REVIEW: Does Mitochondrial Fusion Require Transmembrane Potential?
I. E. Karavaeva1, K. V. Shekhireva1, F. F. Severin2, and D. A. Knorre2*
1Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University, 119992 Moscow, Russia2Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119992 Moscow, Russia; E-mail: knorre@belozersky.msu.ru
* To whom correspondence should be addressed.
Received December 23, 2014; Revision received January 13, 2015
Dissipation of transmembrane potential inhibits mitochondrial fusion and thus prevents reintegration of damaged mitochondria into the mitochondrial network. Consequently, damaged mitochondria are removed by autophagy. Does transmembrane potential directly regulate the mitochondrial fusion machinery? It was shown that inhibition of ATP-synthase induces fragmentation of mitochondria while preserving transmembrane potential. Moreover, mitochondria of the yeast Saccharomyces cerevisiae retain the ability to fuse even in the absence of transmembrane potential. Metazoan mitochondria in some cases retain ability to fuse for a short period even in a depolarized state. It also seems unlikely that transmembrane potential-based regulation of mitochondrial fusion would prevent reintegration of mitochondria with damaged ATP-synthase into the mitochondrial network. Such reintegration could lead to clonal expansion of mtDNAs harboring deleterious mutations in ATP synthase. We speculate that transmembrane potential is not directly involved in regulation of mitochondrial fusion but affects mitochondrial NTP/NDP ratio, which in turn regulates their fusion.
KEY WORDS: mitochondria, fusion, fission, membrane potential, mtDNA, regulationDOI: 10.1134/S0006297915050053
Abbreviations: CCCP, carbonyl cyanide m-chlorophenylhydrazone; DIC, differential interference contrast; FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; mtDNA, mitochondrial DNA; NDP, nucleotide diphosphate; NTP, nucleotide triphosphate; PINK 1, PTEN-induced putative kinase 1.
Mitochondria are present in almost all metabolically active eukaryotic
cells with only a few exceptions, such as mammalian erythrocytes [1], lens fiber cells [2], and
unicellular parasite microsporidia, which have lost their mitochondria
during evolution [3]. In most cell types,
mitochondria form a tubular network with constantly changing
morphology: the network segments fuse and separate continuously. The
balance between these processes determines the structure of the network
[4-6]. Partial or complete
inhibition of the fission results in the formation of hyperfused
mitochondrial network, whereas disruption of fusion results in
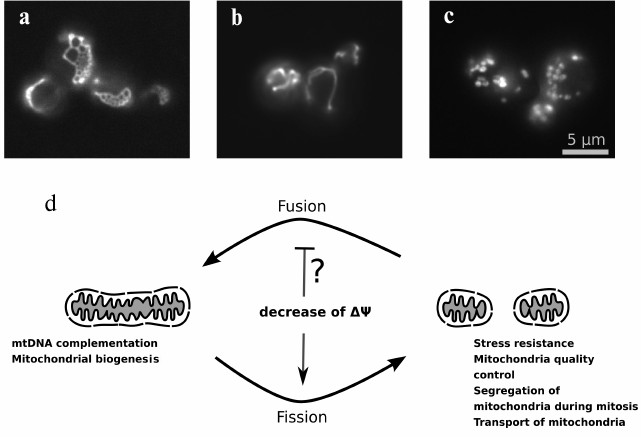
mitochondrial fragmentation (Fig. 1). What is
the biological role of mitochondrial dynamics? Multiple cellular
activities require the processes of mitochondrial fission and
fusion.
Fig. 1. A balance between fission and fusion rates determines mitochondrial network morphology: a) yeast cells with deletion of DNM1 gene (W303 MATa dnm1::KanMX4); b) wild-type cells (W303 MATa); c) yeast cells with deletion of mitofusin gene homolog FZO1 (W303 MATa fzo1::KanMX4). Mitochondrial morphology was visualized using the expression of mitochondrially targeted GFP in yeast cells. d) Scheme illustrating an effect of the inner mitochondrial membrane depolarization and the functional role of fused and fragmented state of mitochondrial network.
1. During cell division, mitochondria have to be distributed between two newly forming cells. Obviously, this process must be preceded by mitochondrial fission. Indeed, mitochondrial network structure changes during the progression through the cell cycle [7, 8]. The average length of mitochondrial filaments is higher in the interphase than in mitosis [9]. Moreover, mitochondrial fragmentation was detected during mitosis in normal rat kidney cells [10], which might be necessary for equal distribution of mitochondria between the two daughter cells [11].
2. Intensive mitochondrial fragmentation is a stage of activation of apoptosis – programmed cell death in multicellular organisms [12, 13]. This is important due to the presence of the apoptotic proteins in the mitochondrial intermembrane space: cytochrome c [14], AIF [15], SMAC and several others [16, 17]. During apoptosis, these proteins are released through the pores in the outer mitochondrial membrane into the cytoplasm where they bind to their receptors and initiate subsequent events of the cascade. However, the release of the proteins can be restrained because most of them are localized within cristae that form as folds of the mitochondrial inner membrane [18]. On one hand, mitochondrial fission can lead to reorganization of the inner mitochondrial membrane structure promoting the release of the apoptotic proteins from cristae. On the other hand, indirect connection between mitochondrial fragmentation and activation of apoptosis is also possible: changes in the mitochondrial structure during fission stimulate the oligomerization of the proapoptotic protein Bax in the outer mitochondrial membrane resulting in permeabilization of the membrane [19].
3. Mitochondrial fusion can follow gamete fusion of some unicellular eukaryotes (i.e. baker’s yeast Saccharomyces cerevisiae) that inherit mitochondria from both parents. After fusion, mtDNA molecules from both parental cells can recombine in the diploid cells [20, 21]. As a result, due to a random segregation of the newly formed mtDNA variants, the subsequent generations of diploid cells may contain all possible combinations of parental mtDNA alleles. Consequently, a functional variant of the mitochondrial genome can be generated from two genomes carrying different loss of function mutations in their coding sequences [22]. Such complementation of mtDNA is also supposed to take place in muscle cells of multicellular animals [23]. Inhibition of mitochondrial fusion was shown to induce muscle atrophy and fast accumulation of mutations in mtDNA in these cells [24].
4. Structural reorganization of mitochondrial morphology can be associated with metabolic changes in the cell. In murine embryonic fibroblasts producing the most of cell’s ATP by means of glycolysis, mitochondria exist as short separate entities, whereas the switch of metabolism to oxidative phosphorylation leads to an increase of the average length of mitochondria in these cells [25]. Also, it can be supposed that under stress the mitochondrial network splits up to localize the damage (e.g. regions with proton leak) in a separate small mitochondrion. Indeed, laser-induced local damage to a mitochondrial membrane was shown to cause the depolarization of an entire filament [26]. In this case, mitochondrial fragmentation has an obvious role as it can prevent uncontrolled energy dissipation in the cell [27].
5. Fission and fusion are necessary for mitochondria quality control [28]. It was shown that periodically mitochondria undergo fission resulting in formation of a short fragment – daughter unit, which subsequently depolarizes [29]. Possibly, the fission process per se leads to a transient increase in conductivity of the inner mitochondrial membrane. Thereafter, the daughter mitochondrion either repolarizes and fuses back to the mitochondrial network or stays depolarized and afterwards undergoes engulfment by an autophagosome and digestion [29-31]. The combination of these processes allows disposing of damaged macromolecules, which cannot be eliminated by other mechanisms [32], and, as it was recently suggested, prevents accumulation and expansion of mutant mtDNA molecules [33, 34].
All these processes require tight regulation of mitochondrial fission and fusion. In this review, we consider bioenergetic aspects of the regulation of mitochondrial dynamics and focus on the role of transmembrane potential in the fission and fusion processes.
MITOCHONDRIAL FISSION: MECHANISM AND REGULATION
In animals, mitochondrial fission is mediated by dynamin-related protein Drp1, which oligomerizes on the surface of the mitochondrial outer membrane [35]. The domain structure of this protein is evolutionarily conserved and contains GTPase, Middle, Variable, and GTPase effector (GED) domains [36]. Earlier, a homolog of Drp1, Dnm1, was found in fungi [37]. Dnm1 is able to associate with lipid membranes, and the association induces the protein self-assembly into a helix wrapping around a membrane vesicle (mitochondria or artificial liposome) several times [38, 39]. Although Dnm1 is able to bind with the membrane surface in the absence of GTP, this is unlikely to happen in vivo due to its high affinity to GTP [36]. Besides, nonhydrolyzable GTP analogs promote Dnm1 polymerization and enhance its interaction with liposomes in vitro. This indicates that in cells Dnm1 binds to the mitochondrial membranes in GTP-bound state [40]. Hydrolysis of GTP induces conformational change of the protein and constriction of the helix: its diameter decreases from ~80 to ~25 nm [39]. It is suggested that such narrowing of the mitochondrial vesicle diameter is sufficient for its division, although the details of the further steps of mitochondrial fission remain unclear. Active transport of mitochondria along the cytoskeleton could be required for later stages of the fission process: stretching of membrane vesicles by kinesin was shown to promote their dynamin-mediated fragmentation [41].
How do dynamin-related proteins select mitochondrial binding sites? It was shown that Drp1 binds to specific receptors localized on the outer mitochondrial membrane: Mid49, Mid51 [42], or Mff [43]. Saccharomyces cerevisiae cells use another receptor – Fis1, which binds Dnm1 through the adapter proteins Caf4 and Mdv1 [44, 45]. In animals, the dynamin-related protein Drp1 can also interact with Fis1 [46], but its deletion does not affect mitochondrial fission [47]. Instead, mammalian Fis1 is involved in the control of organelle degradation [48]. The recruitment of dynamin-related proteins to the mitochondrial membrane surface is not the only function of Drp1 receptors. It was shown that Mid51 and Drp1 copolymerization leads to even greater constriction of the helix diameter (~15 nm) than in case of homo-oligomerization of Drp1 [49]. Besides, according to recent studies, Mid51 protein specifically binds GDP or ADP [50]. This observation suggests a functional link between mitochondrial fission and the metabolic status of the cells, although further investigation is required to provide a complete understanding of this link since a mutation in the nucleotide-binding domain of Mid51 does not inhibit its association with Drp1 [50].
Regulation of Drp1 binding also occurs at the level of its posttranslational modifications. The uncoupler FCCP induces mitochondrial fragmentation in wild-type cells but not in cells expressing nonfunctional mutant variant of Drp1 K38A [51, 52]. It turned out that the uncoupler causes a decrease in Ser637 phosphorylation levels of Drp1, thus activating Drp1 and preventing its binding to the receptors Mid49 and Mid51 [53]. The effect of the uncoupler is due to a release of Ca2+ from mitochondria [54] leading to activation of the protein phosphatase calcineurin, which dephosphorylates Drp1 [55]. As a result, uncouplers induce fragmentation of the mitochondrial network promoting mitochondrial fission.
It is to be noted that the receptors Drp1/Dnm1 colocalize with the endoplasmic reticulum–mitochondria contact sites where fission events usually occur [56].
MITOCHONDRIAL FUSION
Mitochondrial fusion occurs in three stages: 1) docking of two mitochondria via their outer membranes; 2) outer membranes fusion; 3) inner membranes fusion. The first two stages are mediated by the mitofusin proteins: Mfn1 and Mfn2 in animals [57, 58] and Fzo1 in baker’s yeast [59]. Mitofusins (Mfn) and Fzo1 proteins are also dynamin-related GTPases, although the molecular mechanism of their action is studied less extensively than in the case of Drp1. It was shown that Fzo1 integrates into the outer mitochondrial membrane, binds GTP, and then forms a dimer. The dimer is able to interact with another Fzo1 dimer localized in the membrane of the opposing mitochondrion [60]. The formation of such Fzo1 trans-dimers is associated with the appearance of disulfide bonds between the proteins on adjacent outer membranes of fusing mitochondria [61]. Subsequent GTP hydrolysis causes conformational rearrangement of mitofusins, followed by their ubiquitination and proteolytic degradation [60]. Unhydrolyzable GTP analogs inhibit the ability of mitochondria to fuse [62]. The next stage – fusion of the inner mitochondrial membranes – is mediated by the dynamin-related GTPase OPA1 (or Mgm1 in yeast) [63, 64]. This protein binds to the outer surface of the inner mitochondrial membrane and undergoes subsequent processing by proteases localized in the intermembrane space [65]. The processing leads either to protein maturation [25] or to its inactivation [65, 66] depending on the site where proteolysis occurs.
The processes of outer and inner membrane fusion in vitro can be separated in time [25, 62]. In such case, the fusion is accompanied by the appearance of separate mitoplasts encapsulated by a common membrane. Such structures were also detected in intact animal cells [67, 68] and in yeast [27, 69]. The lipid composition of mitochondrial membranes also plays an important role in their fusion. Indeed, simultaneous suppression of cardiolipin and phosphatidylethanolamine synthesis led to inhibition of mitochondrial fusion in yeast [70].
ROLE OF TRANSMEMBRANE POTENTIAL IN MITOCHONDRIAL FUSION
The uncoupling of respiration and oxidative phosphorylation induces mitochondrial fragmentation. This effect is usually explained by inhibition of mitochondrial fusion [71, 72]. Indeed, similar to uncouplers, respiratory chain inhibitors [73-81], mutations in the genes encoding respiratory chain subunits [74, 82, 83], or the loss of mtDNA [72, 81, 84, 85] cause a decrease in mitochondrial transmembrane potential and induce fragmentation of mitochondria. It was shown that uncouplers prevent mitochondrial membrane fusion in vitro [62, 86]. In HeLa cells, mitochondrial fusion was inhibited by addition of the uncoupler CCCP but not by other compounds that decrease cellular ATP levels [72].
All these observations led to a suggestion that the ability of mitochondrial membranes to fuse is regulated by their potential. Indeed, such an explanation seems to be logical: fusion of uncoupled mitochondria with the mitochondrial network might result in uncontrolled dissipation of energy in the cell.
But how can changes in transmembrane potential affect the activity of proteins mediating mitochondrial fusion? At least two possible mechanisms were suggested. (i) Depolarization of mitochondrial membrane inhibits protein import into mitochondria. The inhibition of import of PTEN-induced putative kinase 1 (PINK1) left it anchored on the outer surface of the outer mitochondrial membrane [87]. Subsequently, PINK1 recruits ubiquitin ligase Parkin from the cytoplasm, which ubiquitinates proteins on the outer membrane surface. It was shown that mitofusins could be targets of Parkin [88, 89]. Ubiquitinated mitofusins are proteolytically degraded, which, in turn, prevents mitochondrial outer membrane fusion [90]. Another possibility is that ubiquitination of mitofusins per se can prevent mitochondrial fusion by inhibiting their ability for trans-dimerization. (ii) The second mechanism relies on uncoupler-induced OPA1 proteolysis [91] and its consequent inactivation. Processing of OPA1 is mediated by the proteases Yme1 and OMA1 associated with the outer mitochondrial membrane [65, 91]. This implies existence of a functional connection between their activities and the transmembrane potential. However, a question remains – is ΔΨ a critical parameter for mitochondrial fusion capability? Obviously, the effects of uncouplers in the cell are not limited only to a decrease of ΔΨ. They can also induce changes in NTP/NDP ratios both in the mitochondrial matrix and in the intermembrane space/cytoplasm. The decrease in transmembrane potential should also affect the redox status of the respiratory chain cofactors as well as the distribution of ions and metabolites (which require ΔΨ for their transport) between mitochondrial matrix and intermembrane space. Moreover, a number of experiments do not support the hypothesis that the transmembrane potential directly regulates mitochondrial fusion.
1. The ATP synthase inhibitor oligomycin induces fragmentation of mitochondria in many cell types of multicellular animals [72, 73, 75, 80]. However, inhibition of ATP synthase by oligomycin does not decrease the mitochondrial transmembrane potential if the respiratory chain is active and a significant proton leak is absent. It was suggested that this effect of oligomycin could be explained by its inhibitory action on the enzyme Na+/K+-ATPase [79]. However, the mitochondrial network fragmentation was also detected in cells containing a mutation in a gene of ATP synthase subunit ATP6 encoded in the mitochondrial genome [82]. Moreover, the inhibitory effect of oligomycin was directly shown for the outer membrane fusion in vitro in the absence of an ATP regenerating system [25]. The effect of oligomycin on fusion can be due to OPA1 inactivation, which is induced by ATP decrease. It was shown that proteolytic inactivation of OPA1 could be induced by oligomycin as well as by uncouplers or the respiratory chain inhibitor sodium azide [92]. Thus, nucleotides rather than transmembrane potential play a role in the proteolytic inactivation of OPA1.
2. Oxidized glutathione induces mitochondrial fusion in vitro [61]. An increase in oxidized glutathione concentration indicates oxidative stress in the cell, which may be either a cause or a consequence of mitochondrial deenergization. Conversely, addition of the antioxidant Trolox can shift the equilibrium between the fusion and fission processes towards the formation of elongated mitochondria mediated by induction of Mfn2 expression [93]. It can be suggested that a decrease in transmembrane potential has two opposite effects on mitochondrial dynamics that develop at different rates. The fast effect relies on cellular redox status regulation and promotes mitochondrial fusion, and the slow one promotes fission.
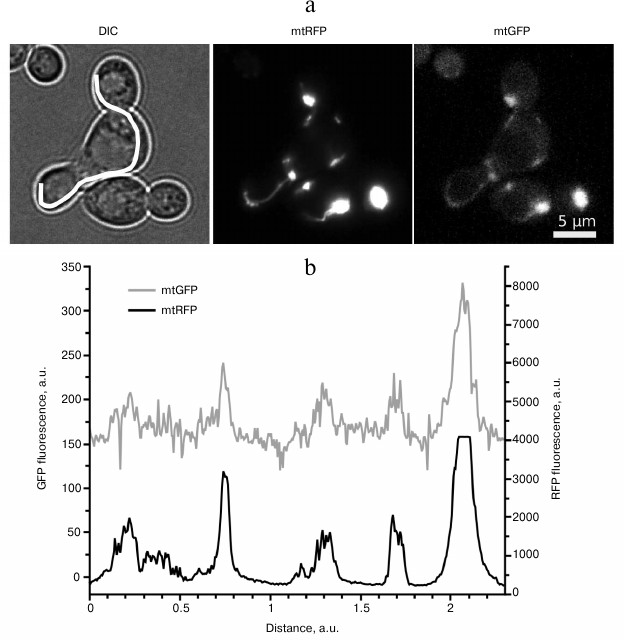
3. Saccharomyces cerevisiae cells lacking mtDNA (Rho0) cannot generate transmembrane potential via respiratory chain function. Instead, they use ATP/ADP antiporter to polarize their mitochondrial membrane up to +55 mV [94]. Nevertheless, such mitochondria are able to fuse, although less effectively than the wild-type mitochondria (Rho+) [95]. Besides, we showed that addition of the uncoupler FCCP does not fully inhibit mitochondrial fusion during mating of two respiratory-competent yeast strains (Fig. 2). The uncoupler was not able to inhibit mitochondrial fusion even in the case when one of the mating strains harbored nonfunctional mtDNA, despite the high sensitivity of such yeast mutants to uncouplers [94]. In the experiments, we used a concentration of FCCP that was sufficient to stimulate respiration of yeast cells more than two-fold [96].
Fig. 2. Uncoupling does not inhibit mitochondrial fusion in Saccharomyces cerevisiae. a) Representative photographs illustrating colocalization of mtGFP and mtRFP signals in a zygote in the presence of 1 µM FCCP. Haploid cells of the opposite mating types expressing either mitochondrially targeted GFP (mtGFP) or mitochondrially targeted RFP (mtRFP) were grown on a selective synthetic medium. Cells were mixed under the conditions with inhibited fluorescent protein expression and in the presence of 1 µM FCCP. The mtGFP and mtRFP proteins were visualized by fluorescence microscopy after 8 h. DIC, differential interference contrast. b) Fluorescence intensity profile of mtGFP and mtRFP proteins. The profile line is shown on the DIC image.
4. Finally, in vivo monitoring of mitochondrial dynamics in animal cells showed that addition of uncouplers induced efflux of potential-sensitive fluorescent dye from mitochondria, whereas mitochondrial fusion was not inhibited and occurred soon after mitochondrial depolarization [97].
Along with the experimental data, there is also a logical argument against transmembrane potential being a key regulator of mitochondrial fusion. Mitochondrial dynamics are essential for mitochondrial quality control. This process provides elimination of nonfunctional mitochondria, which cannot be removed in other ways [28]. It was supposed that fusion and fission events ensure shuffling of mtDNA molecules within the mitochondrial reticulum. This allows elimination of defective mtDNA molecules by the selective degradation of mitochondrial fragments that contain them [33, 34]. Such a mechanism of selection is possible if respiratory chain complexes diffuse slowly in the inner mitochondrial membrane and remain close to the mtDNA molecule that encodes them. Indeed, it was shown that, due to restricted lateral diffusion, the majority of respiratory chain complexes remain within the same cristae where they were synthesized [98]. Another important condition for such a selection mechanism is the inhibition of fusion of mitochondria containing mutant mtDNA with the rest of the mitochondrial network. However, if the fusion process were regulated by the transmembrane potential, then deleterious mutations in the mitochondrially encoded ATP synthase genes would escape such selection because inactivation of ATP synthase inhibition should result in at least transient increase in mitochondrial transmembrane potential. We are not aware of any studies demonstrating the specific accumulation of mutations in the mitochondrial genes encoding ATP synthase subunits.
Thus, the question arises: what parameter of mitochondria controls mitochondrial fusion? We suppose that NTP/NDP ratio in the intermembrane space is the most likely to be the parameter that limits mitochondrial ability to fuse. This type of regulation can be possible due to the diffusion barrier for nucleotide exchange between the cytoplasm and the intermembrane space [99] and high (0.3 mM) Km value of Mgm1 for GTP [100]. Our hypothesis is consistent with the fact that the addition of uncouplers inhibits mitochondrial fusion: a decrease in transmembrane potential in the presence of functional ATP synthase is likely to cause a decrease in the NTP/NDP ratio. At the same time, NTP/NDP ratio is at least as essential as mitochondrial transmembrane potential from the point of view of cell physiology. Mitochondria perform a variety of functions [101], and energy transformation is one of them; therefore, elimination of mitochondria unable to produce ATP does make sense from cell physiology point of view.
Our hypothesis predicts that “mild” uncouplers are inefficient in preventing mitochondrial fusion because the “mild” uncouplers do not target mitochondria with relatively low levels of transmembrane potential. Therefore, while these compounds prevent mitochondrial hyperpolarization, they affect the rate of ATP synthesis to a much lesser extent [102]. Indeed, very recent experiments showed that continuous incubation of cells in the presence of low concentrations of dodecyl triphenylphosphonium or dinitrophenol, which can act as a “mild” uncoupler [103], increases of the average length of mitochondria [104]. We expect that further in vitro experiments with “mild” uncouplers will clarify the role of the transmembrane potential and NTP/NDP ratio in mitochondrial dynamics.
The study was financially supported by the Russian Scientific Foundation (grant No. 14-14-00181).
REFERENCES
1.Mortensen, M., Ferguson, D. J. P., and Simon, A. K.
(2010) Mitochondrial clearance by autophagy in developing erythrocytes:
clearly important, but just how much so? Cell Cycle, 9,
1901-1906.
2.Bassnett, S., and Beebe, D. C. (1992) Coincident
loss of mitochondria and nuclei during lens fiber cell differentiation,
Dev. Dyn., 194, 85-93.
3.Bullerwell, C. E., and Lang, B. F. (2005) Fungal
evolution: the case of the vanishing mitochondrion, Curr. Opin.
Microbiol., 8, 362-369.
4.Westermann, B. (2010) Mitochondrial dynamics in
model organisms: what yeasts, worms and flies have taught us about
fusion and fission of mitochondria, Semin. Cell Dev. Biol.,
21, 542-549.
5.Zhao, J., Lendahl, U., and Nister, M. (2013)
Regulation of mitochondrial dynamics: convergences and divergences
between yeast and vertebrates, Cell. Mol. Life Sci., 70,
951-976.
6.Van der Bliek, A. M., Shen, Q., and Kawajiri, S.
(2013) Mechanisms of mitochondrial fission and fusion, Cold Spring
Harb. Perspect. Biol., 5, a011072.
7.Dewey, W. C., and Fuhr, M. A. (1976) Quantification
of mitochondria during the cell cycle of Chinese hamster cells, Exp.
Cell Res., 99, 23-30.
8.Mitra, K. (2013) Mitochondrial fission-fusion as an
emerging key regulator of cell proliferation and differentiation,
Bioessays, 35, 955-964.
9.Lee, S., Kim, S., Sun, X., Lee, J.-H., and Cho, H.
(2007) Cell cycle-dependent mitochondrial biogenesis and dynamics in
mammalian cells, Biochem. Biophys. Res. Commun., 357,
111-117.
10.Mitra, K., Wunder, C., Roysam, B., Lin, G., and
Lippincott-Schwartz, J. (2009) A hyperfused mitochondrial state
achieved at G1-S regulates cyclin E buildup and entry into S phase,
Proc. Natl. Acad. Sci. USA, 106, 11960-11965.
11.Taguchi, N., Ishihara, N., Jofuku, A., Oka, T.,
and Mihara, K. (2007) Mitotic phosphorylation of dynamin-related GTPase
Drp1 participates in mitochondrial fission, J. Biol. Chem.,
282, 11521-11529.
12.Skulachev, V. P., Bakeeva, L. E., Chernyak,
B. V., Domnina, L. V., Minin, A. A., Pletjushkina, O. Y.,
Saprunova, V. B., Skulachev, I. V., Tsyplenkova, V. G., Vasiliev,
J. M., Yaguzhinsky, L. S., and Zorov, D. B. Thread-grain transition of
mitochondrial reticulum as a step of mitoptosis and apoptosis, Mol.
Cell. Biochem., 256-257, 341-358.
13.Lee, Y., Jeong, S.-Y., Karbowski, M., Smith, C.
L., and Youle, R. J. (2004) Roles of the mammalian mitochondrial
fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis, Mol.
Biol. Cell, 15, 5001-5011.
14.Liu, X., Kim, C. N., Yang, J., Jemmerson, R., and
Wang, X. (1996) Induction of apoptotic program in cell-free extracts:
requirement for dATP and cytochrome c, Cell, 86,
147-157.
15.Susin, S. A., Lorenzo, H. K., Zamzami, N., Marzo,
I., Snow, B. E., Brothers, G. M., Mangion, J., Jacotot, E., Costantini,
P., Loeffler, M., Larochette, N., Goodlett, D. R., Aebersold, R.,
Siderovski, D. P., Penninger, J. M., and Kroemer, G. (1999) Molecular
characterization of mitochondrial apoptosis-inducing factor,
Nature, 397, 441-446.
16.Ola, M. S., Nawaz, M., and Ahsan, H. (2011) Role
of Bcl-2 family proteins and caspases in the regulation of apoptosis,
Mol. Cell. Biochem., 351, 41-58.
17.Gulbins, E., Dreschers, S., and Bock, J. (2003)
Role of mitochondria in apoptosis, Exp. Physiol., 88,
85-90.
18.Scorrano, L., Ashiya, M., Buttle, K., Weiler, S.,
Oakes, S. A., Mannella, C. A., and Korsmeyer, S. J. (2002) A distinct
pathway remodels mitochondrial cristae and mobilizes cytochrome
c during apoptosis, Dev. Cell, 2, 55-67.
19.Montessuit, S., Somasekharan, S. P., Terrones,
O., Lucken-Ardjomande, S., Herzig, S., Schwarzenbacher, R., Manstein,
D. J., Bossy-Wetzel, E., Basanez, G., Meda, P., and Martinou, J. C.
(2010) Membrane remodeling induced by the dynamin-related protein Drp1
stimulates Bax oligomerization, Cell, 142, 889-901.
20.Dujon, B., Slonimski, P. P., and Weill, L. (1974)
Mitochondrial genetics IX: a model for recombination and segregation of
mitochondrial genomes in Saccharomyces cerevisiae,
Genetics, 78, 415-437.
21.Fritsch, E. S., Chabbert, C. D., Klaus, B., and
Steinmetz, L. M. (2014) A genome-wide map of mitochondrial DNA
recombination in yeast, Genetics, 198, 755-771.
22.Kochmak, S. A., Knorre, D. A., Sokolov, S. S.,
and Severin, F. F. (2011) Physiological scenarios of programmed loss of
mitochondrial DNA function and death of yeast, Biochemistry
(Moscow), 76, 167-171.
23.Hom, J., and Sheu, S.-S. (2009) Morphological
dynamics of mitochondria – a special emphasis on cardiac muscle
cells, J. Mol. Cell. Cardiol., 46, 811-820.
24.Chen, H., Vermulst, M., Wang, Y. E., Chomyn, A.,
Prolla, T. A., McCaffery, J. M., and Chan, D. C. (2010) Mitochondrial
fusion is required for mtDNA stability in skeletal muscle and tolerance
of mtDNA mutations, Cell, 141, 280-289.
25.Mishra, P., Carelli, V., Manfredi, G., and Chan,
D. C. (2014) Proteolytic cleavage of Opa1 stimulates mitochondrial
inner membrane fusion and couples fusion to oxidative phosphorylation,
Cell Metab., 19, 630-641.
26.Amchenkova, A. A., Bakeeva, L. E., Chentsov, Y.
S., Skulachev, V. P., and Zorov, D. B. (1988) Coupling membranes as
energy-transmitting cables. I. Filamentous mitochondria in fibroblasts
and mitochondrial clusters in cardiomyocytes, J. Cell Biol.,
107, 481-495.
27.Knorre, D. A., Ojovan, S. M., Saprunova, V. B.,
Sokolov, S. S., Bakeeva, L. E., and Severin, F. F. (2008) Mitochondrial
matrix fragmentation as a protection mechanism of yeast
Saccharomyces cerevisiae, Biochemistry (Moscow),
73, 1254-1259.
28.Youle, R. J., and van der Bliek, A. M. (2012)
Mitochondrial fission, fusion, and stress, Science, 337,
1062-1065.
29.Twig, G., Elorza, A., Molina, A. J. A., Mohamed,
H., Wikstrom, J. D., Walzer, G., Stiles, L., Haigh, S. E., Katz, S.,
Las, G., Alroy, J., Wu, M., Py, B. F., Yuan, J., Deeney, J. T., Corkey,
B. E., and Shirihai, O. S. (2008) Fission and selective fusion govern
mitochondrial segregation and elimination by autophagy, EMBO J.,
27, 433-446.
30.Narendra, D., Tanaka, A., Suen, D.-F., and Youle,
R. J. (2008) Parkin is recruited selectively to impaired mitochondria
and promotes their autophagy, J. Cell Biol., 183,
795-803.
31.Matsuda, N., Sato, S., Shiba, K., Okatsu, K.,
Saisho, K., Gautier, C. A., Sou, Y.-S., Saiki, S., Kawajiri, S., Sato,
F., Kimura, M., Komatsu, M., Hattori, N., and Tanaka, K. (2010) PINK1
stabilized by mitochondrial depolarization recruits Parkin to damaged
mitochondria and activates latent Parkin for mitophagy, J. Cell
Biol., 189, 211-221.
32.Ashrafi, G., and Schwarz, T. L. (2013) The
pathways of mitophagy for quality control and clearance of
mitochondria, Cell Death Differ., 20, 31-42.
33.Vidoni, S., Zanna, C., Rugolo, M., Sarzi, E., and
Lenaers, G. (2013) Why mitochondria must fuse to maintain their genome
integrity, Antioxid. Redox Signal., 19, 379-388.
34.Knorre, D. A., Popadin, K. Y., Sokolov, S. S.,
and Severin, F. F. (2013) Roles of mitochondrial dynamics under
stressful and normal conditions in yeast cells, Oxid. Med. Cell.
Longev., 2013, 139491.
35.Smirnova, E., Griparic, L., Shurland, D. L., and
van der Bliek, A. M. (2001) Dynamin-related protein Drp1 is required
for mitochondrial division in mammalian cells, Mol. Biol. Cell,
12, 2245-2256.
36.Bui, H. T., and Shaw, J. M. (2013) Dynamin
assembly strategies and adaptor proteins in mitochondrial fission,
Curr. Biol., 23, R891-R899.
37.Otsuga, D., Keegan, B. R., Brisch, E., Thatcher,
J. W., Hermann, G. J., Bleazard, W., and Shaw, J. M. (1998) The
dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in
yeast, J. Cell Biol., 143, 333-349.
38.Ingerman, E., Perkins, E. M., Marino, M., Mears,
J. A., McCaffery, J. M., Hinshaw, J. E., and Nunnari, J. (2005) Dnm1
forms spirals that are structurally tailored to fit mitochondria, J.
Cell Biol., 170, 1021-1027.
39.Mears, J. A., Lackner, L. L., Fang, S., Ingerman,
E., Nunnari, J., and Hinshaw, J. E. (2011) Conformational changes in
Dnm1 support a contractile mechanism for mitochondrial fission, Nat.
Struct. Mol. Biol., 18, 20-26.
40.Frohlich, C., Grabiger, S., Schwefel, D.,
Faelber, K., Rosenbaum, E., Mears, J., Rocks, O., and Daumke, O. (2013)
Structural insights into oligomerization and mitochondrial remodeling
of dynamin 1-like protein, EMBO J., 32, 1280-1292.
41.Roux, A., Uyhazi, K., Frost, A., and De Camilli,
P. (2006) GTP-dependent twisting of dynamin implicates constriction and
tension in membrane fission, Nature, 441, 528-531.
42.Palmer, C. S., Osellame, L. D., Laine, D.,
Koutsopoulos, O. S., Frazier, A. E., and Ryan, M. T. (2011) MiD49 and
MiD51, new components of the mitochondrial fission machinery, EMBO
Rep., 12, 565-573.
43.Gandre-Babbe, S., and van der Bliek, A. M. (2008)
The novel tail-anchored membrane protein Mff controls mitochondrial and
peroxisomal fission in mammalian cells, Mol. Biol. Cell,
19, 2402-2412.
44.Tieu, Q., and Nunnari, J. (2000) Mdv1p is a WD
repeat protein that interacts with the dynamin-related GTPase, Dnm1p,
to trigger mitochondrial division, J. Cell Biol., 151,
353-366.
45.Griffin, E. E., Graumann, J., and Chan, D. C.
(2005) The WD40 protein Caf4p is a component of the mitochondrial
fission machinery and recruits Dnm1p to mitochondria, J. Cell
Biol., 170, 237-248.
46.James, D. I., Parone, P. A., Mattenberger, Y.,
and Martinou, J.-C. (2003) hFis1, a novel component of the mammalian
mitochondrial fission machinery, J. Biol. Chem., 278,
36373-36379.
47.Otera, H., Wang, C., Cleland, M. M., Setoguchi,
K., Yokota, S., Youle, R. J., and Mihara, K. (2010) Mff is an essential
factor for mitochondrial recruitment of Drp1 during mitochondrial
fission in mammalian cells, J. Cell Biol., 191,
1141-1158.
48.Shen, Q., Yamano, K., Head, B. P., Kawajiri, S.,
Cheung, J. T. M., Wang, C., Cho, J.-H., Hattori, N., Youle, R. J., and
van der Bliek, A. M. (2014) Mutations in Fis1 disrupt orderly disposal
of defective mitochondria, Mol. Biol. Cell, 25,
145-159.
49.Koirala, S., Guo, Q., Kalia, R., Bui, H. T.,
Eckert, D. M., Frost, A., and Shaw, J. M. (2013) Interchangeable
adaptors regulate mitochondrial dynamin assembly for membrane scission,
Proc. Natl. Acad. Sci. USA, 110, E1342-E1351.
50.Richter, V., Palmer, C. S., Osellame, L. D.,
Singh, A. P., Elgass, K., Stroud, D. A., Sesaki, H., Kvansakul, M., and
Ryan, M. T. (2014) Structural and functional analysis of MiD51, a
dynamin receptor required for mitochondrial fission, J. Cell
Biol., 204, 477-486.
51.Cereghetti, G. M., Stangherlin, A., Martins de
Brito, O., Chang, C. R., Blackstone, C., Bernardi, P., and Scorrano, L.
(2008) Dephosphorylation by calcineurin regulates translocation of Drp1
to mitochondria, Proc. Natl. Acad. Sci. USA, 105,
15803-15808.
52.Stavru, F., Palmer, A. E., Wang, C., Youle, R.
J., and Cossart, P. (2013) Atypical mitochondrial fission upon
bacterial infection, Proc. Natl. Acad. Sci. USA, 110,
16003-16008.
53.Loson, O. C., Song, Z., Chen, H., and Chan, D. C.
(2013) Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in
mitochondrial fission, Mol. Biol. Cell, 24, 659-667.
54.Crompton, M. (1999) The mitochondrial
permeability transition pore and its role in cell death, Biochem.
J., 341, 233-249.
55.Cribbs, J. T., and Strack, S. (2007) Reversible
phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and
calcineurin regulates mitochondrial fission and cell death, EMBO
Rep., 8, 939-944.
56.Friedman, J. R., Lackner, L. L., West, M.,
DiBenedetto, J. R., Nunnari, J., and Voeltz, G. K. (2011) ER tubules
mark sites of mitochondrial division, Science, 334,
358-362.
57.Santel, A., and Fuller, M. T. (2001) Control of
mitochondrial morphology by a human mitofusin, J. Cell Sci.,
114, 867-874.
58.Rojo, M., Legros, F., Chateau, D., and Lombes, A.
(2002) Membrane topology and mitochondrial targeting of mitofusins,
ubiquitous mammalian homologs of the transmembrane GTPase Fzo, J.
Cell Sci., 115, 1663-1674.
59.Hermann, G. J., Thatcher, J. W., Mills, J. P.,
Hales, K. G., Fuller, M. T., Nunnari, J., and Shaw, J. M. (1998)
Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p,
J. Cell Biol., 143, 359-373.
60.Anton, F., Fres, J. M., Schauss, A., Pinson, B.,
Praefcke, G. J. K., Langer, T., and Escobar-Henriques, M. (2011) Ugo1
and Mdm30 act sequentially during Fzo1-mediated mitochondrial outer
membrane fusion, J. Cell Sci., 124, 1126-1135.
61.Shutt, T., Geoffrion, M., Milne, R., and McBride,
H. M. (2012) The intracellular redox state is a core determinant of
mitochondrial fusion, EMBO Rep., 13, 909-915.
62.Meeusen, S., McCaffery, J. M., and Nunnari, J.
(2004) Mitochondrial fusion intermediates revealed in vitro,
Science, 305, 1747-1752.
63.Cipolat, S., Martins de Brito, O., Dal Zilio, B.,
and Scorrano, L. (2004) OPA1 requires mitofusin 1 to promote
mitochondrial fusion, Proc. Natl. Acad. Sci. USA, 101,
15927-15932.
64.Sesaki, H., Southard, S. M., Yaffe, M. P., and
Jensen, R. E. (2003) Mgm1p, a dynamin-related GTPase, is essential for
fusion of the mitochondrial outer membrane, Mol. Biol. Cell,
14, 2342-2356.
65.Song, Z., Chen, H., Fiket, M., Alexander, C., and
Chan, D. C. (2007) OPA1 processing controls mitochondrial fusion and is
regulated by mRNA splicing, membrane potential, and Yme1L, J. Cell
Biol., 178, 749-755.
66.Anand, R., Wai, T., Baker, M. J., Kladt, N.,
Schauss, A. C., Rugarli, E., and Langer, T. (2014) The i-AAA protease
YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission,
J. Cell Biol., 204, 919-929.
67.Malka, F., Guillery, O., Cifuentes-Diaz, C.,
Guillou, E., Belenguer, P., Lombes, A., and Rojo, M. (2005) Separate
fusion of outer and inner mitochondrial membranes, EMBO Rep.,
6, 853-859.
68.Song, Z., Ghochani, M., McCaffery, J. M., Frey,
T. G., and Chan, D. C. (2009) Mitofusins and OPA1 mediate sequential
steps in mitochondrial membrane fusion, Mol. Biol. Cell,
20, 3525-3532.
69.Meeusen, S., DeVay, R., Block, J., Cassidy-Stone,
A., Wayson, S., McCaffery, J. M., and Nunnari, J. (2006) Mitochondrial
inner-membrane fusion and crista maintenance requires the
dynamin-related GTPase Mgm1, Cell, 127, 383-395.
70.Joshi, A. S., Thompson, M. N., Fei, N.,
Huttemann, M., and Greenberg, M. L. (2012) Cardiolipin and
mitochondrial phosphatidylethanolamine have overlapping functions in
mitochondrial fusion in Saccharomyces cerevisiae, J. Biol.
Chem., 287, 17589-17597.
71.Ishihara, N., Jofuku, A., Eura, Y., and Mihara,
K. (2003) Regulation of mitochondrial morphology by membrane potential,
and DRP1-dependent division and FZO1-dependent fusion reaction in
mammalian cells, Biochem. Biophys. Res. Commun., 301,
891-898.
72.Legros, F., Lombes, A., Frachon, P., and Rojo, M.
(2002) Mitochondrial fusion in human cells is efficient, requires the
inner membrane potential, and is mediated by mitofusins, Mol. Biol.
Cell, 13, 4343-4354.
73.Guillery, O., Malka, F., Frachon, P., Milea, D.,
Rojo, M., and Lombes, A. (2008) Modulation of mitochondrial morphology
by bioenergetics defects in primary human fibroblasts, Neuromuscl.
Disord., 18, 319-330.
74.Benard, G., Bellance, N., James, D., Parrone, P.,
Fernandez, H., Letellier, T., and Rossignol, R. (2007) Mitochondrial
bioenergetics and structural network organization, J. Cell Sci.,
120, 838-848.
75.De Vos, K. J., Allan, V. J., Grierson, A. J., and
Sheetz, M. P. (2005) Mitochondrial function and actin regulate
dynamin-related protein 1-dependent mitochondrial fission, Curr.
Biol., 15, 678-683.
76.Barsoum, M. J., Yuan, H., Gerencser, A. A., Liot,
G., Kushnareva, Y., Graber, S., Kovacs, I., Lee, W. D., Waggoner, J.,
Cui, J., White, A. D., Bossy, B., Martinou, J.-C., Youle, R. J.,
Lipton, S. A., Ellisman, M. H., Perkins, G. A., and Bossy-Wetzel, E.
(2006) Nitric oxide-induced mitochondrial fission is regulated by
dynamin-related GTPases in neurons, EMBO J., 25,
3900-3911.
77.Pham, N.-A., Richardson, T., Cameron, J., Chue,
B., and Robinson, B. H. (2004) Altered mitochondrial structure and
motion dynamics in living cells with energy metabolism defects revealed
by real time microscope imaging, Microsc. Microanal., 10,
247-260.
78.Liot, G., Bossy, B., Lubitz, S., Kushnareva, Y.,
Sejbuk, N., and Bossy-Wetzel, E. (2009) Complex II inhibition by 3-NP
causes mitochondrial fragmentation and neuronal cell death via an NMDA-
and ROS-dependent pathway, Cell Death Differ., 16,
899-909.
79.Pletjushkina, O. Y., Lyamzaev, K. G., Popova, E.
N., Nepryakhina, O. K., Ivanova, O. Y., Domnina, L. V., Chernyak, B.
V., and Skulachev, V. P. (2006) Effect of oxidative stress on dynamics
of mitochondrial reticulum, Biochim. Biophys. Acta, 1757,
518-524.
80.Poliakova, I. A., Zorov, D. B., and Leikina, M.
I. (1995) Structure-activity changes of mitochondria from cultured
cells upon disruption of energy metabolism, Dokl. Akad. Nauk,
342, 553-555.
81.Vorobjev, I. A., and Zorov, D. B. (1983) Diazepam
inhibits cell respiration and induces fragmentation of mitochondrial
reticulum, FEBS Lett., 163, 311-314.
82.Szczepanowska, J., Zabłocki, K., and
Duszynski, J. (2004) Influence of a mitochondrial genetic defect on
capacitative calcium entry and mitochondrial organization in the
osteosarcoma cells, FEBS Lett., 578, 316-322.
83.Koopman, W. J. H., Visch, H.-J., Verkaart, S.,
van den Heuvel, L. W. P. J., Smeitink, J. A. M., and Willems, P. H. G.
M. (2005) Mitochondrial network complexity and pathological decrease in
complex I activity are tightly correlated in isolated human complex I
deficiency, Am. J. Physiol. Cell Physiol., 289,
C881-C890.
84.Van den Ouweland, J. M., Maechler, P., Wollheim,
C. B., Attardi, G., and Maassen, J. A. (1999) Functional and
morphological abnormalities of mitochondria harboring the
tRNA(Leu)(UUR) mutation in mitochondrial DNA derived from patients with
maternally inherited diabetes and deafness (MIDD) and progressive
kidney disease, Diabetologia, 42, 485-492.
85.Legros, F., Malka, F., Frachon, P., Lombes, A.,
and Rojo, M. (2004) Organization and dynamics of human mitochondrial
DNA, J. Cell Sci., 117, 2653-2662.
86.Schauss, A. C., Huang, H., Choi, S.-Y., Xu, L.,
Soubeyrand, S., Bilodeau, P., Zunino, R., Rippstein, P., Frohman, M.
A., and McBride, H. M. (2010) A novel cell-free mitochondrial fusion
assay amenable for high-throughput screenings of fusion modulators,
BMC Biol., 8, 100.
87.Jin, S. M., Lazarou, M., Wang, C., Kane, L. A.,
Narendra, D. P., and Youle, R. J. (2010) Mitochondrial membrane
potential regulates PINK1 import and proteolytic destabilization by
PARL, J. Cell Biol., 191, 933-942.
88.Glauser, L., Sonnay, S., Stafa, K., and Moore, D.
J. (2011) Parkin promotes the ubiquitination and degradation of the
mitochondrial fusion factor mitofusin 1, J. Neurochem.,
118, 636-645.
89.Sarraf, S. A., Raman, M., Guarani-Pereira, V.,
Sowa, M. E., Huttlin, E. L., Gygi, S. P., and Harper, J. W. (2013)
Landscape of the PARKIN-dependent ubiquitylome in response to
mitochondrial depolarization, Nature, 496, 372-376.
90.Gegg, M. E., Cooper, J. M., Chau, K.-Y., Rojo,
M., Schapira, A. H., and Taanman, J.-W. (2010) Mitofusin 1 and
mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon
induction of mitophagy, Hum. Mol. Genet., 19,
4861-4870.
91.Head, B., Griparic, L., Amiri, M., Gandre-Babbe,
S., and van der Bliek, A. M. (2009) Inducible proteolytic inactivation
of OPA1 mediated by the OMA1 protease in mammalian cells, J. Cell
Biol., 187, 959-966.
92.Baricault, L., Segui, B., Guegand, L., Olichon,
A., Valette, A., Larminat, F., and Lenaers, G. (2007) OPA1 cleavage
depends on decreased mitochondrial ATP level and bivalent metals,
Exp. Cell Res., 313, 3800-3808.
93.Distelmaier, F., Valsecchi, F., Forkink, M., van
Emst-de Vries, S., Swarts, H. G., Rodenburg, R. J. T., Verwiel, E. T.
P., Smeitink, J. A. M., Willems, P. H. G. M., and Koopman, W. J. H.
(2012) Trolox-sensitive reactive oxygen species regulate mitochondrial
morphology, oxidative phosphorylation and cytosolic calcium handling in
healthy cells, Antioxid. Redox Signal., 17,
1657-1669.
94.Dupont, C. H., Mazat, J. P., and Guerin, B.
(1985) The role of adenine nucleotide translocation in the energization
of the inner membrane of mitochondria isolated from rho+ and
rho° strains of Saccharomyces cerevisiae, Biochem.
Biophys. Res. Commun., 132, 1116-1123.
95.Sauvanet, C., Duvezin-Caubet, S., Salin, B.,
David, C., Massoni-Laporte, A., di Rago, J.-P., and Rojo, M. (2012)
Mitochondrial DNA mutations provoke dominant inhibition of
mitochondrial inner membrane fusion, PLoS One, 7,
e49639.
96.Antonenko, Y. N., Khailova, L. S., Knorre, D. A.,
Markova, O. V., Rokitskaya, T. I., Ilyasova, T. M., Severina, I.
I., Kotova, E. A., Karavaeva, Y. E., Prikhodko, A. S., Severin, F. F.,
and Skulachev, V. P. (2013) Penetrating cations enhance uncoupling
activity of anionic protonophores in mitochondria, PLoS One,
8, e61902.
97.Liu, X., Weaver, D., Shirihai, O., and Hajnoczky,
G. (2009) Mitochondrial “kiss-and-run”: interplay between
mitochondrial motility and fusion-fission dynamics, EMBO J.,
28, 3074-3089.
98.Muster, B., Kohl, W., Wittig, I., Strecker, V.,
Joos, F., Haase, W., Bereiter-Hahn, J., and Busch, K. (2010)
Respiratory chain complexes in dynamic mitochondria display a patchy
distribution in life cells, PLoS One, 5, e11910.
99.Gellerich, F. N., Trumbeckaite, S., Opalka, J.
R., Seppet, E., Rasmussen, H. N., Neuhoff, C., and Zierz, S. (2000)
Function of the mitochondrial outer membrane as a diffusion barrier in
health and diseases, Biochem. Soc. Trans., 28,
164-169.
100.Meglei, G., and McQuibban, G. A. (2009) The
dynamin-related protein Mgm1p assembles into oligomers and hydrolyzes
GTP to function in mitochondrial membrane fusion, Biochemistry,
48, 1774-1784.
101.Zorov, D. B., Krasnikov, B. F., Kuzminova, A.
E., Vysokikh, M. Yu., and Zorova, L. D. (1997) Mitochondria revisited.
Alternative functions of mitochondria, Biosci. Rep., 17,
507-520.
102.Skulachev, V. P. (1996) Role of uncoupled and
non-coupled oxidations in maintenance of safely low levels of oxygen
and its one-electron reductants, Q. Rev. Biophys., 29,
169-202.
103.Severin, F. F., Severina, I. I., Antonenko, Y.
N., Rokitskaya, T. I., Cherepanov, D. A., Mokhova, E. N., Vyssokikh, M.
Y., Pustovidko, A. V., Markova, O. V., Yaguzhinsky, L. S.,
Korshunova, G. A., Sumbatyan, N. V., Skulachev, M. V., and Skulachev,
V. P. (2010) Penetrating cation/fatty acid anion pair as a
mitochondria-targeted protonophore, Proc. Natl. Acad. Sci. USA,
107, 663-668.
104.Romaschenko, V. P., Zinovkin, R. A., Galkin, I.
I., Zakharova, V. V., Panteleeva, A. A., Tokarchuk, A. V., Lyamzaev, K.
G., Pletyushkina, O. Yu., Chernyak, B. V., and Popova, E. N. (2015)
Uncouplers of oxidative phosphorylation at low concentrations prevent
inflammatory activation of endothelial cells by tumor necrosis factor,
Biochemistry (Moscow), 80, 610-619.