Ancient Systems of Sodium/Potassium Homeostasis as Predecessors of Membrane Bioenergetics
D. V. Dibrova1, M. Y. Galperin2, E. V. Koonin2, and A. Y. Mulkidjanian1,3,4*
1Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119992 Moscow, Russia2National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland 20894, USA
3School of Physics, Osnabrueck University, 49069 Osnabrueck, Germany; E-mail: amulkid@uos.de
4School of Bioengineering and Bioinformatics, Lomonosov Moscow State University, 119992 Moscow, Russia
* To whom correspondence should be addressed.
Received December 22, 2014; Revision received January 26, 2015
Cell cytoplasm of archaea, bacteria, and eukaryotes contains substantially more potassium than sodium, and potassium cations are specifically required for many key cellular processes, including protein synthesis. This distinct ionic composition and requirements have been attributed to the emergence of the first cells in potassium-rich habitats. Different, albeit complementary, scenarios have been proposed for the primordial potassium-rich environments based on experimental data and theoretical considerations. Specifically, building on the observation that potassium prevails over sodium in the vapor of inland geothermal systems, we have argued that the first cells could emerge in the pools and puddles at the periphery of primordial anoxic geothermal fields, where the elementary composition of the condensed vapor would resemble the internal milieu of modern cells. Marine and freshwater environments generally contain more sodium than potassium. Therefore, to invade such environments, while maintaining excess of potassium over sodium in the cytoplasm, primordial cells needed means to extrude sodium ions. The foray into new, sodium-rich habitats was the likely driving force behind the evolution of diverse redox-, light-, chemically-, or osmotically-dependent sodium export pumps and the increase of membrane tightness. Here we present a scenario that details how the interplay between several, initially independent sodium pumps might have triggered the evolution of sodium-dependent membrane bioenergetics, followed by the separate emergence of the proton-dependent bioenergetics in archaea and bacteria. We also discuss the development of systems that utilize the sodium/potassium gradient across the cell membranes.
KEY WORDS: ATP synthase, sodium-motive force, proton-motive force, sodium symporter, membrane efflux pumps, abiogenesis, anoxic geothermal fieldsDOI: 10.1134/S0006297915050016
Abbreviations: KREEP, Potassium [K], Rare Earth Elements and Phosphorus; LUCA, Last Universal Cellular Ancestor; P-loop, phosphate-binding loop – amino acid sequence (in nucleotide-binding proteins) also known as a Walker A-motif; Δψ, transmembrane potential.
We dedicate this paper to Professor Vladimir P. Skulachev on the
occasion of his 80th birthday and in appreciation of his seminal
contribution to the field of science that, many years ago, has been
named bioenergetics at his suggestion.
Some 40 years ago, Vladimir Skulachev has suggested that prokaryotic cells might use the Na+/K+ gradient across the cell membrane for storing energy [1]. By that time, the chemiosmotic hypothesis of Peter Mitchell [2] was already generally accepted, owing largely to the experimental evidence provided by Skulachev and his co-workers. The transmembrane proton gradient was recognized as a major convertible energy source within the cell [3]. However, the energy-buffering capacity of proton gradient was clearly very low. Because of the much higher physiological concentrations of Na+ and K+ ions compared to the H+ ions, the energy storage capacity of the Na+/K+ gradient could be greater than that of the proton gradient by a factor of 105 [1, 4]. In support of this idea, the ability of several bacteria and archaea to maintain cell motility and the ATP levels have been shown by Skulachev and co-workers to last for much shorter time intervals under anoxic conditions in a K+-rich medium, in the absence of a Na+/K+ gradient, than in a Na+-rich medium. In these experiments, the cellular ATP stock was depleted completely in a K+-rich medium but only halved in a Na+-rich medium [4].
Subsequently, in the 1980s, Skulachev published a series of seminal articles in which he argued that in certain bacteria, the Na+ gradient, similarly to the H+ gradient in others, could perform the role of the membrane-linked convertible energy currency [5, 6]. In the following years, the phenomenon of sodium-dependent bioenergetics had been firmly established as one of the pillars of biological energy conversion [7, 8]. More recently, based on structural and phylogenomic analyses, we reported observations indicating that sodium-dependent bioenergetics is not an exotic adaptation found in some alkaliphilic and/or thermophilic prokaryotes but a widespread and likely primordial mode of membrane bioenergetics [9-12]. Given the fundamental difference in the mechanisms of sodium and proton transfer across the lipid bilayer [13, 14] and following the insightful suggestion by Skulachev that sodium-tight membranes could be easier to build than proton-tight membranes [15], we reasoned that sodium-tight membranes evolved earlier than proton-tight membranes, accounting for the evolutionary primacy of the sodium-dependent bioenergetics [10-12]. This conjecture was validated by the reconstruction of the ancestral sequences of bacterial and archaeal sodium- and proton-translocating rotary membrane ATPases that supported the ancestral status of the amino acid ligands required for sodium translocation [10].
While the Na+/K+ gradient across the cell membrane is universal for all cellular organisms including eukaryotes, Na+-dependent energetics, which is characteristic only of a limited number of bacteria and archaea, had been traditionally considered an exotic property of alkaliphiles and/or hyperthermophiles [16]. Accordingly, Na+-dependent energetics and maintenance of the cellular Na+/K+ homeostasis have been usually treated as two separate phenomena. Not surprisingly, the Na+/K+ homeostasis, e.g. in an animal cell, used to be considered separately from the mitochondrial proton-dependent energetics of the same cell. In this paper, we argue that all these phenomena have a common origin in ancient membrane systems that were responsible for extruding sodium ions out of the primordial cells. First, we discuss how interplay between several, initially independent sodium pumps that were involved in the maintenance of the Na+/K+ gradient across the membrane of the primordial cells could have yielded Na+-dependent membrane bioenergetics, which later evolved into the proton-dependent bioenergetics. Then, we briefly consider how cells “learned” to utilize the Na+/K+ gradient for their needs.
PREVALENCE OF K+ IONS OVER Na+ IONS IN THE
CYTOPLASM
It is well known that the cytoplasm of living cells, generally, contains more potassium ions than sodium ions [17-19]. This trait seems to be universal: [K+]/[Na+] values of less than unity have been reported only for two types of organisms, namely marine picobacteria [20] and halophilic bacteria [21]. The causes of these deviations from the general trend have been analyzed [20, 22]. Several factors have been noted that could have led to an underestimation of the cellular [K+]/[Na+] ratios. First, the measured values of the [K+]/[Na+] ratios could be underestimated owing to the contribution from the sodium ions bound on the cell surface, which could not be fully excluded as long as the bacteria were grown at high sodium levels. Second, [K+]/[Na+] > 1 is a property of actively metabolizing cells in the exponential growth phase; when the cells reach the stationary phase, this ratio drops dramatically and can even be reversed [22]. Keeping the cells actively metabolizing during their separation from the medium to ensure preservation of their content is a common problem for all measurements of membrane transport; in the case of potassium and sodium ions, this problem is even more complicated than for sugars or amino acids. When the cell metabolism is compromised by centrifugation, fixation, and other treatments, the cytosolic levels of K+ and Na+ would tend to equilibrate with those in the surrounding milieu. Importantly, the contribution of these two factors increases with the increase of the surface-to-volume ratio, i.e. is the greatest for smaller-sized cells. It is hardly surprising that the [K+]/[Na+] ratios of less than unity have been reported only for smallest, halotolerant prokaryotic cells. Hence, [K+]/[Na+] ratios in the 0.1-1.0 range, as reported for the halophilic and marine bacteria [20-22], can be safely assumed to be underestimations of the true magnitude of the K+/Na+ gradient in active cells (A. Oren, personal communication). Generally, there is overwhelming evidence that K+ levels in all actively growing cells are substantially higher than the levels of Na+ ions.
EMERGENCE OF THE FIRST CELLS IN K+-RICH
ENVIRONMENTS
The evolutionary importance of the potassium prevalence in the cytosol had been first noted by Canadian biochemist Archibald Macallum in his seminal paper where he formulated the principles of evolutionary chemistry (paleochemistry) of living organisms with specific application to the inorganic biochemistry of the cell [17]. Macallum argued that: “As the cell is older than its media as at present constituted, the relative proportions of the inorganic elements which obtain in it are of more ancient origin than the relative proportions of the same elements which prevail in the media, blood plasmas, and lymph or in the ocean and river water of today”. Based on the data on the prevalence of potassium over sodium in cellular tissues, Macallum suggested that the first cells might have emerged in K+-rich habitats [17]. Further studies have shown that K+ ions prevail over Na+ ions not only in eukaryotic cells studied by Macallum, but also, as discussed above, in the cytoplasm of bacteria and archaea ([18] and references therein).
The high [K+]/[Na+] ratio in the vast majority of prokaryotes implies prevalence of K+ over Na+ also in the cytoplasm of the common ancestor of archaea and bacteria, the radiation of which dates back more than 3.5 Gy [23]. Importantly, the cytoplasmic [K+]/[Na+] > 1 does not appear to be a simple “frozen” evolutionary accident. It is well established that prevalence of K+ ions is crucial for the activity of numerous (nearly) universal, key enzymes, including the translation factor EF-Tu [24], recombinase RadA [25], chaperonin GroEL/Hsp60 [26], diol dehydratases, including glycerol dehydratase [27], pyruvate kinase [28], S-adenosylmethionine synthase [29], dialkylglycine decarboxylase [30], membrane pyrophosphatase [31], CDP-diglyceride synthase [32], and many others. Accordingly, K+ ions are required for some core cellular processes, most importantly, for protein synthesis [33]. Potassium cations have been shown to help ribosomes to attain functional conformation [34], which might be related to the specific ability of K+ ions to stabilize certain RNA folds ([35] and references therein). In addition, the X-ray structure of the ribosome has revealed a K+ ion within the peptidyl transferase center [36, 37], which, according to several independent reconstructions, makes the evolutionarily oldest part of the ribosome [38-40]. The protein synthesis in the very first cells must have already depended on the presence of K+ ions as the peptidyl transferase machinery must have already evolved by that stage [39, 41], well before the Last Universal Cellular Ancestor (LUCA).
The LUCA is the life form that existed immediately before the separation of the cellular life into bacterial and archaeal domains. The number of protein-coding genes that are common to (nearly) all extant cellular life forms and thus most likely were represented in LUCA initially decreased with more genomes being sequenced but stopped at the mark of about 60 [41-43]. The functional distribution of these genes is remarkably different from uniform and provides some information about the LUCA (see Table 1). The great majority of the ubiquitous proteins are involved in translation [41, 44]. These include proteins of large and small ribosome subunits, translation factors, and aminoacyl-tRNA synthetases of all specificities [42, 43]. Thus, it appears that the translation machinery of the LUCA was (almost) as complex as the translation systems of modern bacteria and archaea. Furthermore, the translation system of the LUCA most likely produced large (including multidomain) proteins because the molecular chaperone GroEL/Hsp60 that is essential for the folding of such proteins also belongs to the ubiquitous protein set. The functions of most of the other genes in the ubiquitous set are connected with nucleic acids (RecA/RadA recombinase, transcription antitermination factor NusG, transcription pausing factor NusA, 5′-3′ exonuclease, topoisomerase IA, clamp loader ATPase, pseudouridylate synthase) or their building blocks, nucleotides (thymidylate kinase, catalytic subunits of the rotary membrane ATPase).
Table 1. Products of ubiquitous genes and
their association with essential inorganic cations and anions
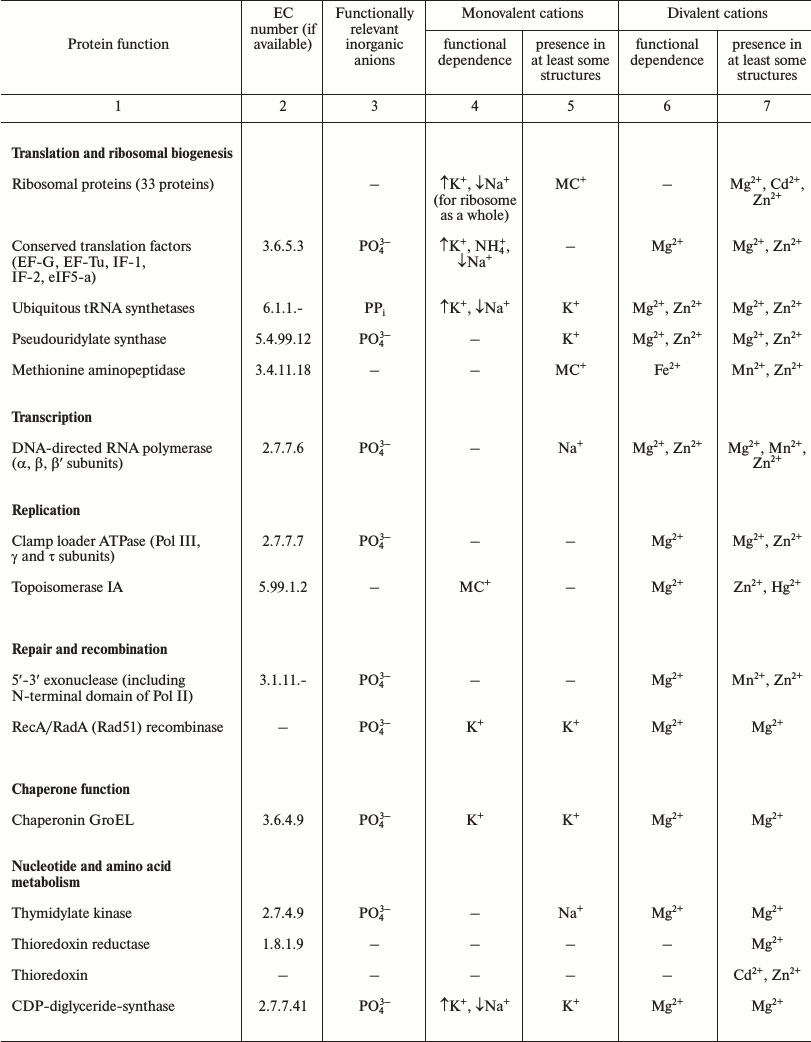
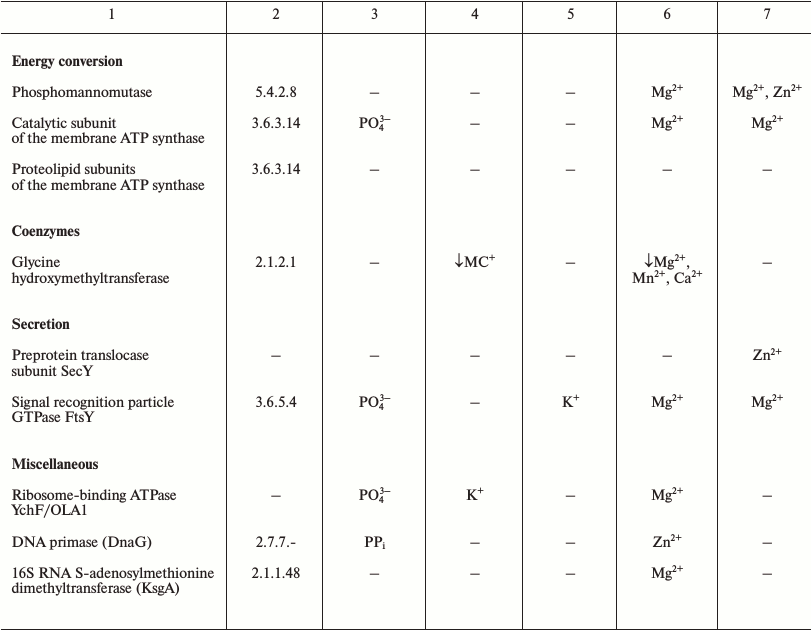
Note: The lists of ubiquitous genes were extracted from refs. [42, 43]. The data on the
dependence of functional activity on particular metals were taken from
the BRENDA database [155]. According to the
BRENDA database, the enzymatic activity of most
Mg2+-dependent enzymes could be routinely restored by
Mn2+. As concentration of Mg2+ ions in the cell
is ca. 10–2 M, whereas that of Mn2+
ions is ca. 10–6 M, the data on the functional
importance of Mn2+ were not included in the table for many
enzymes. The presence of metals in protein structures is as listed in
the respective PDB entries. MC stands for “monovalent
cation”. The table includes all enzymes represented by orthologs
in all cellular life forms as well as several cases when the function
is ubiquitous (e.g. DNA polymerase, DNA primase) but the enzymes
responsible for that function are represented by two or more
non-orthologous forms [41]. Upward arrows indicate
activation by the particular ion and downward arrows indicate
inhibition by this ion. For the references and further details, see the
supplementary material to [18].
The available information on the dependence of the activities of these ancient proteins on different inorganic ions shows that, in addition to the preference for Zn2+ and Mn2+ as transition metals, which has been detected and discussed previously [45, 46], several proteins and functional systems specifically require K+, whereas none of the surveyed ancestral proteins specifically requires Na+ (see Table 1 and [18]).
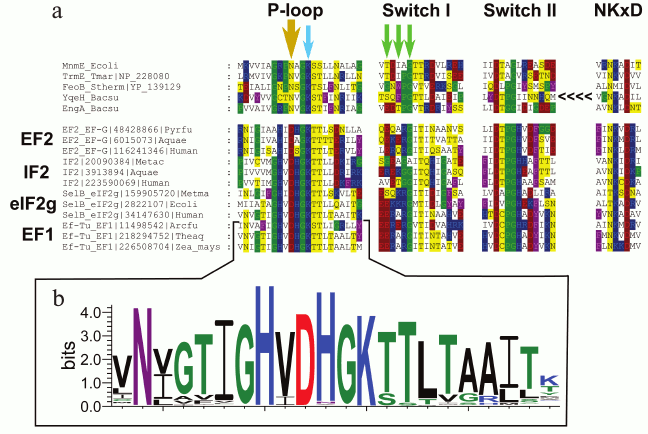
In particular, comparative structural analysis of several translation factors that belong to the P-loop GTPase superfamily allowed prediction of a conserved aspartate residue in the P-loop region, which likely plays a major role in potassium binding. This residue is absolutely conserved in the inspected translation factors including EF-Tu/EF1, which is known to be K+-dependent [24] (Fig. 1). The corresponding residue in other P-loop GTPases with known potassium dependence [47-51] is asparagine (see Fig. 1a, yellow arrow, and Fig. 2), but mutation of this residue to aspartate in the Bacillus subtilis GTPase YqeH has been shown to affect GTPase activity only slightly [47].
Fig. 1. Multiple alignment of potassium-dependent GTPases and translation factors from archaea, bacteria, and eukaryotes. a) Amino acid residues are colored according to the chemical nature of their side chains (basic residues are in blue, acidic residues are in red, polar residues are in yellow, nonpolar aliphatic residues are in white, aromatic residues are in violet, proline and glycine residues are in green). The bold yellow arrow shows the key residue that is important for K+ binding (the Asn→Asp substitution in this position in Bacillus subtilis YqeH did not affect its GTPase activity, whereas Asn→Leu or Asn→Gln replacements abolished both the activity and its potassium-dependence [47]). One of the green arrows shows the glycine residue that is not directly involved in the binding, but seems to provide the required flexibility of the chain in this region. The two residues marked with further green arrows participate in binding through their backbone oxygen atoms, which does not constrain the nature of the respective residues. The YqeH protein is circularly permutated and its NKxD motif is located at the N-terminus of the protein (shown by the <<<< sign). The multiple alignment was constructed using MUSCLE [147] and visualized with GeneDoc. b) The Logo diagram for 174 EF-Tu and EF1 proteins sampled from the members of COG0050 (EF-Tu) and COG5256 (EF1) in the COG database [148]. The logo was constructed for the positions from the –12 to +9 relative to the position of the conserved lysine residue in the Walker A motif using the WebLogo tool [149]. The aspartic acid residue in the position –3 is absolutely conserved within this family.
In a previous phylogenetic analysis of the P-loop GTPase superfamily [52], several protein families, including the translation factors, have been predicted to descend from the LUCA. The Asn residue that corresponds to the potassium-binding Asp residue is mostly conserved in these proteins including, in addition to the translation factors, Obg, YyaF/Ygr210, HflX, YyaW families (see Fig. 2 in [52]). Notably, one of these ancient GTPase families, the Obg-like proteins, has been subsequently shown to play an essential role in ribosome maturation [53]. Thus, our comparative analysis of the sequences and structures of the P-loop GTPases indicates that in this ancient protein superfamily, the activation of catalysis by potassium ions is the ancestral trait that was independently supplanted in several lineages by substrate activation via positively charged amino acid side chains.
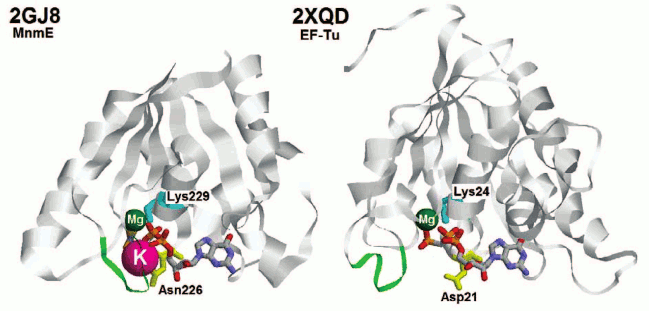
Fig. 2. Structures of the P-loop GTPase domains of MnmE protein (left) and translation elongation factor Tu (right). Proteins are shown in ribbon representation. GDP and AlF3 (together forming a GTP analog) in the PDB entry 2GJ8 [48] and GCP (guanosine 5′-(α,β-methylene) triphosphate, a GTP analog) in the PDB entry 2XQD [150] are colored by atoms while the metal ions are shown as spheres (Mg2+ in green, K+ in pink). The green loops correspond to the region marked with green arrows in Fig. 1 (Switch I region). The lysine residues of the P-loops are shown in cyan. The Asn residue responsible for the binding of the K+ ion in the MnmE structure is shown in yellow. The corresponding Asp residue in the EF-Tu structure is also colored yellow.
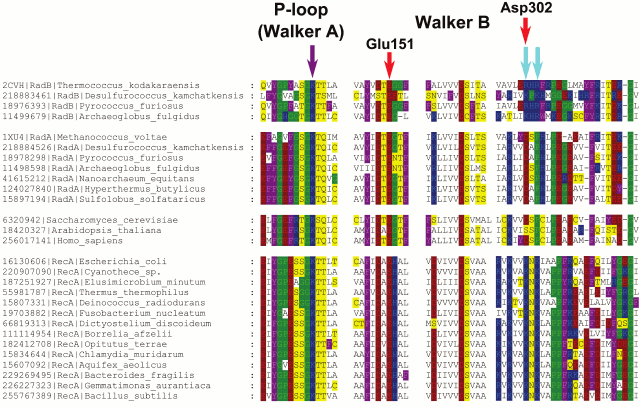
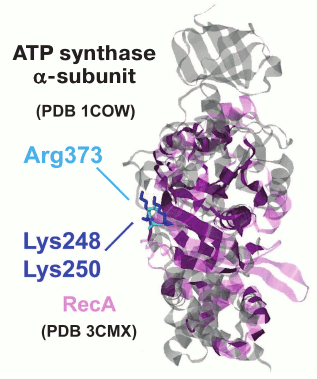
We performed phylogenomic analysis for several additional enzymes from the group of potassium-dependent ubiquitous proteins. One example of such proteins includes RecA/RadA ATPases, which belong to a distinct superfamily of P-loop NTPases that is only distantly related to the P-loop GTPases. This superfamily of ATPases has been denoted ASCE (Additional Strand, Catalytic E) ATPases because they encompass an additional β-strand between the P-loop (Walker A) and the Walker B motifs and possess a conserved, proton-abstracting glutamate residue that is involved in phosphoanhydride bond hydrolysis [54]. The ASCE superfamily also includes the catalytic subunits of the membrane rotary ATPases. The activity of the RadA protein from the archaeon Methanococcus voltae shows strong potassium dependence [25]. The crystal structure of RadA (PDB entry 1XU4) contains two binding sites for potassium ions, one of which is bound in the same orientation relative to ATP and Mg2+ as in the P-loop GTPases. Interestingly, in the absence of K+, the binding sites are occupied by water molecules, as observed previously for FeoB GTPase [50]. The bacterial homolog of RadA is the RecA recombinase. A comparison of the crystal structures of RadA from Methanococcus maripaludis (PDB ID 3EW9) and E. coli RecA [55] shows that, after superposition of the P-loops, the K+ ions bound by RadA overlay the lysine residues K248 and K250 of RecA, which are important for catalysis [56]. Although these residues are located on the surface of the RecA monomer that is far away from the ATP binding site, they are inserted into the ATP binding site of the next monomer in the 3D structure of the complex with DNA.
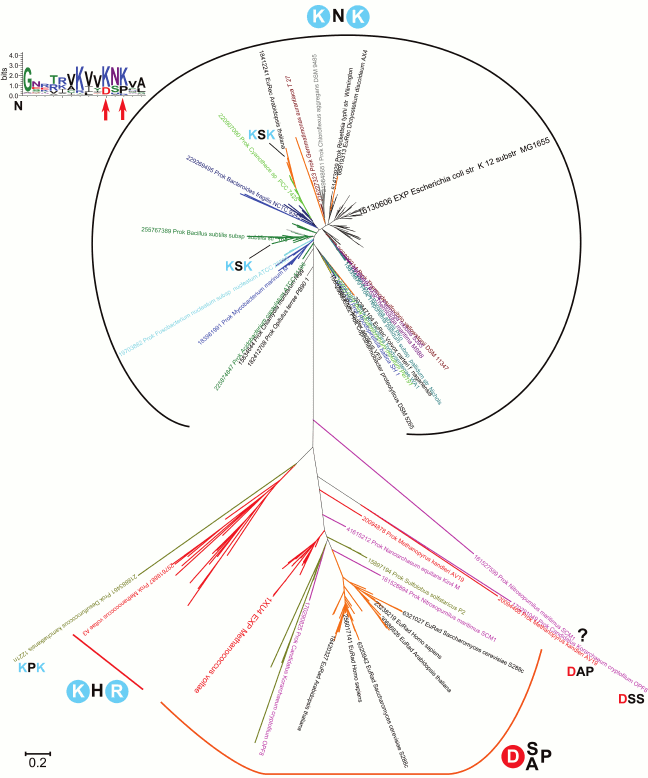
A phylogenetic tree for the RecA/RadA superfamily shows that most of the bacterial proteins contain the pair of lysine residues, whereas archaea and eukaryotes (except for those few that apparently received this protein as a result of lateral gene transfer from bacteria) have a conserved aspartic acid (Asp302 in M. voltae) in the same position in the alignment (see Fig. 3 for the schematic representation of the tree and Fig. 4 for a multiple alignment). This residue has been shown to directly bind one potassium ion with its side chain [25]. The strict distinction between the bacterial and archaea/eukaryotic clades precludes unequivocal inference of the ancestral state, but there is at least one indication in favor of the primacy of the potassium-binding site. All euryarchaeal genomes encode two proteins of this family, one of which groups with eukaryotic and other archaeal sequences (RadA), whereas the other forms a distinct branch in the tree (RadB [57, 58]). Thus, these paralogous archaeal ATPases seem to have evolved as a result of an ancient gene duplication [59]. In the RadB proteins, the potassium-binding aspartate is replaced with two positively charged residues; in contrast to the conserved pair of lysines in all bacterial RecAs, the RadB proteins can accommodate any combination of lysines and arginines. The catalytic subunits of the rotary membrane ATP synthase are distant homologs of the RecA/RadA proteins (see Fig. 5 for the structural superposition). The pair of lysines, which in RecA occupies the same place as potassium ions in RadA, is located in the same position as the “arginine finger” residue of the rotary ATP synthase α-subunit. This arginine is strictly conserved in both α- and β-subunits of the ATP synthase, compatible with the apparent lack of potassium dependence for this thoroughly studied enzyme.
Fig. 3. Phylogenetic tree of the RadA/RecA subfamily. Sequences for the alignment and the phylogenetic tree were taken from COG0468 [148] and represent a set of 179 genomes from all bacterial and archaeal phyla and RadA/RecA orthologs (Rad51, Dmc1) from 35 eukaryotic species. The phylogenetic tree was constructed with the PhyML software under default parameters [151] and visualized in MEGA [152].
Fig. 4. Multiple alignment of RecA/RadA/RadB and their Rad51-like eukaryotic homologs. Amino acid residues are colored as in Fig. 1. The violet arrow indicates the P-loop lysine residue, and the two cyan arrows indicate two lysine residues in RecA sequences, which are inserted into the ATP-binding site of the adjacent monomer. Red arrows mark two acidic residues involved in potassium binding in M. voltae (the numbers correspond to the M. voltae sequence). The multiple alignment was constructed using Muscle [147] and visualized with GeneDoc.
Fig. 5. Superposition of the ATP synthase α-subunit and RecA protein. The α-subunit of bovine rotary ATP synthase (PDB 1COW [153], shown in gray) and the RecA protein from E. coli (PDB 3CMX [55], shown in light violet) were superimposed with the PDBeFold software [154]. The superimposed part is shown in dark violet.
Thus, in homologous proteins of the RecA/RadA/Rad51 family and α/β-subunits of the ATP synthase, different possible arrangements of positively charged residues are observed (the KNK motif in bacterial RecA, KHR motif in RadB, and a K+-replacing arginine residue in α- and β-subunits of the ATP synthase), whereas the K+-binding motif with a key Asp residue remains conserved in archaeal RadA and eukaryotic Rad51.
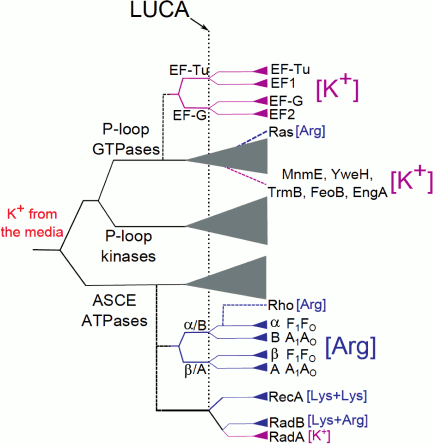
Hence, several ubiquitous proteins, which apparently belong to the LUCA gene set, show an apparent dependence on K+ ions. For some of these enzymes (the P-loop GTPase family, the RecA/RadA family, the GroEL family), we obtained indications of K+-dependence of the ancestral forms (see above and [18, 60]). The early, seminal results of Lowenstein concerning the effect of monovalent cations on the rate of nonenzymatic transphosphorylation reaction [61] suggest that large monovalent cations, such as K+ and NH4+, strongly enhance transphosphorylation, in contrast to the weak impact from Na+ ions. Thus, utilization of K+ in reactions that involve phosphate group transfer is likely to have preceded the utilization of amino acid side chains (Arg or Lys “fingers”) in the same capacity. In Fig. 6, we present a putative scheme of the evolution of the catalytic site of P-loop NTPases.
Fig. 6. A possible scheme of evolution of the catalytic site of P-loop NTPases. The solid lines show the general classification of the P-loop NTPases superfamily [52], the dashed lines show putative events of the emergence of particular protein families. The vertical dotted line marks the presumed position of the LUCA; the lengths of horizontal lines do not represent a real time scale. The magenta color in the branch shows appearance and further spreading of the specific K+-binding site in the protein family while the blue color indicates the appearance and further spreading of lysine and arginine finger(s).
In those experiments where Na+ and K+ ions were both included in the reaction mix for K+-dependent cellular systems, Na+ ions have been shown to act as inhibitors [33, 62] (with a single known exception of eukaryotic dynamin that can be activated by both K+ and Na+ ions [63]). Specifically, type II chaperonin from M. maripaludis (Mm-cpn), the archaeal homolog of GroEL, has been shown to be strongly inhibited when K+ and Na+ ions were present in same concentrations of 100 mM [62]. Thus, many K+-dependent enzymes require [K+]/[Na+] >> 1 for optimal activity.
POTASSIUM-RICH ENVIRONMENTS OF THE PRIMORDIAL EARTH
In modern prokaryotic cells, the difference in the ionic composition between the interior and the surrounding medium is maintained by ion-tight cellular membranes. Because these membranes are not completely impermeable, various ions are continuously leaking across the membranes in accordance with their concentration gradients. In prokaryotes, the influx of both K+ and Na+ ions into the cell is driven by the electric potential over the cell membrane (negative inside). Thereby, prokaryotic cells bring the [K+]/[Na+] ratio above unity by selectively extruding Na+ ions. Indeed, owing to the transmembrane potential (Δψ) of about –150 mV, the concentration of K+ within the cell can be higher than the concentration outside by 2-3 orders of magnitude [64, 65]. In most cases, this mechanism is sufficient to keep the cytosolic [K+] at the physiological level of about 100 mM. In cases of K+ limitation, prokaryotic cells express additional systems to actively pump K+ ions into the cell. These systems are quite different in different lineages (see [65, 66] for reviews).
As discussed previously, it is unlikely that modern-type ion-tight membranes, not to mention a plethora of ion-pumping machines, were present in the very first cells [12, 67, 68]. As Macallum wrote 90 years ago, “the very earliest organisms must have been of the micellar or ultramicroscopic kind... These had as yet no nuclei and an enclosing membrane could have been only of the most elementary character. From the first there must have been an adjustment in the composition of these very simple organisms to that of their medium, ... which diffusing into each minute multi-micellar mass brought into it the inorganic elements in the proportions in which they obtained in the external medium. In consequence, ... all the processes of metabolism, synthetic, assimilative and oxidative, were based on the physicochemical conditions thus obtaining in the internal medium” [17].
The cell membranes of modern archaea and bacteria are built of two-tail lipids, so that the tightly packed hydrophobic “double tails” make the membrane ion- and electrically tight [69]. The two-tail lipids of archaea and bacteria, however, are fundamentally different [70-72]. In both domains, phospholipids consist of two hydrophobic hydrocarbon chains that are attached to glycerol phosphate (GP). The GP moieties, however, are different: while bacteria use sn-glycerol-3-phosphate (G3P), archaea utilize its optical isomer sn-glycerol-1-phosphate (G1P). The hydrophobic chains, with a few exceptions, differ as well, being based on fatty acids in bacteria and on isoprenoids in archaea. In bacterial lipids, the hydrophobic tails are routinely linked to the glycerol moiety by ester bonds whereas archaeal lipids contain ether bonds. The difference extends beyond the chemical structures of the phospholipids, to the evolutionary provenance of the enzymes involved in synthesis of phospholipids: most of these are either non-homologous or distantly related, but not orthologous, in bacteria and archaea [70-74]. The only lipid biosynthetic pathways common for bacteria and archaea are the mevalonate pathway of isoprenoid biosynthesis and the enzymatic machinery for attaching polar heads to the protruding phosphate groups of lipid biosynthesis intermediates [70, 74, 75].
Given the drastic difference between the structures and biosynthetic pathways of the archaeal and bacterial membranes, it has been argued that the membranes of the earliest cellular life forms all the way to the LUCA could have been built of single-chain isoprenoid lipids [76-80]. However, membranes made of single-tail lipids are by many orders of magnitude more leaky to monovalent cations than modern-type membranes made of two-tail lipids [67, 68, 81, 82]. Thus, in accordance with the insightful suggestion of Macallum, the monovalent ion content of the cytoplasm of the first cells would have to be equilibrated with the environment. This equilibration does not necessarily imply an overall thermodynamic equilibrium because even highly permeable lipid vesicles made of single-chain lipids can securely occlude polymer molecules such as proteins and nucleic acids [67, 68, 81, 82]. The leakiness of the first membranes, however, could have been a key advantage to the primordial cells [68, 83, 84]. In the absence of membrane-embedded transport proteins that apparently could emerge only at a relatively late step of evolution, after the emergence of water-soluble proteins [12], the membrane leakiness should have enabled the “consumption” of diverse small molecules by the first cells, in a process that had been experimentally demonstrated [84]. This would favor the evolution of systems that could trap small molecules by attaching them to intracellular polymers and thus preventing their escape. Hence, leaky membranes could have favor the emergence of diverse polymerases, including the translation system.
As argued in the preceding section, early cells should have already possessed numerous K+-dependent enzymes including the K+-dependent components of the translation system. The aforementioned inhibitory effect of Na+ on many of these K+-dependent enzymes does not seem compatible with the evolution of the respective cellular systems in environments with high sodium levels. Thus, the emergence of such systems could have been possible only if the [K+]/[Na+] ratio in the habitats of the first cells was greater than unity (the alternatives to this possibility are considered in more detail elsewhere [85]).
On modern Earth, the excess of Na+ over K+ is observed not only in marine, but also in freshwater environments. The primordial ocean should have been salty as well; this could be judged from the chemical composition of geologically trapped water [86]. Specifically, primary fluid inclusions from the Pilbara craton, Western Australia (3.49 Ga), contain more than 1 M of Na+, twice as much as the water of modern ocean [87]. In search of natural habitats with a [K+]/[Na+] ratio > 1, a variety of environments have been examined [18]. The ionic composition conducive to the origin of cells was found to be compatible only with the emissions of vapor-dominated zones of inland geothermal systems. A major distinctive feature of such systems is the separation of the vapor phase from the liquid phase due to the boiling of the ascending hot hydrothermal fluids. The ascending vapor, after reaching the surface of the rock, discharges via numerous fumaroles and mud pots, which make a geothermal field. The chemical compositions of the two phases differ dramatically: the liquid phase contains large amounts of Na+ and Cl– ions, whereas the vapor phase is specifically enriched in H2S, CO2, and NH3 [18, 88-92]. The vapor phase is also enriched in K+ and shows [K+]/[Na+] > 1, apparently owing to the higher volatility of K+ ions compared to Na+ [18]. Based on this analysis, it has been proposed that the first cells could have emerged in the pools and puddles at the periphery of primordial anoxic geothermal fields, where the elementary composition of the condensed vapor, in the absence of atmospheric oxygen, would resemble the internal milieu of modern cells (see [18, 85] for details).
After Macallum [17], the problem of [K+]/[Na+] > 1 in the habitats of the first cells was considered in several papers, see e.g. [93, 94]. Specifically, an additional source of the vitally important K+ ions is invoked in the recent origin of life scenario of Maruyama and coworkers [93]. These authors built on the data on the 4.1-4.4 Gy old zircon grains, the only geologically informative remnants from the Hadean Earth. Analysis of these grains indicated that they were formed within re-melted granitic rocks related to the Earth protocrust that likely resembled the so-called KREEP-enriched basalts of the Moon [95], where KREEP stands for Potassium [K], Rare Earth Elements and Phosphorus. The KREEP basalts, widespread on the modern Moon, are believed to have formed upon the cooling of the lunar magma ocean [96]. By analogy, it has been speculated that the cooling of the terrestrial magma ocean would result in the formation of the KREEP-enriched protocrust, which later got lost owing to plate tectonics (see [95] and references therein). Maruyama and coworkers noted that two of the KREEP elements, namely potassium and phosphorus, are critically important for life and hypothesized that life could have emerged in the lakes of the first KREEP-enriched continent that should have contained high levels of potassium and phosphate ions [93].
The KREEP-rich continent scenario does not contradict the scenario of anoxic geothermal fields. Because the young Earth would produce much more heat than modern Earth, geothermal systems should have been widespread on the surface of the first continents, whether they were KREEP-enriched or not. In a way, the two scenarios even seem to be complementary. The KREEP scenario does not provide enough details on the chemistry of the first continental water basins. In contrast, the geothermal field scenario implies that the chemical separation should have led to the enrichment of the vapor phase not only with already mentioned H2S, CO2, NH3 and K+ ions, but also borate, transition metals, and geothermally formed organic molecules [18]. All these compounds should have been crucial for building up the first biomolecules (see [97] for a review). Conversely, the scenario of the anoxic geothermal fields is inconclusive regarding the time of the emergence of the first cells. Geochemical fields apparently remained anoxic for almost 2 billion years – from the emergence of the first continents until the oxygenation of the atmosphere some 2.5 Gy ago. The KREEP scenario is more precise in this respect, i.e. it attributes the emergence of life to the very first continent(s) [93], which emerged upon the cooling of the magma ocean some 4.4 Gy ago and probably lasted for only few hundred My [93-95].
THE ROLE OF THE Na+-EXPORTING MACHINERY
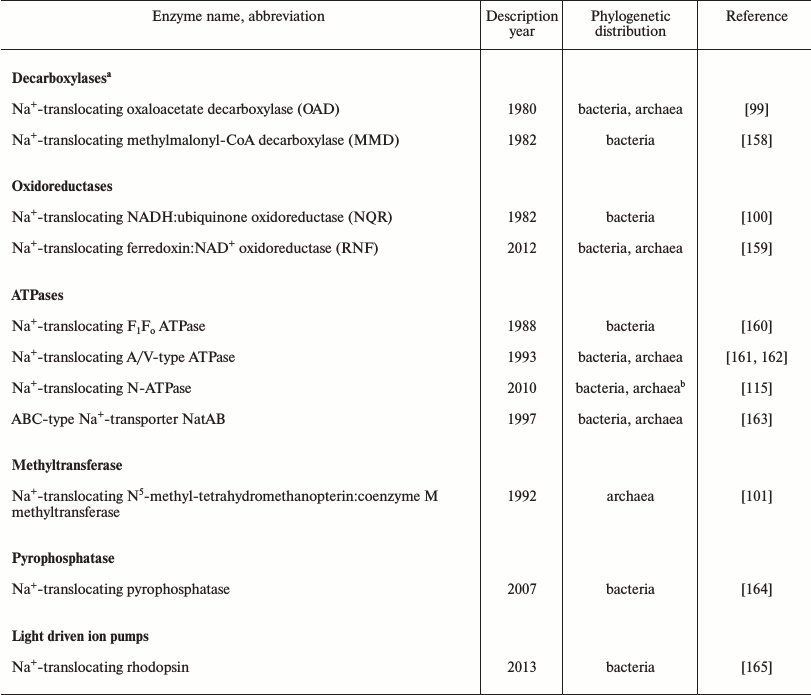
The evidence summarized above indicates that the first cells emerged in K+-rich habitats. However, marine and freshwater environments generally contain much less potassium than sodium, so that primordial K+-rich environments should have been constrained in space and time [18, 93, 98]. Accordingly, the spread of the first organisms from their K+-rich cradle into potential new habitats would encounter a serious challenge in the form of the sodium excess over potassium in the surrounding media. To keep protein synthesis and other K+-dependent processes running, these cells would have to be able to maintain the cytoplasmic [K+]/[Na+] ratio at >1. In modern prokaryotes, this outcome is achieved by a variety of Na+-extruding and K+-uptake systems embedded in ion-tight membranes. Thus, as argued previously [10, 12, 98], the foray into new, Na+-rich habitats can be considered a major evolutionary transition that benefited from and was the driving force behind the emergence of diverse sodium export pumps. While sodium export in many modern organisms is carried out by Na+/H+ antiporters, energized by the proton-motive force, there is also a variety of proton-motive force-independent primary Na+ pumps. Three classes of primary Na+ pumps, namely the Na+-translocating oxaloacetate decarboxylase, Na+-translocating NADH:ubiquinone oxidoreductase, and Na+-translocating coenzyme M methylase have been known for more than 20 years [99-101]. In the past several years, this list has been complemented by another redox enzyme, Na+-translocating ferredoxin:NAD+ oxidoreductase (RNF), as well as Na+-translocating pyrophosphatase and Na+-translocating bacterial rhodopsin (see Table 2). In addition, there is growing evidence for an “embedded” Na+/H+ antiporter module in some members of the enzyme superfamily that includes the complex I-like membrane NADH-dehydrogenase, the electrogenic membrane hydrogenase, and the formate oxidase. Owing to such a module, these enzymes, seemingly, could function as de facto redox-driven Na+ pumps [102-106]. With the current interest in anoxic prokaryotes isolated from diverse habitats, this list can be expected to grow even further. The diversity of the Na+ pumps, some of apparent ancient origin [107, 108], shows that the first cells could possess a variety of independently acting redox-, light-, chemically-, or osmotically-dependent Na+ export pumps that would enable their survival in Na+-rich environments. The redundancy of such pumps and the diversity of energy sources they could harness would be important for survival, particularly in a marine environment, and could compensate for the imperfection of the primitive membranes that most likely were made from single-tail lipids.
To conclude this section, continuous cell growth in environments with [K+]/[Na+] < 1 required maintaining the cytosolic [K+]/[Na+] > 1 through constant extrusion of Na+ ions, which therefore became a key function in the energetics of the primordial cell. Thus, explicit consideration of primordial Na+ export mechanisms provides an evolutionary and structural framework that helps to explain the gradual increase in the impermeability (ion-tightness) of the lipid bilayer. Obviously, organisms with less leaky membranes would have a substantial evolutionary advantage by requiring less energy for pumping sodium ions out of the cell.
EMERGENCE OF MEMBRANE BIOENERGETICS
So far, for the sake of simplicity, we have not discussed the rotary ATP synthases (ATPases), the major source of ATP in modern cells. These enzyme complexes are reversible, rotary molecular machines that couple ion transfer across the membrane with the synthesis or hydrolysis of ATP (see [109-115] and references therein). The rotary ATPases fall into two distinct types, namely the F/N-type that is present in bacteria, a few archaea, mitochondria, and chloroplasts, and A/V-type, which is represented in archaea, some bacteria, and in the cytoplasmic and vacuolar membranes of eukaryotes. The rotary ATPases couple transfer of protons or sodium cations across the respective membrane with hydrolysis or synthesis of ATP via a binding change mechanism in their protruding catalytic parts. These enzyme complexes are “dynamo machines” in which sequential hydrolysis of ATP molecules by the protruding catalytic hexamer drives the rotation of the central stalk together with the ring of the so-called proteolipid subunits (from 8 to 15, depending on the species). The ring (rotor of the dynamo) is thought to slide along the interface with the membrane subunits that make part of the stator, so that the sliding movement is coupled to the transmembrane ion transfer and generation of the membrane potential. When the enzyme functions as an ATP synthase, the translocation of protons or sodium ions between the ring of proteolipid subunits and the membrane stator subunits drives the rotation of the ring, and accordingly, of the central stalk. The sequential interaction of the rotating stalk with the three catalytic centers of the hexameric ring drives the formation of ATP molecules in these centers. The ion specificity of the rotary ATP synthase defines the nature of the bioenergetic cycle in any organism given that this appears to be the only enzyme capable of synthesizing ATP at the expense of the membrane potential. Whether the ATP synthase is H+- or Na+-driven depends solely on whether the proteolipid subunits possess a full set of ligands for binding Na+ ions. If at least one ligand from this set is absent, the enzyme loses the ability to translocate Na+ ions but usually retains the ability to translocate protons (which ultimately requires only the active-site Asp/Glu residue) [9, 10, 116, 117].
The rotary ATPase is the only energy-converting complex the subunits of which belong to the set of ubiquitous proteins attributable to LUCA [41, 42]. It has even been hypothesized that that this enzyme could precede the emergence of membranes and operate within the walls of inorganic bubbles harboring the communities of first replicators [62, 118] or within primitive, leaky membranes [119]. However, this appears to be an unlikely possibility. The prerequisite for ATP synthesis by membrane rotary ATP synthases is its operation within a dielectric membrane capable of maintaining transmembrane proton or sodium potential, the electric component of which must be over 100-150 mV, which corresponds to an electric field of >2·107 V/m. Even modern, sophisticated membranes become leaky at high voltage since at Δψ > 150 mV the conductivity increases in a nonlinear fashion [120, 121]. During the last decades, it has been experimentally established that any defect in the membrane dielectric that increases its ion conductivity leads to an arrest in ATP production by the membrane rotary ATP synthase (the ions escape through the defect(s) instead of passing under very high load through the ATP synthase). The membrane can become uncoupled owing, for example, to the presence of amphiphilic ion carriers (ionophores), as first shown by Skulachev and coworkers [122, 123], or of proton-carrying single-tail lipids, such as fatty acids [124], the physiological importance of which has been clarified by Skulachev and coworkers [125, 126], or lysophospholipids that have one of the tails removed by hydrolysis [127]. Because of the strict constraint on membrane tightness, neither inorganic membrane-like precipitates, which could not be dielectrically tight and defect-free by default, nor even membranes made of single-chain lipids could support ATP synthesis by a rotary ATP synthase.
Still, the presence of two subunits of the rotary ATP synthase, namely, the ATP-binding, catalytic subunit and the proteolipid subunit that forms the membrane ring, in the set of (nearly) ubiquitous genes assignable to the LUCA calls for an explanation. Since the proteolipid ring is essentially a lipid-filled membrane pore, it has been argued that the ancient version of the enzyme might have served not as an ATP synthase, but as an ATP-driven protein translocase that could push protein molecules through a primitive membrane pore [12, 110]. Transition to an ion-translocating enzyme could have been triggered by a mutation that hindered protein translocation. Protein molecules would then get transiently stuck in the pore and eventually cause its rotation; this scenario could ultimately result in coupling between the ATP hydrolysis and ion translocation along the interface of the rotating membrane ring. Considering the revealed evolutionary primacy of Na+-dependent membrane energetics, we hypothesized that the function of the first ion-translocating rotary ATPase was to expel sodium ions out of the cell [10, 12]. In a search for relics of such primordial rotary Na+ export pump, we have identified, by means of phylogenomic analysis, a distinct family of rotary ATPases (N-ATPases), almost all members of which, as judged from the sequence of their proteolipid subunits, appeared to be Na+-translocating ATPases [115]. These enzymes are encoded by a highly mobile operon, which always co-occurs with the operon coding for the “main” F-type or A/V-type ATP synthase typical of the respective prokaryotic group. Based on these observations, we speculated that N-ATPases, which carry several primitive features and which are found in many marine bacteria and archaea, might function as Na+ export pumps in the modern organisms [115]. And indeed, the N-ATPase of the halotolerant cyanobacterium Aphanothece halophytica has been experimentally demonstrated to translocate Na+ and to increase tolerance to salt stress in the freshwater cyanobacterium Synechococcus elongatus PCC 7942 [128, 129]. These data prompted us to include the N-ATPase in the list of prokaryotic sodium exporters (Table 2).
Table 2. Sodium export pumps in bacteria and
archaea
a This enzyme family also includes malonate decarboxylase and
glutaconyl-CoA decarboxylase, see [156, 157].
b The N-ATPase operons found in the genomes of two archaea,
Methanosarcina acetivorans and Methanosarcina barkeri,
likely have bacterial origin and have been acquired by lateral gene
transfer [115].
As discussed in the preceding section, gradual increase of membrane tightness should have been beneficial for cells because it allowed them to survive in Na+-rich environments at progressively decreasing costs. At some point, the cell membranes would become capable of maintaining substantial voltage. It seems likely that this advance was coupled with the emergence of two-tail lipids. At that point, under the conditions of the aforementioned high salinity of the primordial ocean, the Na+-translocating rotary ATPase, which initially functioned as a sodium export pump, could change the direction of rotation and start to synthesize ATP at the expense of the transmembrane difference in the electrochemical potential of sodium ions (sodium-motive force). In marine environments, the sodium gradient of three orders of magnitude would have enabled ATP synthesis at the voltage of only 100 mV. This switch of a rotary ATPase from Na+ export to ATP synthesis would lead to the emergence of sodium-dependent membrane bioenergetics, the essence of which is the coupling between membrane enzymes that pump sodium ions out of the cell and the “consumers” that use the sodium gradient to perform useful work. The most important type of such work is the synthesis of ATP by the rotary ATP synthase. The aforementioned rotary N-ATPase that functions as a sodium exporter but seems to be capable of ATP synthesis in vitro [129] can be considered a “living fossil” that marks the path from simple outward pumping of Na+ ions to the full Na+-based bioenergetic cycle.
One could ask what would be the evolutionary advantage of reversing the rotary ATPase. As argued by Boiteau and Pascal, it is the rotary scaffold of the membrane ATP synthase that determines its pivotal role in the cell [130]. This kind of rotary mechanism was initially proposed by Glagolev and Skulachev for the bacterial flagella motor [131], then considered by Skulachev for the rotary membrane ATPase [15] and, finally, elaborated in detail for the proton-translocating rotary ATP synthase by Vick and Antonio [132] and Junge and coworkers [133, 134]. According to this mechanism, to synthesize a single ATP molecule, several ions have to be sequentially translocated across the membrane; thereby the free energy of ion translocation is stored stepwise in the elastic deformation of the enzyme until enough free energy to drive ATP synthesis is accumulated [134]. Hence, the rotary ATP synthase is a unique machine that can store small portions of free energy and use them for ATP synthesis [130]. The magnitude of the transmembrane sodium (or proton) potential at the cell membrane could hardly exceed 250 mV, which determines the maximum free energy needed to export a sodium ion or a proton out of the cell, so that sodium efflux could be accomplished by redox-driven sodium pumps, such as NQR or RNF.
The free energy sufficient to expel a sodium ion out of the cell and generate sodium-motive force could be even less if the Na+ ion is accompanied by an anion. Anions of diverse carbonic acids, such as acetate, lactate, or butyrate, are usually the end-products of cell fermentation pathways that, most likely, made the core of heterotrophic metabolism on the ancient anoxic Earth. On one hand, accumulation of acid end-products within the cell would hamper the fermentation process via product inhibition and, eventually, kill the cell by acidification. On the other hand, the negatively charged organic acids would not be able to escape the cell easily by crossing the hydrophobic membrane barrier. In modern anaerobes, acidic end-products are carried out of the cell by symport with positively charged cations, usually sodium ions or protons, which leads to the generation of the sodium- or proton-motive force, respectively [135]. The possibility of generating membrane potential through this mechanism has been experimentally established both for organisms with proton-dependent energetics [135] and sodium-dependent energetics [136]. Under modern conditions, this mechanism is relevant only for anaerobes. Aerobic organisms, rather than expel acidic end-products, typically oxidize them to yield additional energy via respiration. However, on the primordial anoxic Earth, organic acids should have been the end-products in most of prokaryotic fermentation pathways; the respective symporters should have been widespread and capable of generating sodium potential for ATP synthesis (almost) “for free”. Hence, the rotary ATP synthases of primordial organisms could be driven just by expelling “garbage” out of the cell, which would have been strongly beneficial for these organisms.
The proposed scenario implies that membrane bioenergetics emerged relatively late, being limited by the evolution of membrane lipids, and could be preceded by an evolutionary stage when various, independent sodium export pumps enabled the survival of the early cells in habitats with [K+]/[Na+] ratio < 1. This scenario contradicts the widespread view on the evolutionary antiquity of chemiosmotic coupling, see e.g. [137]. However, the chemiosmotic mechanism of ATP synthesis is absent from the cell membranes of eukaryotic cells, where proton-dependent chemiosmosis is practiced only by bacterial endosymbionts, mitochondria and chloroplasts. Because electrically-tight membranes built of two-tail lipids seem to have emerged independently in bacteria and archaea, it seems likely that the chemiosmosis might have also emerged independently in bacteria and archaea.
The secondary, derived nature of the membrane bioenergetics, as well as its relation to the Na+ export, is illustrated by the seminal early work of Harold and Van Brunt of 1977 [138], the full significance of which had not been appreciated at that time. These authors have shown that bacterial cells could still grow – in a rich medium – in the presence of ionophoric compounds that fully collapsed the membrane ion potential – but only if the growth medium contained at least 100 mM K+. In a similarly rich but Na+-based medium, bacteria could not grow in the presence of ionophores. Notably, even the growth rate in the K+-rich medium was not affected by the collapse of the membrane potential, which indicates that the energy from substrate-level phosphorylation reactions was fully sufficient to support the growth of the cells. The authors concluded that membrane bioenergetics was not needed, at least under certain conditions, for the well-being of the cell. Indeed, available estimates indicate that maintaining the [K+]/[Na+] disequilibrium demands up to 20-50% of cellular energy resources (see [15, 139] and references therein), so that K+-based media with no Na+, such as M63, are often used in biotechnology to increase cell yield. The cells that happily grew – in spite of their leaky membranes – in a K+-rich medium, as shown by Harold and Van Brunt [138], may be considered a “reincarnation” of the ancient cells, whose primitive leaky membranes would enable cell proliferation in the primordial K+-rich habitats but could not sustain cell growth in the high-sodium media.
UTILIZATION OF THE K+/Na+ GRADIENT BY THE
CELL
The observations of Harold and Van Brunt [138] seem to contradict the aforementioned data of Skulachev and coworkers that energy-depleted prokaryotic cells remained mobile for a longer time in a Na+-based medium than in a K+-based medium [4]. One could expect that in a K+-rich medium, where energy is not needed to pump Na+ ions out of the cell, more energy would be available to drive bacterial motility. A detailed consideration of processes involved, perhaps, could offer a solution of this conundrum. In the Na+-rich medium, which is physiologically relevant for E. coli, the discharge of the Na+/K+ gradient would cause two major effects, namely (i) transient formation of proton potential, which could drive the rotation of bacterial flagella, and (ii) a decrease of the cytoplasmic [K+]/[Na+] ratio followed by its reversal, as shown for energy- or nutrient-limited cells in the stationary phase of growth [22]. Upon the reversal of the [K+]/[Na+] ratio the entire cellular metabolism would be essentially shut-off by Na+ ions because blocking of any enzyme that is a part of a metabolic pathway would switch off the respective pathway. Hence, on one hand, the reversal of the [K+]/[Na+] ratio would block the activity of numerous K+-dependent enzymes and thereby prevent the complete depletion of the cellular stock of ATP and GTP, as observed by Skulachev and co-workers [4]. On the other hand, the few autonomous nucleoside triphosphate hydrolases that are not involved in metabolic pathways and are, in addition, K+-independent, would have the entire ATP/GTP stock at their disposal. At low [K+]/[Na+] ratio, the membrane rotary ATPase, which is K+-independent (see above), could then hydrolyze ATP, generate proton potential and drive flagellar motility until the membrane potential dropped below threshold value of ca. –100 mV under which the flagellar motor stops [140]. In contrast, in a K+-rich medium, energy depletion would not lead to the blocking of the cellular metabolism by Na+ ions. Na+-sensitive hydrolases would still run at a high speed and promptly deplete the stock of ATP in the cell (also in line with experimental observations [4]). In this case, the flagellar rotation could not be supported either by the discharge of the Na+/K+ gradient or by the long-lasting generation of proton potential by the membrane rotary ATPase.
Hence, under the conditions of energy depletion, discharging of the Na+/K+ gradient at the cell membranes might transiently maintain the membrane proton (or sodium) potential, as suggested by Skulachev and co-workers [1, 4, 141]. In addition, the collapse of the Na+/K+ gradient, by decreasing the [K+]/[Na+] ratio in the cell and the resulting inhibition of numerous hydrolases, prevents the full depletion of the cellular stock of ATP/GTP [4]. During the stationary (dormant) phase, the saved ATP/GTP stock could be used for the basic maintenance and repair functions, thus keeping the cell alive. In addition, upon the change to better conditions, the retained ATP/GTP stock would drive the awakening of the cell.
Thus, on one hand, the incomplete replacement of the “primordial” cofactor K+ ions by Arg or Lys residues in the course of evolution forces the cells to invest up to 50% of their energy into maintaining the Na+/K+ gradient at the cell membrane. On the other hand, the residual dependence of many enzymes on K+ ions as catalytic cofactors and the inhibition of such enzymes by Na+ ions seem to secure the survival of the prokaryotic cell under unfavorable, energy-limiting conditions when the [K+]/[Na+] ratio in the cell drops below unity.
Upon the transition to multicellularity, the survival of the whole organism became the evolutionary priority, which led to the development of diverse apoptotic mechanisms that eliminate nonfunctional cells in a controlled way. It has been shown that the apoptotic machinery in the cells of vertebrates might be activated in response to the drop in the cytoplasmic [K+]/[Na+] ratio (see [142] and references therein). The inability of the cell to maintain a physiologically high [K+]/[Na+] ratio might reflect an ATP shortage and, hence, indicate malfunctioning mitochondria. Since damaged mitochondria are known to produce reactive oxygen species at high yield, the elimination of energy-compromised cells becomes an urgent task, as suggested by Skulachev [143]. Apparently, at least in some cases, the elimination might be triggered by impairments in the Na+/K+ homeostasis in the cell.
OUTLOOK
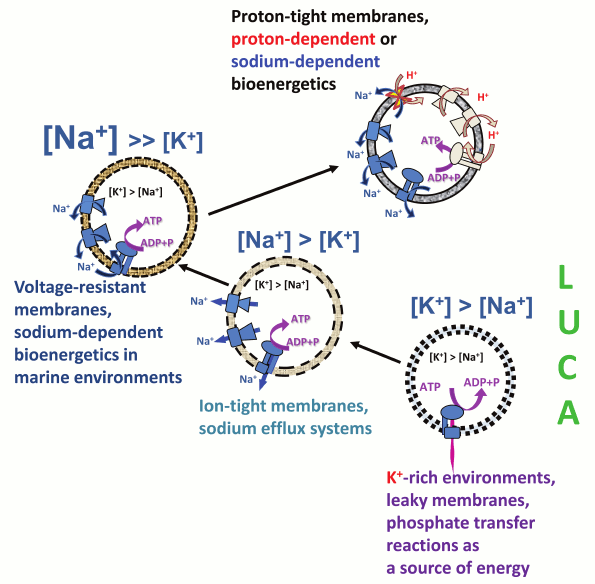
The evolutionary framework described here and schematically depicted in Fig. 7 reveals the intrinsic coupling between the Na+/K+ disequilibrium at the cell membrane and membrane bioenergetics. The Na+/K+ gradient most likely appeared first, concomitant with the escape of the primordial cellular organisms from geochemically highly specialized K+-rich habitats. The Na+/K+ gradient could have been initially maintained by a plethora of sodium export pumps, driven by the energy of chemical and redox reactions, energy of light, as well as, in the case of Na+-dependent symporters, the energy of osmotic gradients. Once the cell membranes evolved to maintain ion gradients of sufficient magnitude, one of such export pumps, namely the rotary Na+-translocating ATPase, switched to ATP synthesis at the expense of the energy of the transmembrane sodium gradient, thus giving start to membrane bioenergetics that functionally connected the initially independent sodium pumps with each other and enabled harvesting of small energy quanta for ATP synthesis.
Fig. 7. Proposed scenario for the evolution of Na+/K+ homeostasis and membrane bioenergetics. The proposed scenario involves transition from primitive membranes that were leaky to both Na+ and H+ (dotted lines), via membranes that could support sodium efflux pumps and membranes that were tight enough to support Na+-dependent membrane bioenergetics (dashed lines), to the modern-type membranes impermeable to H+ and Na+ ions (solid lines).
With further evolution of cell membranes and after the oxygenation of the atmosphere, the enzymes of membrane bioenergetics gradually became decoupled from the machinery responsible for maintaining the Na+/K+ disequilibrium. On one hand, prokaryotic membranes became largely impermeable not only to sodium ions but also to protons, and the proton-dependent bioenergetics, which should have been more beneficial under oxidizing conditions [11], became prevalent. Still in many organisms sodium pumps are functioning concurrently with proton pumps, being functionally connected by Na+/H+ antiporters (Fig. 7 and [9, 11]). On the other hand, the ancestors of eukaryotes evolved a specialized, extremely efficient enzyme to maintain the Na+/K+ disequilibrium, namely the Na+/K+-ATPase, which couples hydrolysis of a single ATP molecule with the transport of 3 Na+ ions out of the cell and 2 K+ ions into the cell [98, 144]. This Na+/K+-ATPase belongs to the large superfamily of P-type ATPases, which includes a variety of other members that are involved in detoxification, particularly in pumping out of the cell heavy metal ions such as Cd2+, Hg2+, and Cu+, rather than in energy transformation [145, 146]. As a result, in animal and plant cells, the major systems of Na+/K+ exchange (Na+/K+-ATPase and ion channels) became uncoupled from the membrane energy-converting machinery (the H+-translocating enzymes of endosymbiotically obtained mitochondria and chloroplasts). It is not surprising that Na+/K+ homeostasis and membrane bioenergetics are commonly treated as independent phenomena. In animal cells, the only relics from the ancient reign of Na+-export pumps are numerous Na+-coupled membrane transporters, as well as the cell membrane itself which, unlike the membranes of modern bacteria and archaea, is impermeable to Na+ ions but leaky to protons, as insightfully noted by Skulachev many years ago [15].
The authors are grateful to Drs. A. V. Bogachev, A. Y. Bychkov, Y. I. Wolf and K. S. Makarova for useful suggestions and interest in this work.
This work was supported in part by Grants of the Russian Science Foundation (14-50-00029, DVD, phylogenomic analysis of K+-binding sites in nucleoside triphosphatases; and 14-14-00592, AYM, reconstruction of evolutionary events that could lead to the emergence of membrane bioenergetics) and by the Intramural Research Program of the National Institutes of Health at the National Library of Medicine, USA (MYG and EVK).
REFERENCES
1.Skulachev, V. P. (1978) Membrane-linked energy
buffering as the biological function of Na+/K+
gradient, FEBS Lett., 87, 171-179.
2.Mitchell, P. (1961) Coupling of phosphorylation to
electron and hydrogen transfer by a chemi-osmotic type of mechanism,
Nature, 191, 144-148.
3.Skulachev, V. P. (1977) Transmembrane
electrochemical H+-potential as a convertible energy source
for the living cell, FEBS Lett., 74, 1-9.
4.Brown, I. I., Galperin, M. Y., Glagolev, A. N., and
Skulachev, V. P. (1983) Utilization of energy stored in the form of
Na+ and K+ ion gradients by bacterial cells,
Eur. J. Biochem., 134, 345-349.
5.Skulachev, V. P. (1984) Sodium bioenergetics,
Trends Biochem. Sci., 9, 483-485.
6.Skulachev, V. P. (1989) The sodium cycle: a novel
type of bacterial energetics, J. Bioenerg. Biomembr., 21,
635-647.
7.Cramer, W. A., and Knaff, D. B. (1990) Energy
Transduction in Biological Membranes: A Textbook of Bioenergetics,
Springer-Verlag.
8.Dimroth, P. (1994) Bacterial sodium ion-coupled
energetics, Antonie Van Leeuwenhoek, 65, 381-395.
9.Hase, C. C., Fedorova, N. D., Galperin, M. Y., and
Dibrov, P. A. (2001) Sodium ion cycle in bacterial pathogens: evidence
from cross-genome comparisons, Microbiol. Mol. Biol. Rev.,
65, 353-370.
10.Mulkidjanian, A. Y., Galperin, M. Y., Makarova,
K. S., Wolf, Y. I., and Koonin, E. V. (2008) Evolutionary primacy of
sodium bioenergetics, Biol. Direct., 3, 13.
11.Mulkidjanian, A. Y., Dibrov, P., and Galperin, M.
Y. (2008) The past and present of sodium energetics: may the
sodium-motive force be with you, Biochim. Biophys. Acta,
1777, 985-992.
12.Mulkidjanian, A. Y., Galperin, M. Y., and Koonin,
E. V. (2009) Co-evolution of primordial membranes and membrane
proteins, Trends Biochem. Sci., 34, 206-215.
13.Deamer, D. W. (1987) Proton permeation of lipid
bilayers, J. Bioenerg. Biomembr., 19, 457-479.
14.Van de Vossenberg, J. L., Ubbink-Kok, T.,
Elferink, M. G., Driessen, A. J., and Konings, W. N. (1995) Ion
permeability of the cytoplasmic membrane limits the maximum growth
temperature of bacteria and archaea, Mol. Microbiol., 18,
925-932.
15.Skulachev, V. P. (1989) Membrane
Bioenergetics, Springer Verlag, Heidelberg.
16.Konings, W. N. (2006) Microbial transport:
adaptations to natural environments, Antonie Van Leeuwenhoek,
90, 325-342.
17.Macallum, A. B. (1926) The paleochemistry of the
body fluids and tissues, Physiol. Rev., 6, 316-357.
18.Mulkidjanian, A. Y., Bychkov, A. Y., Dibrova, D.
V., Galperin, M. Y., and Koonin, E. V. (2012) Origin of first cells at
terrestrial, anoxic geothermal fields, Proc. Natl. Acad. Sci.
USA, 109, E821-830.
19.Williams, R. J. P., and Frausto da Silva, J. J.
R. (1991) The Biological Chemistry of the Elements, Clarendon
Press, Oxford.
20.Heldal, M., Scanlan, D. J., Norland, S.,
Thingstad, F., and Mann, N. H. (2010) Elemental composition of single
cells of various strains of marine prochlorococcus and synechococcus
using X-ray microanalysis, Limnol. Oceanogr., 48,
1732-1743.
21.Ventosa, A., Nieto, J. J., and Oren, A. (1998)
Biology of moderately halophilic aerobic bacteria, Microbiol. Mol.
Biol. Rev., 62, 504-544.
22.Fagerbakke, K. M., Norland, S., and Heldal, M.
(1999) The inorganic ion content of native aquatic bacteria, Can. J.
Microbiol., 45, 304-311.
23.Battistuzzi, F. U., Feijao, A., and Hedges, S. B.
(2004) A genomic timescale of prokaryote evolution: insights into the
origin of methanogenesis, phototrophy, and the colonization of land,
BMC Evol. Biol., 4, 44.
24.Fasano, O., De Vendittis, E., and Parmeggiani, A.
(1982) Hydrolysis of GTP by elongation factor Tu can be induced by
monovalent cations in the absence of other effectors, J. Biol.
Chem., 257, 3145-3150.
25.Wu, Y., Qian, X., He, Y., Moya, I. A., and Luo,
Y. (2005) Crystal structure of an ATPase-active form of Rad51 homolog
from Methanococcus voltae. Insights into potassium dependence,
J. Biol. Chem., 280, 722-728.
26.Viitanen, P. V., Lubben, T. H., Reed, J.,
Goloubinoff, P., O’Keefe, D. P., and Lorimer, G. H. (1990)
Chaperonin-facilitated refolding of ribulosebisphosphate carboxylase
and ATP hydrolysis by chaperonin 60 (groEL) are K+
dependent, Biochemistry, 29, 5665-5671.
27.Toraya, T., and Fukui, S. (1977) Immunochemical
evidence for the difference between coenzyme-B12-dependent diol
dehydratase and glycerol dehydratase, Eur. J. Biochem.,
76, 285-289.
28.Laughlin, L. T., and Reed, G. H. (1997) The
monovalent cation requirement of rabbit muscle pyruvate kinase is
eliminated by substitution of lysine for glutamate 117, Arch.
Biochem. Biophys., 348, 262-267.
29.McQueney, M. S., and Markham, G. D. (1995)
Investigation of monovalent cation activation of S-adenosylmethionine
synthetase using mutagenesis and uranyl inhibition, J. Biol.
Chem., 270, 18277-18284.
30.Toney, M. D., Hohenester, E., Keller, J. W., and
Jansonius, J. N. (1995) Structural and mechanistic analysis of two
refined crystal structures of the pyridoxal phosphate-dependent enzyme
dialkylglycine decarboxylase, J. Mol. Biol., 245,
151-179.
31.Belogurov, G. A., and Lahti, R. (2002) A lysine
substitute for K+. A460K mutation eliminates K+
dependence in H+-pyrophosphatase of Carboxydothermus
hydrogenoformans, J. Biol. Chem., 277,
49651-49654.
32.McCaman, R. E., and Finnerty, W. R. (1968)
Biosynthesis of cytidine diphosphate-diglyceride by a particulate
fraction from Micrococcus cerificans, J. Biol. Chem.,
243, 5074-5080.
33.Spirin, A. S., and Gavrilova, L. P. (1969) The
Ribosome, Springer, New York.
34.Miskin, R., Zamir, A., and Elson, D. (1970)
Inactivation and reactivation of ribosomal subunits: the peptidyl
transferase activity of the 50 S subunit of Escherihia coli,
J. Mol. Biol., 54, 355-378.
35.Sigel, R. K. O., and Pyle, A. M. (2007)
Alternative roles for metal ions in enzyme catalysis and the
implications for ribozyme chemistry, Chem. Rev., 107,
97-113.
36.Ban, N., Nissen, P., Hansen, J., Moore, P. B.,
and Steitz, T. A. (2000) The complete atomic structure of the large
ribosomal subunit at 2.4 Å resolution, Science,
289, 905-920.
37.Klein, D. J., Moore, P. B., and Steitz, T. A.
(2004) The contribution of metal ions to the structural stability of
the large ribosomal subunit, RNA, 10, 1366-1379.
38.Bokov, K., and Steinberg, S. V. (2009) A
hierarchical model for evolution of 23S ribosomal RNA, Nature,
457, 977-980.
39.Davidovich, C., Belousoff, M., Bashan, A., and
Yonath, A. (2009) The evolving ribosome: from non-coded peptide bond
formation to sophisticated translation machinery, Res.
Microbiol., 160, 487-492.
40.Fox, G. E., Tran, Q., and Yonath, A. (2012) An
exit cavity was crucial to the polymerase activity of the early
ribosome, Astrobiology, 12, 57-60.
41.Koonin, E. V. (2003) Comparative genomics,
minimal gene-sets and the last universal common ancestor, Nat. Rev.
Microbiol., 1, 127-136.
42.Koonin, E. V. (2000) How many genes can make a
cell: the minimal-gene-set concept, Annu. Rev. Genom. Hum.
Genet., 1, 99-116.
43.Charlebois, R. L., and Doolittle, W. F. (2004)
Computing prokaryotic gene ubiquity: rescuing the core from extinction,
Genome Res., 14, 2469-2477.
44.Mushegian, A. (2005) Protein content of minimal
and ancestral ribosome, RNA, 11, 1400-1406.
45.Mulkidjanian, A. Y., and Galperin, M. Y. (2009)
On the origin of life in the zinc world. 2. Validation of the
hypothesis on the photosynthesizing zinc sulfide edifices as cradles of
life on Earth, Biol. Direct., 4, 27.
46.Mulkidjanian, A. Y., and Galperin, M. Y. (2010)
On the abundance of zinc in the evolutionarily old protein domains,
Proc. Natl. Acad. Sci. USA, 107, E137.
47.Anand, B., Surana, P., and Prakash, B. (2010)
Deciphering the catalytic machinery in 30S ribosome assembly GTPase
YqeH, PLoS One, 5, e9944.
48.Scrima, A., and Wittinghofer, A. (2006)
Dimerisation-dependent GTPase reaction of MnmE: how potassium acts as
GTPase-activating element, EMBO J., 25, 2940-2951.
49.Yamanaka, K., Hwang, J., and Inouye, M. (2000)
Characterization of GTPase activity of TrmE, a member of a novel GTPase
superfamily, from Thermotoga maritima, J. Bacteriol.,
182, 7078-7082.
50.Ash, M. R., Guilfoyle, A., Clarke, R. J., Guss,
J. M., Maher, M. J., and Jormakka, M. (2010) Potassium-activated GTPase
reaction in the G protein-coupled ferrous iron transporter B, J.
Biol. Chem., 285, 14594-14602.
51.Foucher, A. E., Reiser, J. B., Ebel, C., Housset,
D., and Jault, J. M. (2012) Potassium acts as a GTPase-activating
element on each nucleotide-binding domain of the essential Bacillus
subtilis EngA, PLoS One, 7, e46795.
52.Leipe, D. D., Wolf, Y. I., Koonin, E. V., and
Aravind, L. (2002) Classification and evolution of P-loop GTPases and
related ATPases, J. Mol. Biol., 317, 41-72.
53.Kint, C., Verstraeten, N., Hofkens, J., Fauvart,
M., and Michiels, J. (2014) Bacterial Obg proteins: GTPases at the
nexus of protein and DNA synthesis, Crit. Rev. Microbiol.,
40, 207-224.
54.Leipe, D. D., Koonin, E. V., and Aravind, L.
(2003) Evolution and classification of P-loop kinases and related
proteins, J. Mol. Biol., 333, 781-815.
55.Li, Y., He, Y., and Luo, Y. (2009) Conservation
of a conformational switch in RadA recombinase from Methanococcus
maripaludis, Acta Crystallogr. D Biol. Crystallogr.,
65, 602-610.
56.Chen, Z., Yang, H., and Pavletich, N. P. (2008)
Mechanism of homologous recombination from the RecA-ssDNA/dsDNA
structures, Nature, 453, 489-484.
57.Haldenby, S., White, M. F., and Allers, T. (2009)
RecA family proteins in archaea: RadA and its cousins, Biochem. Soc.
Trans., 37, 102-107.
58.Sandler, S. J., Hugenholtz, P., Schleper, C.,
DeLong, E. F., Pace, N. R., and Clark, A. J. (1999) Diversity of
radA genes from cultured and uncultured archaea: comparative
analysis of putative RadA proteins and their use as a phylogenetic
marker, J. Bacteriol., 181, 907-915.
59.Lin, Z., Kong, H., Nei, M., and Ma, H. (2006)
Origins and evolution of the recA/RAD51 gene family: evidence for
ancient gene duplication and endosymbiotic gene transfer, Proc.
Natl. Acad. Sci. USA, 103, 10328-10333.
60.Dibrova, D. V. (2013) Phylogenomic Analysis of
Energy Converting Enzymes, Osnabrueck University.
61.Lowenstein, J. M. (1960) The stimulation of
transphosphorylation by alkali-metal ions, Biochem. J.,
75, 269-274.
62.Kusmierczyk, A. R., and Martin, J. (2003) Nested
cooperativity and salt dependence of the ATPase activity of the
archaeal chaperonin Mm-cpn, FEBS Lett., 547, 201-204.
63.Ash, M. R., Maher, M. J., Mitchell Guss, J., and
Jormakka, M. (2012) The cation-dependent G-proteins: in a class of
their own, FEBS Lett., 586, 2218-2224.
64.Harold, F. M., and Altendorf, K. (1974) in
Current Topics in Membranes and Transport (Bronner, F., and
Kleinzeller, A., eds.) Academic Press, New York, pp. 1-51.
65.Corratge-Faillie, C., Jabnoune, M., Zimmermann,
S., Very, A. A., Fizames, C., and Sentenac, H. (2010) Potassium and
sodium transport in non-animal cells: the Trk/Ktr/HKT transporter
family, Cell Mol. Life Sci., 67, 2511-2532.
66.Haupt, M., Bramkamp, M., Coles, M., Kessler, H.,
and Altendorf, K. (2005) Prokaryotic Kdp-ATPase: recent insights into
the structure and function of KdpB, J. Mol. Microbiol.
Biotechnol., 10, 120-131.
67.Deamer, D. W. (1997) The first living systems: a
bioenergetic perspective, Microbiol. Mol. Biol. Rev., 61,
239-261.
68.Deamer, D. W. (2008) Origins of life: how leaky
were primitive cells? Nature, 454, 37-38.
69.Paula, S., Volkov, A. G., Van Hoek, A. N.,
Haines, T. H., and Deamer, D. W. (1996) Permeation of protons,
potassium ions, and small polar molecules through phospholipid bilayers
as a function of membrane thickness, Biophys. J., 70,
339-348.
70.Boucher, Y., Kamekura, M., and Doolittle, W. F.
(2004) Origins and evolution of isoprenoid lipid biosynthesis in
archaea, Mol. Microbiol., 52, 515-527.
71.Pereto, J., Lopez-Garcia, P., and Moreira, D.
(2004) Ancestral lipid biosynthesis and early membrane evolution,
Trends Biochem. Sci., 29, 469-477.
72.Koonin, E. V., and Martin, W. (2005) On the
origin of genomes and cells within inorganic compartments, Trends
Genet., 21, 647-654.
73.Koga, Y., and Morii, H. (2007) Biosynthesis of
ether-type polar lipids in archaea and evolutionary considerations,
Microbiol. Mol. Biol. Rev., 71, 97-120.
74.Dibrova, D. V., Galperin, M. Y., and
Mulkidjanian, A. Y. (2014) Phylogenomic reconstruction of archaeal
fatty acid metabolism, Environ. Microbiol., 16,
907-918.
75.Smit, A., and Mushegian, A. (2000) Biosynthesis
of isoprenoids via mevalonate in Archaea: the lost pathway, Genome
Res., 10, 1468-1484.
76.Ourisson, G., and Nakatani, Y. (1994) The
terpenoid theory of the origin of cellular life: the evolution of
terpenoids to cholesterol, Chem. Biol., 1, 11-23.
77.Gotoh, M., Sugawara, A., Akiyoshi, K., Matsumoto,
I., Ourisson, G., and Nakatani, Y. (2007) Possible molecular evolution
of biomembranes: from single-chain to double-chain lipids, Chem.
Biodivers., 4, 837-848.
78.Nakatani, Y., Ribeiro, N., Streiff, S., Desaubry,
L., and Ourisson, G. (2012) Search for the most primitive membranes:
some remaining problems, Orig. Life Evol. Biosph., 42,
497-501.
79.Dibrova, D. V., Chudetsky, M. Y., Galperin, M.
Y., Koonin, E. V., and Mulkidjanian, A. Y. (2012) The role of energy in
the emergence of biology from chemistry, Orig. Life Evol.
Biosph., 42, 459-468.
80.Mulkidjanian, A. Y., and Galperin, M. Y. (2010)
in Structural Bioinformatics of Membrane Proteins (Frishman, D.,
ed.) Springer, Viena, pp. 1-28.
81.Chen, I. A., and Szostak, J. W. (2004) Membrane
growth can generate a transmembrane pH gradient in fatty acid vesicles,
Proc. Natl. Acad. Sci. USA, 101, 7965-7970.
82.Mansy, S. S. (2010) Membrane transport in
primitive cells, Cold Spring Harb. Perspect. Biol., 2,
a002188.
83.Szostak, J. W., Bartel, D. P., and Luisi, P. L.
(2001) Synthesizing life, Nature, 409, 387-390.
84.Mansy, S. S., Schrum, J. P., Krishnamurthy, M.,
Tobe, S., Treco, D. A., and Szostak, J. W. (2008) Template-directed
synthesis of a genetic polymer in a model protocell, Nature,
454, 122-125.
85.Mulkidjanian, A. Y., Bychkov, A. Y., Dibrova, D.
V., Galperin, M. Y., and Koonin, E. V. (2012) Open questions on the
origin of life at anoxic geothermal fields, Orig. Life Evol.
Biosph., 42, 507-516.
86.Pinti, D. L. (2005) in Lectures in
Astrobiology (Gargaud, M., Barbier, B., Martin, H., and Reisse, J.,
eds.) Springer-Verlag, Berlin, pp. 83-111.
87.Foriel, J., Philippot, P., Rey, P., Somogyi, A.,
Banks, D., and Menez, B. (2004) Biological control of Cl/Br and low
sulfate concentration in a 3.5-Gyr-old seawater from North Pole,
Western Australia, Earth Planet. Sci. Lett., 228,
451-463.
88.Aver’ev, V. V. (1961) Proc. Volcanol.
Lab. Rus. Acad. Sci. (Moscow), Issue 19, pp. 90-98.
89.White, D. E., Muffler, L. J. P., and Truesdell,
A. N. (1971) Vapor-dominated hydrothermal systems compared with
hot-water systems, Econ. Geol., 66, 75-97.
90.Fournier, R. O. (2004) Geochemistry and
Dynamics of the Yellowstone National Park Hydrothermal System, US
Geological Survey, Menlo Park, California.
91.Bortnikova, S. B., Gavrilenko, G. M., Bessonova,
E. P., and Lapukhov, A. S. (2009) The hydrogeochemistry of thermal
springs on Mutnovskii Volcano, southern Kamchatka, J. Volcanol.
Seismol., 3, 388-404.
92.Bychkov, A. Y. (2009) Geochemical Model of
Present-Day Ore Formation in the Uzon Caldera [in Russian], GEOS,
Moscow.
93.Maruyama, S., Ikoma, M., Genda, H., Hirose, K.,
Yokoyama, T., and Santosh, M. (2013) The naked planet Earth: most
essential pre-requisite for the origin and evolution of life,
Geosci. Front., 4, 141-165.
94.Galimov, E. M., Natochin, Yu. V., Ryzhenko, B.
N., and Cherkasova, E. V. (2012) Chemical composition of the primary
aqueous phase of the Earth and origin of life, Geochem. Int.,
50, 1048-1068.
95.Guitreau, M., Blichert-Toft, J., Martin, H.,
Mojzsis, S. J., and Albarede, F. (2012) Hf isotope evidence from
Archean granitic rocks for deep-mantle origin of continental crust,
Earth Planet. Sci. Lett., 337/338, 211-223.
96.Warren, P. H., and Wasson, J. T. (1979) The
origin of KREEP, Rev. Geophys. Space Phys., 17,
73-88.
97.Benner, S. A., Kim, H. J., and Carrigan, M. A.
(2012) Asphalt, water, and the prebiotic synthesis of ribose,
ribonucleosides, and RNA, Acc. Chem. Res., 45,
2025-2034.
98.Natochin, Y. V. (2007) The physiological
evolution of animals: sodium is the clue to resolving contradictions,
Herald Russ. Acad Sci., 77, 581-591.
99.Dimroth, P. (1980) A new sodium-transport system
energized by the decarboxylation of oxaloacetate, FEBS Lett.,
122, 234-236.
100.Tokuda, H., and Unemoto, T. (1982)
Characterization of the respiration-dependent Na+ pump in
the marine bacterium Vibrio alginolyticus, J. Biol.
Chem., 257, 10007-10014.
101.Becher, B., Muller, V., and Gottschalk, G.
(1992) N5-methyl-tetrahydromethanopterin:coenzyme M
methyltransferase of Methanosarcina strain Go1 is an
Na+-translocating membrane protein, J. Bacteriol.,
174, 7656-7660.
102.Roberts, P. G., and Hirst, J. (2012) The
deactive form of respiratory complex I from mammalian mitochondria is a
Na+/H+ antiporter, J. Biol. Chem.,
287, 34743-34751.
103.Mayer, F., and Muller, V. (2014) Adaptations of
anaerobic archaea to life under extreme energy limitation, FEMS
Microbiol. Rev., 38, 449-472.
104.Gorecki, K., Hagerhall, C., and Drakenberg, T.
(2014) The Na+ transport in gram-positive bacteria defect in
the Mrp antiporter complex measured with 23Na nuclear
magnetic resonance, Anal. Biochem., 445, 80-86.
105.Moparthi, V. K., Kumar, B., Al-Eryani, Y.,
Sperling, E., Gorecki, K., Drakenberg, T., and Hagerhall, C. (2014)
Functional role of the MrpA- and MrpD-homologous protein subunits in
enzyme complexes evolutionary related to respiratory chain complex I,
Biochim. Biophys. Acta, 1837, 178-185.
106.Lim, J. K., Mayer, F., Kang, S. G., and Muller,
V. (2014) Energy conservation by oxidation of formate to carbon dioxide
and hydrogen via a sodium ion current in a hyperthermophilic archaeon,
Proc. Natl. Acad. Sci. USA, 111, 11497-11502.
107.Luoto, H. H., Belogurov, G. A., Baykov, A. A.,
Lahti, R., and Malinen, A. M. (2011) Na+-translocating
membrane pyrophosphatases are widespread in the microbial world and
evolutionarily precede H+-translocating pyrophosphatases,
J. Biol. Chem., 286, 21633-21642.
108.Baykov, A. A., Malinen, A. M., Luoto, H. H.,
and Lahti, R. (2013) Pyrophosphate-fueled Na+ and
H+ transport in prokaryotes, Microbiol. Mol. Biol.
Rev., 77, 267-276.
109.Weber, J., and Senior, A. E. (2003) ATP
synthesis driven by proton transport in F1F0-ATP
synthase, FEBS Lett., 545, 61-70.
110.Mulkidjanian, A. Y., Makarova, K. S., Galperin,
M. Y., and Koonin, E. V. (2007) Inventing the dynamo machine: the
evolution of the F-type and V-type ATPases, Nat. Rev.
Microbiol., 5, 892-899.
111.Boyer, P. D. (1997) The ATP synthase – a
splendid molecular machine, Annu. Rev. Biochem., 66,
717-749.
112.Pogoryelov, D., Krah, A., Langer, J. D.,
Yildiz, O., Faraldo-Gomez, J. D., and Meier, T. (2010) Microscopic
rotary mechanism of ion translocation in the Fo complex of
ATP synthases, Nat. Chem. Biol., 6, 891-899.
113.Nakanishi-Matsui, M., Sekiya, M., and Futai, M.
(2013) Rotating proton pumping ATPases: subunit/subunit interactions
and thermodynamics, IUBMB Life, 65, 247-254.
114.Walker, J. E. (2013) The ATP synthase: the
understood, the uncertain and the unknown, Biochem. Soc. Trans.,
41, 1-16.
115.Dibrova, D. V., Galperin, M. Y., and
Mulkidjanian, A. Y. (2010) Characterization of the N-ATPase, a
distinct, laterally transferred Na+-translocating form of
the bacterial F-type membrane ATPase, Bioinformatics, 26,
1473-1476.
116.Muller, V., and Gruber, G. (2003) ATP
synthases: structure, function and evolution of unique energy
converters, Cell Mol. Life Sci., 60, 474-494.
117.Meier, T., Krah, A., Bond, P. J., Pogoryelov,
D., Diederichs, K., and Faraldo-Gomez, J. D. (2009) Complete
ion-coordination structure in the rotor ring of
Na+-dependent F-ATP synthases, J. Mol. Biol.,
391, 498-507.
118.Martin, W., and Russell, M. J. (2007) On the
origin of biochemistry at an alkaline hydrothermal vent, Philos.
Trans. R. Soc. Lond. B Biol. Sci., 362, 1887-1925.
119.Sojo, V., Pomiankowski, A., and Lane, N. (2014)
A bioenergetic basis for membrane divergence in archaea and bacteria,
PLoS Biol., 12, e1001926.
120.Krishnamoorthy, G., and Hinkle, P. C. (1984)
Non-ohmic proton conductance of mitochondria and liposomes,
Biochemistry, 23, 1640-1645.
121.Blicher, A., Wodzinska, K., Fidorra, M.,
Winterhalter, M., and Heimburg, T. (2009) The temperature dependence of
lipid membrane permeability, its quantized nature, and the influence of
anesthetics, Biophys. J., 96, 4581-4591.
122.Liberman, E. A., Topaly, V. P., Tsofina, L. M.,
Jasaitis, A. A., and Skulachev, V. P. (1969) Mechanism of coupling of
oxidative phosphorylation and the membrane potential of mitochondria,
Nature, 222, 1076-1078.
123.Skulachev, V. P., Sharaf, A. A., and Liberman,
E. A. (1967) Proton conductors in the respiratory chain and artificial
membranes, Nature, 216, 718-719.
124.Lardy, H. A., and Pressman, B. C. (1956) Effect
of surface active agents on the latent ATPase of mitochondria,
Biochim. Biophys. Acta, 21, 458-466.
125.Korshunov, S. S., Korkina, O. V., Ruuge, E. K.,
Skulachev, V. P., and Starkov, A. A. (1998) Fatty acids as natural
uncouplers preventing generation of O2.- and
H2O2 by mitochondria in the resting state,
FEBS Lett., 435, 215-218.
126.Skulachev, V. P. (1991) Fatty acid circuit as a
physiological mechanism of uncoupling of oxidative phosphorylation,
FEBS Lett., 294, 158-162.
127.Mills, J. K., and Needham, D. (2005) Lysolipid
incorporation in dipalmitoylphosphatidylcholine bilayer membranes
enhances the ion permeability and drug release rates at the membrane
phase transition, Biochim. Biophys. Acta, 1716,
77-96.
128.Soontharapirakkul, K., and Incharoensakdi, A.
(2010) Na+-stimulated ATPase of alkaliphilic halotolerant
cyanobacterium Aphanothece halophytica translocates
Na+ into proteoliposomes via Na+ uniport
mechanism, BMC Biochem., 11, 30.
129.Soontharapirakkul, K., Promden, W., Yamada, N.,
Kageyama, H., Incharoensakdi, A., Iwamoto-Kihara, A., and Takabe, T.
(2011) Halotolerant cyanobacterium Aphanothece halophytica
contains an Na+-dependent F1F0-ATP
synthase with a potential role in salt-stress tolerance, J. Biol.
Chem., 286, 10169-10176.
130.Boiteau, L., and Pascal, R. (2011) Energy
sources, self-organization, and the origin of life, Orig. Life Evol.
Biosph., 41, 23-33.
131.Glagolev, A. N., and Skulachev, V. P. (1978)
The proton pump is a molecular engine of motile bacteria,
Nature, 272, 280-282.
132.Vik, S. B., and Antonio, B. J. (1994) A
mechanism of proton translocation by F1F0 ATP
synthases suggested by double mutants of the a subunit, J. Biol.
Chem., 269, 30364-30369.
133.Engelbrecht, S., and Junge, W. (1997) ATP
synthase: a tentative structural model, FEBS Lett., 414,
485-491.
134.Cherepanov, D. A., Mulkidjanian, A. Y., and
Junge, W. (1999) Transient accumulation of elastic energy in proton
translocating ATP synthase, FEBS Lett., 449, 1-6.
135.Otto, R., Sonnenberg, A. S., Veldkamp, H., and
Konings, W. N. (1980) Generation of an electrochemical proton gradient
in Streptococcus cremoris by lactate efflux, Proc. Natl.
Acad. Sci. USA, 77, 5502-5506.
136.Michel, T. A., and Macy, J. M. (1990)
Generation of a membrane potential by sodium-dependent succinate efflux
in Selenomonas ruminantium, J. Bacteriol., 172,
1430-1435.
137.Lane, N., Allen, J. F., and Martin, W. (2010)
How did LUCA make a living? Chemiosmosis in the origin of life,
Bioessays, 32, 271-280.
138.Harold, F. M., and Van Brunt, J. (1977)
Circulation of H+ and K+ across the plasma
membrane is not obligatory for bacterial growth, Science,
197, 372-373.
139.Silver, I. A., and Erecinska, M. (1997)
Energetic demands of the Na+/K+ ATPase in
mammalian astrocytes, Glia, 21, 35-45.
140.Khan, S., Dapice, M., and Humayun, I. (1990)
Energy transduction in the bacterial flagellar motor. Effects of load
and pH, Biophys. J., 57, 779-796.
141.Skulachev, V. P. (1978) Membrane-linked energy
buffering as the biological function of Na+/K+
gradient, FEBS Lett., 87, 171-179.
142.Bortner, C. D., and Cidlowski, J. A. (2007)
Cell shrinkage and monovalent cation fluxes: role in apoptosis,
Arch. Biochem. Biophys., 462, 176-188.
143.Skulachev, V. P. (1996) Why are mitochondria
involved in apoptosis? Permeability transition pores and apoptosis as
selective mechanisms to eliminate superoxide-producing mitochondria and
cell, FEBS Lett., 397, 7-10.
144.Morth, J. P., Pedersen, B. P., Toustrup-Jensen,
M. S., Sorensen, T. L., Petersen, J., Andersen, J. P., Vilsen, B., and
Nissen, P. (2007) Crystal structure of the sodium-potassium pump,
Nature, 450, 1043-1049.
145.Aravind, L., Galperin, M. Y., and Koonin, E. V.
(1998) The catalytic domain of the P-type ATPase has the haloacid
dehalogenase fold, Trends Biochem. Sci., 23, 127-129.
146.Chan, H., Babayan, V., Blyumin, E., Gandhi, C.,
Hak, K., Harake, D., Kumar, K., Lee, P., Li, T. T., Liu, H. Y., et
al. (2010) The P-type ATPase superfamily, J. Mol. Microbiol.
Biotechnol., 19, 5-104.
147.Edgar, R. C. (2004) MUSCLE: a multiple sequence
alignment method with reduced time and space complexity, BMC
Bioinformatics, 5, 113.
148.Galperin, M. Y., Makarova, K. S., Wolf, Y. I.,
and Koonin, E. V. (2015) Expanded microbial genome coverage and
improved protein family annotation in the COG database, Nucleic
Acids Res., 43, D261-D269.
149.Crooks, G. E., Hon, G., Chandonia, J. M., and
Brenner, S. E. (2004) WebLogo: a sequence logo generator, Genome
Res., 14, 1188-1190.
150.Voorhees, R. M., Schmeing, T. M., Kelley, A.
C., and Ramakrishnan, V. (2010) The mechanism for activation of GTP
hydrolysis on the ribosome, Science, 330, 835-838.
151.Guindon, S., Dufayard, J. F., Lefort, V.,
Anisimova, M., Hordijk, W., and Gascuel, O. (2010) New algorithms and
methods to estimate maximum-likelihood phylogenies: assessing the
performance of PhyML 3.0, Syst. Biol., 59, 307-321.
152.Tamura, K., Peterson, D., Peterson, N.,
Stecher, G., Nei, M., and Kumar, S. (2011) MEGA5: molecular
evolutionary genetics analysis using maximum likelihood, evolutionary
distance, and maximum parsimony methods, Mol. Biol. Evol.,
28, 2731-2739.
153.Van Raaij, M. J., Abrahams, J. P., Leslie, A.
G., and Walker, J. E. (1996) The structure of bovine
F1-ATPase complexed with the antibiotic inhibitor aurovertin
B, Proc. Natl. Acad. Sci. USA, 93, 6913-6917.
154.Krissinel, E., and Henrick, K. (2004)
Secondary-structure matching (SSM), a new tool for fast protein
structure alignment in three dimensions, Acta Cryst.,
D60, 2256-2268.
155.Schomburg, I., Chang, A., Placzek, S., Sohngen,
C., Rother, M., Lang, M., Munaretto, C., Ulas, S., Stelzer, M., Grote,
A., et al. (2013) BRENDA in 2013: integrated reactions, kinetic
data, enzyme function data, improved disease classification: new
options and contents in BRENDA, Nucleic Acids Res., 41,
D764-772.
156.Dimroth, P. (1997) Primary sodium ion
translocating enzymes, Biochim. Biophys. Acta, 1318,
11-51.
157.Dimroth, P., and von Ballmoos, C. (2008) ATP
synthesis by decarboxylation phosphorylation, Results Probl. Cell
Differ., 45, 153-184.
158.Hilpert, W., and Dimroth, P. (1982) Conversion
of the chemical energy of methylmalonyl-CoA decarboxylation into a
Na+ gradient, Nature, 296, 584-585.
159.Biegel, E., and Muller, V. (2010) Bacterial
Na+-translocating ferredoxin:NAD+ oxidoreductase,
Proc. Natl. Acad. Sci. USA, 107, 18138-18142.
160.Laubinger, W., and Dimroth, P. (1988)
Characterization of the ATP synthase of Propionigenium modestum
as a primary sodium pump, Biochemistry, 27,
7531-7537.
161.Takase, K., Yamato, I., and Kakinuma, Y. (1993)
Cloning and sequencing of the genes coding for the A and B subunits of
vacuolar-type Na+-ATPase from Enterococcus hirae.
Coexistence of vacuolar- and F0F1-type ATPases in
one bacterial cell, J. Biol. Chem., 268, 11610-11616.
162.Solioz, M., and Davies, K. (1994) Operon of
vacuolar-type Na+-ATPase of Enterococcus hirae, J.
Biol. Chem., 269, 9453-9459.
163.Cheng, J., Guffanti, A. A., and Krulwich, T. A.
(1997) A two-gene ABC-type transport system that extrudes
Na+ in Bacillus subtilis is induced by ethanol or
protonophore, Mol. Microbiol., 23, 1107-1120.
164.Malinen, A. M., Belogurov, G. A., Baykov, A.
A., and Lahti, R. (2007) Na+-pyrophosphatase: a novel
primary sodium pump, Biochemistry, 46, 8872-8878.
165.Inoue, K., Ono, H., Abe-Yoshizumi, R.,
Yoshizawa, S., Ito, H., Kogure, K., and Kandori, H. (2013) A
light-driven sodium ion pump in marine bacteria, Nat. Commun.,
4, 1678.