Proteorhodopsin from Dokdonia sp. PRO95 Is a Light-Driven Na+-Pump
Y. V. Bertsova, A. V. Bogachev, and V. P. Skulachev*
Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119992 Moscow, Russia; fax: +7 (495) 939-0338; E-mail: skulach@belozersky.msu.ru; bogachev@belozersky.msu.ru* To whom correspondence should be addressed.
Received November 23, 2014
The gene encoding proteorhodopsin AEX55013 from Dokdonia sp. PRO95 was cloned and expressed in Escherichia coli cells. Illumination of the proteorhodopsin-producing E. coli cells in Na+-containing media resulted in alkalinization of the media. This response was accelerated by uncoupler CCCP and inhibited by penetrating anion SCN–. Illumination of the cells in a sodium-free medium (made by substituting Na+ with K+) resulted in SCN–-stimulated and CCCP-sensitive acidification of the medium. Illumination of the proteorhodopsin-containing E. coli cells caused CCCP-resistant transmembrane sodium export from these cells. We conclude that the proteorhodopsin from the marine flavobacterium Dokdonia sp. PRO95 is a primary light-driven Na+-pump. A high level of the heterologous production in E. coli cells as well as stability and purity of the isolated protein makes this proteorhodopsin an attractive model for studying the mechanism of active sodium transmembrane translocation.
KEY WORDS: Na+-translocating proteorhodopsin, transmembrane sodium transport, flavobacteriaDOI: 10.1134/S0006297915040082
Abbreviations: CCCP, carbonyl cyanide m-chlorophenylhydrazone; DDM, n-dodecyl β-D-maltoside; R-E. coli, E. coli BL21/pRhod_10.1 cells induced for proteorhodopsin synthesis in the presence of retinal; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; ΔpH, transmembrane pH difference; ΔµH+ and ΔµNa+, transmembrane differences in H+ and Na+ electrochemical potentials, respectively; Δψ, transmembrane difference in electric potentials.
The majority of bacteria utilize ΔµH+ as
energetic currency on energy conservation. Catalytic activity of
different complexes of respiratory and photosynthetic chains as well as
other energy-transducing membrane-associated enzymes is coupled with
transmembrane translocation of H+. The
ΔµH+ formed is then used to drive various
energy-consuming processes.
However, enzymatic activity of some bacterial proteins is coupled with transmembrane translocation of Na+ instead of protons. In 1980, Peter Dimroth reported the discovery of a Na+-translocating oxaloacetate decarboxylase from the enterobacterium Klebsiella aerogenes [1]. It was further established that all membrane-associated bacterial decarboxylases decarboxylating different carbonic acids use only Na+ as a coupling ion [2]. Later, other prokaryotic primary sodium pumps were described: Na+-translocating NADH:quinone-oxidoreductase [3, 4] and homologous Na+-translocating NADH:ferredoxin-oxidoreductase [5, 6], Na+-motive methyltransferase complex [7] and Na+-formylmethanofuran dehydrogenase [8], Na+-translocating ATPases (ATP synthases) [9, 10], and Na+-translocating pyrophosphatases [11]. The ΔµNa+ established by these enzymes can be used to drive chemical [10], osmotic [12], and mechanical [13] work, thus forming a sodium cycle of energy transduction [14].
Analysis of prokaryotic genomes reveals that the above-mentioned Na+-translocating enzymes are widely distributed among bacteria and archaea [15], especially among marine, pathogenic, and methanogenic microorganisms. It is not clear yet why the coupling ion is changed from H+ to Na+ in these organisms. It was supposed that the sodium cycle is advantageous for bacterial growth at alkaline pH as well as at other conditions decreasing ΔµH+ level [14]. The primacy of a sodium cycle in the course of the origin and evolution of energy-transducing enzymes is an alternative explanation [15].
The set of primary sodium pumps has been recently supplemented by an additional enzyme. It was earlier considered that different bacteriorhodopsins could function as either light-dependent H+ and Cl– pumps or light receptors [16]. However, it has been established recently that one of the proteorhodopsins of the marine flavobacterium Krokinobacter eikastus is capable of light-driven Na+ export from the cytoplasm to the external medium [17]. In the absence of sodium ions, this protein pumps protons with the same transport direction, while with significantly slower rate of the photocycle [17]. This specificity to coupling ion is similar to that of some Na+-translocating ATPases and pyrophosphatases [18]. Later, such Na+-proteorhodopsin was described in another marine flavobacterium, Nonlabens marinus [19]. Analysis of primary structures of Na+-proteorhodopsins reveals that they form a phylogenetically distinct group among all known retinal-binding proteins [20].
The study of Na+-translocating enzymes has some advantages compared to proton pumps. First of all, with Na+ pumps it is possible to manipulate freely by concentration of the coupling ion without influence on the stability of the enzyme, which is impossible with H+-translocating enzymes. Thus, it is possible to study the catalytic cycle of a Na+-dependent enzyme on limitation of its turnover rate by low Na+ concentration and to reveal the stages that are specifically activated by sodium ion. Accordingly, investigation of Na+-pumping proteorhodopsins is important not only for studying of sodium cycle, but it can also lead to significant progress in determination of the functioning mechanism of retinal-dependent energy transducing enzymes.
MATERIALS AND METHODS
Bacterial strains and growth conditions. Dokdonia sp. PRO95 was obtained from the Leibniz Institute collection of microorganisms and cell cultures (DSMZ). The Dokdonia sp. PRO95 cells were grown at 25°C in Difco™ Marine Broth 2216 as described in [21]. Escherichia coli cells were grown at 37°C in LB or TB medium. When necessary, the media were supplemented with 125 µg/ml ampicillin.
Construction of an expression vector bearing the gene encoding the C-terminal 6×His-tagged proteorhodopsin AEX55013 from Dokdonia sp. PRO95. The gene sequence encoding the AEX55013 protein was amplified by PCR with High Fidelity Tersus polymerase (Evrogen, Russia) and primer pair rhod_dir (5′-GCCATGGCACAAGAACTAGGA) and rhod_rev (5′-CGAGCTCCTATTTGCTTCGACTAG) using genomic DNA of Dokdonia sp. PRO95 as a template. The amplified 849-bp fragment was cloned into a pBAD-TOPO™ vector (Invitrogen, USA), resulting in the pTopo_Rhod_10 plasmid. To remove the N-terminal leader sequence provided to the cloned gene by the pBAD-TOPO vector, the pTopo_Rhod_10 construct was digested by NcoI and self-ligated accordingly to the pBAD-TOPO TA expression kit manual, resulting in the pRhod_10.1 plasmid. The cloned fragment was verified by DNA sequencing. The final plasmid pRhod_10.1 was transformed into E. coli BL21 cells.
Isolation of recombinant 6×His-tagged proteorhodopsin. For proteorhodopsin synthesis induction, E. coli/pRhod_10.1 cells were grown at 37°C in the TB medium to late exponential phase (A600 ≈ 1.5), after which the growth medium was supplemented with 0.15% (w/v) L-arabinose and 1.5 mg/liter all-trans retinal. The cells were further grown for 16 h at 15°C [22]. The cells were harvested by centrifugation (10,000g, 10 min) and washed twice with medium containing 300 mM KCl, 10 mM Tris-HCl, and 5 mM MgSO4 (pH 8.0). The cell pellet was suspended in medium A containing 300 mM KCl, 20 mM Tris-HCl, 5 mM MgSO4, 1 mM phenylmethylsulfonyl fluoride, and 5 mM imidazole-HCl (pH 8.0), and the suspension was passed twice through a French press (16,000 psi). Cell debris and unbroken cells were removed by centrifugation at 22,500g (10 min), and the supernatant was further centrifuged at 200,000g (75 min). The pellet was suspended in medium A, solubilized with 1.5% n-dodecyl β-D-maltoside (DDM), and centrifuged at 200,000g for 60 min. The 6×His-tagged proteorhodopsin was purified from the supernatant using affinity chromatography. This was accomplished by loading the supernatant onto a Ni-NTA column equilibrated with solution B containing 300 mM KCl, 10 mM Tris-HCl, 0.05% DDM, and 10 mM imidazole-HCl (pH 8.0); washing the column twice, first with solution B and then with solution B containing 100 mM imidazole-HCl; and eluting the 6×His-tagged protein with solution B containing 250 mM imidazole-HCl. The protein obtained was concentrated and kept frozen at –80°C until use. Protein concentration was determined by the bicinchoninic acid method [23] using bovine serum albumin as a standard.
Measurement of light-driven proton transport in E. coli cells. The induced E. coli/pRhod_10.1 cells (R-E. coli) were collected by centrifugation (10,000g, 10 min), washed twice with medium containing 100 mM Na2SO4 (or K2SO4), 10 mM Tris-HCl, and 5 mM MgSO4 (pH 8.0), suspended in the same medium, and kept anaerobically at 4°C for 16 h. This procedure resulted in depletion of endogenous substrates and in loading of the cells by the desired cation (Na+ or K+). The depleted cells were washed twice with 100 mM Na2SO4 (or K2SO4) and resuspended in the same solvent at protein concentration 2 mg/ml. The suspension was placed in a 1.4-ml chamber equipped with a pH electrode, and the pH level was adjusted to pH 7.0 with NaOH (or KOH). The chamber was placed in darkness and then illuminated for 3 min using a 100 W halogen lamp. The pH electrode was calibrated with argon-saturated H2SO4 solution.
Measurement of light-driven Na+ transport in R-E. coli cells. The depleted, Na+-loaded R-E. coli cells were prepared as described above. The suspension was placed in darkness and then illuminated using a 100 W halogen lamp. Then the cell suspension was filtered (45 µm; Millipore, USA), the filter was washed with 5 ml 0.5 M mannitol solution, and the sodium content on the filter was measured by flame photometry [24].
RESULTS AND DISCUSSION
We chose “putative xanthorhodopsin” from the marine flavobacterium Dokdonia sp. PRO95 (GenBank accession number AEX55013) as a model object for studying Na+-translocating proteorhodopsin. The primary structure of this protein is highly similar to those of two known Na+-pumping proteorhodopsins of K. eikastus (98% identity) and N. marinus (84% identity). The amino acid sequence of the protein from Dokdonia sp. PRO95 as well as sequences of known Na+-pumping proteorhodopsins contains characteristic substitutions of conserved glutamate residues involved in transmembrane proton translocation in H+-pumping bacteriorhodopsins, Glu85Asn and Glu96Gln (numbering is in accordance to bacteriorhodopsin from Halobacterium salinarium), i.e. the so-called NQ-motif [20]. Thus, analysis of the primary structure of proteorhodopsin AEX55013 points to Na+ as the coupling ion of this light-driven pump. This along with growth characteristics of Dokdonia sp. PRO95 and availability of the complete genome sequence of this bacterium [21] makes the protein an attractive object for study of the mechanism of light-driven transmembrane Na+ translocation.
To study proteorhodopsin from Dokdonia sp. PRO95, the gene of this protein was cloned into expression vector pBAD-TOPO™. The resulting construct was transformed into E. coli BL21 cells. Upon induction of the cloned gene expression in the presence of all-trans retinal, the cells became purple-colored, which indicated a high production level of a retinal-binding protein.
Ion-pumping properties of the produced protein were studied by measuring light-dependent transmembrane H+ translocation. Earlier, on studying Na+-translocating NADH:quinone-oxidoreductase from Vibrio alginolyticus, it was shown that addition of O2 to an anaerobic cell suspension of this bacterium in the presence of an uncoupler gives rise to an alkalization of the medium [25]. This effect was interpreted as electrophoretic H+ uptake supported by Δψ generated by the transmembrane Na+ transport. Later, this method was successfully applied for studying various ion-translocating enzymes including different rhodopsins [17, 19, 26, 27].
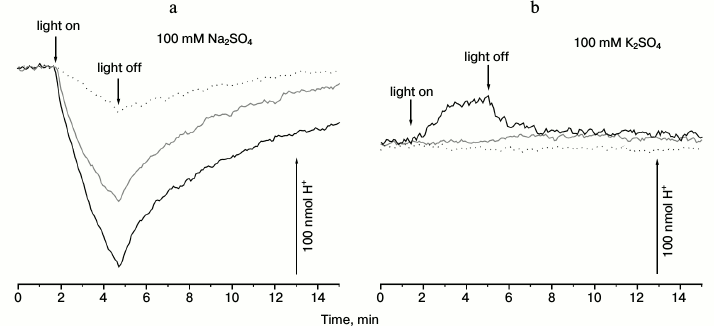
As can be seen from Fig. 1a (gray line), 3-min illumination of an anaerobic suspension of R-E. coli cells in Na2SO4 solution resulted in progressive alkalinization of the medium, which decayed to the initial pH values after switching the light off. Illumination of control E. coli cells, which did not contain the pRhod_10.1 plasmid, did not lead to any pH changes (data not shown). Both the rate and amplitude of the light-dependent response of R-E. coli cells were increased significantly by the uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) (Fig. 1a, black line). Addition of the penetrating anion SCN– resulted in substantial inhibition of the pH changing (Fig. 1a, dotted line), indicating that the alkalization is a passive Δψ-driven H+ uptake. These results suggest the ability of proteorhodopsin from Dokdonia sp. PRO95 to pump some other ion different from H+.
Fig. 1. Light-dependent ΔpH generation by E. coli cells producing proteorhodopsin from Dokdonia sp. PRO95. Anaerobic suspension of R-E. coli cells (2 mg protein per ml) in 100 mM Na2SO4 (a) or 100 mM K2SO4 (b) was illuminated using a 100 W halogen lamp for 3 min. a) Gray line, without addition; black line, in the presence of 20 µM CCCP; dotted line, in the presence of 20 µM CCCP and 25 mM KSCN. b) Gray line, without addition; black line, in the presence of 25 mM KSCN; dotted line, in the presence of 25 mM KSCN and 20 µM CCCP.
In contrast, illumination of the R-E. coli suspension in a sodium-free medium (in K2SO4 solution) did not lead to pH changes (Fig. 1b, gray line). Addition of penetrating anion SCN– resulted in a small light-dependent acidification of the medium (Fig. 1b, black line). This response was fully inhibited by the uncoupler CCCP (Fig. 1b, dotted line). As the media used (Fig. 1a versus Fig. 1b) differed only in the cation (Na+ or K+), we conclude that the proteorhodopsin from Dokdonia sp. PRO95 pumps Na+ in sodium-containing media. In the absence of sodium ions, this protein is able to use the proton as the coupling ion. The H+-translocating activity of the proteorhodopsin from Dokdonia sp. PRO95 was less expressed compared to the Na+-translocating proteorhodopsin from K. eikastus [17]; in this respect, the studied protein is more similar to its analog from N. marinus [19].
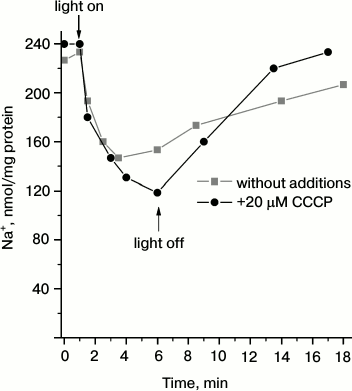
To test the conclusion on Na+-pumping activity of Dokdonia sp. PRO95 proteorhodopsin, we measured light-dependent sodium transport in R-E. coli cells. As shown in Fig. 2, illumination of the R-E. coli cells resulted in progressive decrease in cytoplasmic Na+ concentration. This process was slightly accelerated by CCCP, or it was at least resistant to this uncoupler. Thus, it is impossible to attribute the measured transport to any secondary process like activity of a Na+/H+-antiporter. It is noteworthy that either medium alkalization in the presence of CCCP (Fig. 1a) or Na+ transport (Fig. 2) had similar kinetics. Moreover, both these processes had similar amplitudes for the first 3 min of illumination (~90 nmol H+ per mg protein and ~110 nmol Na+ per mg protein, respectively). This supports the assumption that light-dependent CCCP-activating alkalinization of the medium (Fig. 1a) is caused by active transmembrane Na+ transport. Thus, the data indicate that proteorhodopsin from Dokdonia sp. PRO95 is a primary sodium pump.
Fig. 2. Light-dependent Na+ transport in E. coli cells producing proteorhodopsin from Dokdonia sp. PRO95. An anaerobic suspension of Na+-loaded R-E. coli cells in 100 mM Na2SO4, 10 mM Tris-HCl (pH 8.0), and 5 mM MgSO4 was illuminated using a 100 W halogen lamp in the absence (squares) or presence (circles) of 20 µM CCCP.
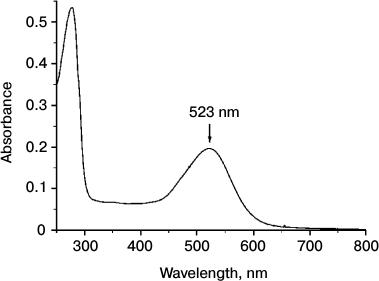
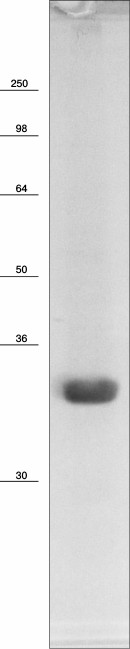
Na+-translocating proteorhodopsin was isolated from R-E. coli cells by one-step affinity chromatography purification yielding about 3 mg of purified protein per liter of cell culture. As shown in Fig. 3, the electronic absorption spectrum of the preparation showed maxima at 279 and 523 nm. The short-wavelength peak corresponds to absorption by aromatic amino acid residues, while the long-wavelength peak is due to light absorption by the protein-bound retinal. The maximum for the latter peak at 523 nm is quite similar to the spectral properties of known Na+-translocating proteorhodopsins [17, 19]. Relatively low ratio of amplitudes of the peaks (A279/A523 = 2.7) indicates high purity of the preparation [22]. It is noteworthy that the absorption spectrum of the proteorhodopsin preparation did not contain any additional peaks, in particular in the area of γ-peaks of cytochromes (400-450 nm). This indicates absence of the bo-type quinol oxidase in the preparation, which is the main contaminant at 6×His-tagged proteins purification from E. coli membranes. High purity of the proteorhodopsin preparation was confirmed by SDS-PAGE analysis. As seen in Fig. 4, SDS-PAGE revealed only one protein band with apparent molecular mass of about 33 kDa (calculated mass of the 6×His-tagged proteorhodopsin, 34.8 kDa). These data demonstrate both spectral and protein content purity of the preparation, which is sufficient for further study.
Fig. 3. Electronic absorption spectrum of recombinant proteorhodopsin from Dokdonia sp. PRO95 purified from E. coli cells. The spectrum was determined in medium containing 20 mM KH2PO4 (pH 7.0) and 0.05% DDM.
Fig. 4. SDS-PAGE analysis of recombinant proteorhodopsin from Dokdonia sp. PRO95 purified from E. coli cells. Coomassie Blue-stained gel, the bars with numbers on the left side denote positions and molecular masses of marker proteins (in kDa).
Thus, we conclude that proteorhodopsin AEX55013 from the marine flavobacterium Dokdonia sp. PRO95 is a primary light-dependent sodium pump. A high level of heterologous production of this protein in E. coli cells as well as stability and purity of the purified preparations makes this protein a useful model for investigation of the mechanism of active transmembrane Na+ translocation.
This work was supported by the RSCF research project 14-14-00128.
REFERENCES
1.Dimroth, P. (1980) A new sodium-transport system
energized by the decarboxylation of oxaloacetate, FEBS Lett.,
122, 234-236.
2.Dimroth, P., Jockel, P., and Schmid, M. (2001)
Coupling mechanism of the oxaloacetate decarboxylase Na+
pump, Biochim. Biophys. Acta, 1505, 1-14.
3.Unemoto, T., and Hayashi, M. (1979) NADH:quinone
oxidoreductase as a site of Na+-dependent activation in the
respiratory chain of marine Vibrio alginolyticus, J.
Biochem., 85, 1461-1467.
4.Verkhovsky, M. I., and Bogachev, A. V. (2010)
Sodium-translocating NADH:quinone oxidoreductase as a redox-driven ion
pump, Biochim. Biophys. Acta, 1797, 738-746.
5.Schmehl, M., Jahn, A., Meyer zu Vilsendorf, A.,
Hennecke, S., Masepohl, B., Schuppler, M., Marxer, M., Oelze, J., and
Klipp, W. (1993) Identification of a new class of nitrogen fixation
genes in Rhodobacter capsulatus: a putative membrane complex
involved in electron transport to nitrogenase, Mol. Gen. Genet.,
241, 602-615.
6.Muller, V., Imkamp, F., Biegel, E., Schmidt, S.,
and Dilling, S. (2008) Discovery of a
ferredoxin:NAD+-oxidoreductase (Rnf) in Acetobacterium
woodii. A novel potential coupling site in acetogens, Ann. N. Y.
Acad. Sci., 1125, 137-146.
7.Gottschalk, G., and Thauer, R. K. (2001) The
Na+-translocating methyltransferase complex from
methanogenic archaea, Biochim. Biophys. Acta, 1505,
28-36.
8.Kaesler, B., and Schonheit, P. (1989) The role of
sodium ions in methanogenesis. Formaldehyde oxidation to CO2
and 2H2 in methanogenic bacteria is coupled with primary
electrogenic Na+ translocation at a stoichiometry of 2-3
Na+/CO2, Eur. J. Biochem., 184,
223-232.
9.Heefner, D. L., and Harold, F. M. (1982) ATP-driven
sodium pump in Streptococcus faecalis, Proc. Natl. Acad. Sci.
USA, 79, 2798-2802.
10.Kluge, C., Laubinger, W., and Dimroth, P. (1992)
The Na+-translocating ATPase of Propionigenium
modestum, Biochem. Soc. Trans., 20, 572-577.
11.Malinen, A. M., Belogurov, G. A., Baykov, A. A.,
and Lahti, R. (2007) Na+-pyrophosphatase: a novel primary
sodium pump, Biochemistry, 46, 8872-8878.
12.Kakinuma, Y., and Unemoto, T. (1985) Sucrose
uptake is driven by the Na+ electrochemical potential in the
marine bacterium Vibrio alginolyticus, J. Bacteriol.,
163, 1293-1295.
13.Dibrov, P. A., Kostyrko, V. A., Lazarova, R. L.,
Skulachev, V. P., and Smirnova, I. A. (1986) The sodium cycle. I.
Na+-dependent motility and modes of membrane energization in
the marine alkalotolerant Vibrio alginolyticus, Biochim.
Biophys. Acta, 850, 449-457.
14.Skulachev, V. P. (1989) The sodium cycle: a novel
type of bacterial energetics, J. Bioenerg. Biomembr., 21,
635-647.
15.Mulkidjanian, A. Y., Dibrov, P., and Galperin, M.
Y. (2008) The past and present of the sodium energetics: may the
sodium-motive force be with you, Biochim. Biophys. Acta,
1777, 985-992.
16.Grote, M., Engelhard, M., and Hegemann, P. (2014)
On ion pumps, sensors and channels – perspectives on
microbial rhodopsins between science and history, Biochim. Biophys.
Acta, 1837, 533-545.
17.Inoue, K., Ono, H., Abe-Yoshizumi, R., Yoshizawa,
S., Ito, H., Kogure, K., and Kandori, H. (2013) A light-driven sodium
ion pump in marine bacteria, Nat. Commun., 4, 1678.
18.Luoto, H. H., Nordbo, E., Baykov, A. A., Lahti,
R., and Malinen, A. M. (2013) Membrane Na+-pyrophosphatases
can transport protons at low sodium concentrations, J. Biol.
Chem., 288, 35489-35499.
19.Yoshizawa, S., Kumagai, Y., Kim, H., Ogura, Y.,
Hayashi, T., Iwasaki, W., DeLong, E. F., and Kogure, K. (2014)
Functional characterization of flavobacteria rhodopsins reveals a
unique class of light-driven chloride pump in bacteria, Proc. Natl.
Acad. Sci. USA, 111, 6732-6737.
20.Kwon, S. K., Kim, B. K., Song, J. Y., Kwak, M.
J., Lee, C. H., Yoon, J. H., Oh, T. K., and Kim, J. F. (2013) Genomic
makeup of the marine flavobacterium Nonlabens
(Donghaeana) dokdonensis and identification of a novel
class of rhodopsins, Genome Biol. Evol., 5, 187-199.
21.Riedel, T., Gomez-Consarnau, L., Tomasch, J.,
Martin, M., Jarek, M., Gonzalez, J. M., Spring, S., Rohlfs, M.,
Brinkhoff, T., Cypionka, H., Goker, M., Fiebig, A., Klein, J.,
Goesmann, A., Fuhrman, J. A., and Wagner-Dobler, I. (2013) Genomics and
physiology of a marine flavobacterium encoding a proteorhodopsin and a
xanthorhodopsin-like protein, PLoS One, 8, e57487.
22.Gourdon, P., Alfredsson, A., Pedersen, A.,
Malmerberg, E., Nyblom, M., Widell, M., Berntsson, R., Pinhassi, J.,
Braiman, M., Hansson, O., Bonander, N., Karlsson, G., and Neutze, R.
(2008) Optimized in vitro and in vivo expression of
proteorhodopsin: a seven-transmembrane proton pump, Protein Expr.
Purif., 58, 103-113.
23.Smith, P. K., Krohn, R. I., Hermanson, G. T.,
Mallia, A. K., Gartner, F. H., Provenzano, M. D., Fujimoto, E. K.,
Goeke, N. M., Olson, B. J., and Klenk, D. C. (1985) Measurement of
protein using bicinchoninic acid, Anal. Biochem., 150,
76-85.
24.Verkhovskaya, M. L., Verkhovsky, M. I., and
Wikstrom, M. (1996) The respiration-driven active sodium transport
system in E. coli does not function with lithium, FEBS
Lett., 388, 217-218.
25.Tokuda, H., and Unemoto, T. (1982)
Characterization of the respiration-dependent Na+ pump in
the marine bacterium Vibrio alginolyticus, J. Biol.
Chem., 257, 10007-10014.
26.Bogachev, A. V., Murtasina, R. A., and Skulachev,
V. P. (1997) The Na+/e– stoichiometry of
the Na+-motive NADH:quinone oxidoreductase in Vibrio
alginolyticus, FEBS Lett., 409, 475-477.
27.Avetisyan, A. V., Kaulen, A. D., Skulachev, V.
P., and Feniouk, B. A. (1998) Photophosphorylation in alkalophilic
halobacterial cells containing halorhodopsin: chloride-ion cycle?
Biochemistry (Moscow), 63, 625-628.