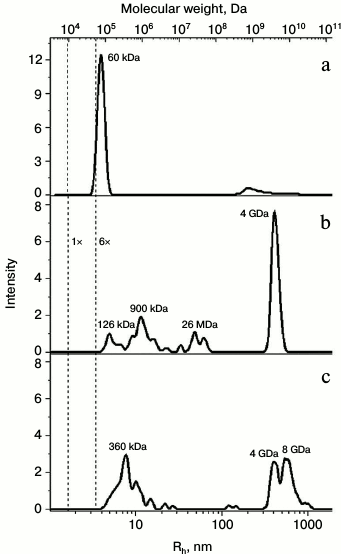
The oligomerization state of the proteins was confirmed by dynamic light scattering (Fig. 3). The data demonstrated that wild-type Pae Hfq in solution forms hexamers with molecular weight of about 56 kDa (Fig. 3a). Pae Hfq YKHA/RGDT and Mja SmAP were present as heterogeneous populations of molecules with different molecular weights: Pae Hfq YKHA/RGDT particles ranged from 126 kDa to 26 MDa with a peak at 4 GDa (Fig. 3b); Mja SmAP had peaks at 360 kDa and at 4-8 GDa (Fig. 3c). Thus, the YKHA/RGDT substitution in Pae Hfq increased its ability to form large molecular weight oligomers, whereas wild-type protein did not have this ability. Meanwhile, Mja SmAP possessing YKHA consensus in the Sm2 motif was able to form oligomers with high molecular weight.
Fig. 3. Dependence of hydrodynamic radii (bottom) and molecular weight (top) from intensity of light scattering for Pae Hfq wt (a), with YKHA/RGDT substitution (b), and for Mja SmAP (c). Vertical dotted lines mark calculated peak positions for globular proteins with molecular weights corresponding to monomer and hexamer of Pae Hfq (9.5 and 57 kDa). The molecular weight estimation for the largest peaks according to the hydrodynamic radii of the particles is marked.
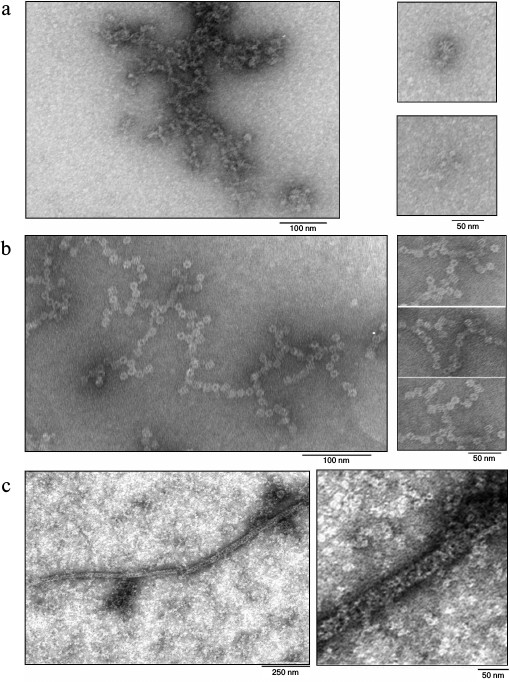
Electron microscopy of the proteins. For detailed analysis of the oligomers, the protein samples were studied by transmission electron microscopy. Attempts to get fibers of Pae Hfq according the earlier applied procedure with Hfq from E. coli (Eco Hfq) [17] were not successful. Under these conditions, as well as upon dilution of the protein samples to 0.2 mg/ml by 50 mM NaCl in 50 mM Tris-HCl, pH 8.0, there are only isolated hexamers (rings with 8 nm diameter), and disorderly aggregates of various sizes were observed (Fig. 4a).
Fig. 4. Electron micrographs for samples of Pae Hfq wt (a) (irregular aggregates), Pae Hfq YKHA/RGDT (b) (“barrels”), and Mja SmAP (c) (extended fibers). Large fragments of the EM field and enlarged parts of the characteristic particles are shown. In all cases, isolated small particles corresponding to protein hexamers of 10 nm diameters are visible near the oligomers.
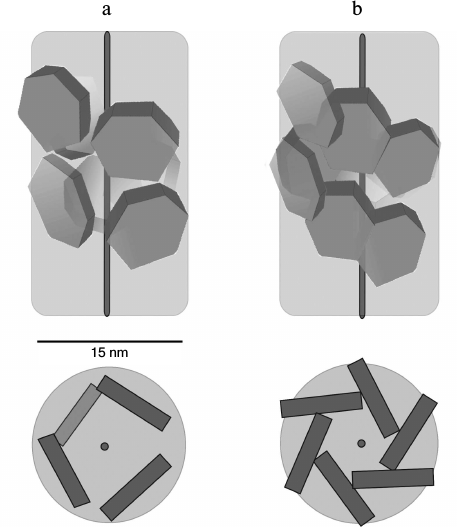
In contrast to the wild-type protein, Pae Hfq YKHA/RGDT at 0.2 mg/ml in 50 mM NaCl and 50 mM Tris-HCl, pH 8.0, formed uniform oligomer barrel-like particles of about 15 × 15-nm size (Fig. 4b). These particles had pronounced 3-fold symmetry from their faces. The diameter of the particles differs from that of SmAP fibers (about 8 nm, corresponding to the diameter of the protein heptamer) and is less than the diameter of Eco Hfq fibers (~17 nm) [17]. As in the central part of the particles darkening can be seen, we assume that the packing of the protein hexamers is not continuous, but is similar to that observed for the Eco Hfq fibers. The difference in the diameter of the fibers could be explained by the following: 1) reduction in Pae Hfq dimensions compared with Eco Hfq due to shortening the C-terminal part of the protein (Fig. 1), or 2) large protein hexamers incline plane to the fibril axis. Thus, these “protofibrils” can be modeled by several helix turns of spirally arranged protein hexamers and, in accordance with the 3-fold symmetry, each turn should contain three or six hexamers (Fig. 5).
Fig. 5. Modeled arrangement of Pae Hfq YKHA/RGDT hexamers in nanoparticles (top and side views). According to 3-fold axis, one turn of the helix contains three (a) or six (b) hexamers. In case of three hexamers per turn, the packing of the nanoparticles is spongier, so the first hexamer of the second turn should be visible from the face of the barrel.
Mja SmAP protein at 0.2 mg/ml in 0.1 M NaCl and 50 mM Tris-HCl, pH 8.0, formed long (more than 1 µm) fibers with diameters of about 25-30 nm (Fig. 4c). Since on the surface of the fibers Mja SmAP hexamers located as a helix can be seen, it can be assumed that they are arranged strictly on the cylinder surface. This may explain the larger diameter of the Mja SmAP fibers in comparison with fibers of Pae Hfq YKHA/RGDT and Eco Hfq.
Since the studied proteins are mostly β-proteins, we tested whether Hfq and SmAP formed amyloid fibrils. X-Ray scattering patterns of Pae Hfq YKHA/RGDT and Mja SmAP fibers were collected as described previously [21]. These patterns revealed the presence of diffuse scattering only without any reflection from the samples (data not shown), which corresponds to absence of amyloid in the fibers.
Thus, we have shown that Hfq proteins, despite the differences in their amino acid sequences, form fibers of similar architecture: the protein hexamers are arranged spirally on the surface of a hollow cylinder, and the hexamer planes are parallel to the cylinder plane; the fibers have an inner cavity passing along the cylinder axis. Apparently, the main difference between fibers formed by different proteins is the number of hexamers per turn and the inclination angle between the hexamer planes and the fibril axis. These parameters have to define the diameter of the fibers and the packing parameters of the protein hexamers. The replacement of conserved YKHA sequence in the Sm2 motif of Pae Hfq with archaeal RGDT seems to effect a local change of the surface charge distribution that leads to formation of barrel-like nanoparticles with more compact packaging of the protein hexamers. As the result, a new form of protein nanoparticles was found that is not similar with previously obtained Eco Hfq and archaeal SmAP fibers.
The obtained fibrils and nanoparticles can be of interest for application in nanobiotechnology because they can be reproduced easily under physiological conditions, and the corresponding proteins are produced in large quantity. An example of such application is the use of archaeal flagella as a scaffold to produce high-performance materials for electrodes of batteries [22, 23].
This work was supported by the Russian Scientific Foundation (project 14-14-00496). The DLS experiment was done by A. S. Kazakov with support from the Molecular and Cell Biology Program of the Russian Academy of Sciences. EM experiments were done by O. S. Selivanova with support from the Russian Scientific Foundation (project 14-14-00536).
REFERENCES
1.Vogel, J., and Luisi, B. F. (2011) Hfq and its
constellation of RNA, Nat. Rev. Microbiol., 9,
578-589.
2.Valentin-Hansen, P., and Eriksen, M. (2004)
Micro-review: the bacterial Sm-like protein Hfq: a key player in RNA
transactions, Mol. Microbiol., 51, 1525-1533.
3.Brennan, R. G., and Link, T. M. (2007) Hfq
structure, function and ligand binding, Curr. Opin. Microbiol.,
10, 125-133.
4.Beggs, J. D. (2005) Lsm proteins and RNA
processing, Biochem. Soc. Trans., 33, 433-438.
5.Spiller, M. P., Boon, K.-L., Reijns, M. M., and
Beggs, J. D. (2007) The Lsm2-8 complex determines nuclear localization
of the spliceosomal U6 snRNA, Nucleic Acids Res., 35,
923-929.
6.Mura, C., Randolph, P. S., Patterson, J., and
Cozen, A. E. (2013) A structural and evolutionary perspective on Sm
function archaeal and eukaryotic homologs of Hfq, RNA Biol.,
10, 636-651.
7.Fischer, S., Benz, J., Spath, B., Maier, L.-K.,
Straub, J., Granzow, M., Raabe, M., Urlaub, H., Hoffmann, J., Brutschy,
B., Allers, T., Soppa, J., and Marchfelder, A. (2010) The archaeal Lsm
protein binds to small RNAs, J. Biol. Chem., 285,
34429-34438.
8.Murina, V. N., and Nikulin, A. D. (2011)
RNA-binding Sm-like proteins of bacteria and archaea: similarity and
difference in structure and function, Biochemistry (Moscow),
76, 1434-1449.
9.Khusial, P., Plaag, R., and Zieve, G. W. (2005) LSm
proteins form heptameric rings that bind to RNA via repeating motifs,
Trends Biochem. Sci., 30, 522-528.
10.Robinson, K. E., Orans, J., Kovach, A. R., Link,
T. M., and Brennan, R. G. (2014) Mapping Hfq-RNA interaction surfaces
using tryptophan fluorescence quenching, Nucleic Acids Res.,
42, 2736-2749.
11.Sauter, C. (2003) Sm-like proteins in Eubacteria:
the crystal structure of the Hfq protein from Escherichia coli,
Nucleic Acids Res., 31, 4091-4098.
12.Salgado-Garrido, J., Bragado-Nilsson, E.,
Kandels-Lewis, S., and Seraphin, B. (1999) Sm and Sm-like proteins
assemble in two related complexes of deep evolutionary origin, EMBO
J., 18, 3451-3462.
13.Nikulin, A., Stolboushkina, E., Perederina, A.,
Vassilieva, I., Blaesi, U., Moll, I., Kachalova, G., Yokoyama, S.,
Vassylyev, D., Garber, M., and Nikonov, S. (2005) Structure of
Pseudomonas aeruginosa Hfq protein, Acta Crystallogr. D.
Biol. Crystallogr., 61, 141-146.
14.Moskaleva, O., Melnik, B., Gabdulkhakov, A.,
Garber, M., Nikonov, S., Stolboushkina, E., and Nikulin, A. (2010) The
structures of mutant forms of Hfq from Pseudomonas aeruginosa
reveal the importance of the conserved His57 for the protein hexamer
organization, Acta Crystallogr. F, 66, 760-764.
15.Murina, V. N., Melnik, B. S., Filimonov, V. V.,
Uhlein, M., Weiss, M. S., Muller, U., and Nikulin, A. D. (2014) Effect
of conserved intersubunit amino acid substitutions on Hfq protein
structure and stability, Biochemistry (Moscow), 79,
469-477.
16.Mura, C., Kozhukhovsky, A., Gingery, M., and
Phillips, M. (2003) The oligomerization and ligand-binding properties
of Sm-like archaeal proteins (SmAPs), Protein Sci., 12,
832-847.
17.Arluison, V., Mura, C., Guzman, M. R., Liquier,
J., Pellegrini, O., Gingery, M., Regnier, P., and Marco, S. (2006)
Three-dimensional structures of fibrillar Sm proteins: Hfq and other
Sm-like proteins, J. Mol. Biol., 356, 86-96.
18.Nielsen, J. S., Boggild, A., Andersen, C. B. F.,
Nielsen, G., Boysen, A., Brodersen, D. E., and Valentin-Hansen, P.
(2007) An Hfq-like protein in archaea: crystal structure and functional
characterization of the Sm protein from Methanococcus
jannaschii, RNA, 13, 2213-2223.
19.Calderone, T. L., Stevens, R. D., and Oas, T. G.
(1996) High-level misincorporation of lysine for arginine at AGA codons
in a fusion protein expressed in Escherichia coli, J. Mol.
Biol., 262, 407-412.
20.Brinkmann, U., Mattes, R. E., and Buckel, P.
(1989) High-level expression of recombinant genes in Escherichia
coli is dependent on the availability of the dnaY gene product,
Gene, 85, 109-114.
21.Guryanov, S. G., Selivanova, O. M., Nikulin, A.
D., Enin, G. A., Melnik, B. S., Kretov, D. A., Serdyuk, I. N., and
Ovchinnikov, L. P. (2012) Formation of amyloid-like fibrils by Y-box
binding protein 1 (YB-1) is mediated by its cold shock domain and
modulated by disordered terminal domains, PLoS One, 7,
e36969.
22.Beznosov, S. N., Pyatibratov, M. G., and Fedorov,
O. V. (2009) Archaeal flagellas as new nanomaterial creation matrixes,
Russ. Nanotechnol., 4, 94-98.
23.Beznosov, S. N., Pyatibratov, M. G., Fedorov, O.
V., Kulova, T. L., and Skundin, A. M. (2011) Electrochemical
characteristics of nanostructured material based on modified flagella
of halophilic archaea Halobacterium salinarum for the negative
electrode of lithium-ion battery, Russ. Nanotechnol., 6,
43-47.