Pure Mitochondrial DNA Does Not Activate Human Neutrophils in vitro
A. S. Prikhodko1, A. K. Shabanov2, L. A. Zinovkina1, E. N. Popova3, M. A. Aznauryan1, N. O. Lanina1, M. V. Vitushkina4, and R. A. Zinovkin3,4*
1Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University, 119991 Moscow, Russia2Sklifosovsky Research Institute of Emergency Care, 129010 Moscow, Russia
3Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia
4Faculty of Biology, Lomonosov Moscow State University, 119234 Moscow, Russia; fax: +7 (495) 939-0338; E-mail: roman.zinovkin@gmail.com
* To whom correspondence should be addressed.
Received December 22, 2014; Revision received January 22, 2015
Excessive activation of the innate immune system often leads to fatal consequences and can be considered as one of the phenoptotic events. After traumatic injury, various components of mitochondria are released into the circulation and stimulate myeloid cells of the innate immunity. Presumably, mitochondrial DNA (mtDNA) might activate immune cells (Zhang, Q., et al. (2010) Nature, 464, 104-107). In the present study, we investigated the role of mtDNA as a direct activator of human neutrophils, as well as a prognostic marker in patients with severe trauma. Quantitative determination of mtDNA in the plasma of these patients revealed its significant increase (p < 0.02) in the group of survivors compared to non-survivors. Highly purified mtDNA was not able to induce activation of human neutrophils, thus possibly indicating the existence of additional factor(s) ensuring the recognition of mtDNA as a damage-associated molecular pattern.
KEY WORDS: trauma, extracellular DNA, mitochondrial DNA, neutrophil activation, damage-associated molecular patterns (DAMPs)DOI: 10.1134/S0006297915050168
Abbreviations: DAMP, damage-associated molecular patterns; eDNA, extracellular DNA; fMLP, formyl-Met-Leu-Phe; ISS, injury severity score; MMP9, matrix metalloproteinase 9 (gelatinase); MTD, mitochondrial DAMP; mtDNA, mitochondrial DNA; nDNA, nuclear DNA; P-p38, phosphorylated form of p38 MAPK.
Injury is known to release into the circulation disrupted cell
fragments, including mitochondrial components. These compounds may
function as endogenous damage-associated molecular patterns (DAMPs) and
initiate inflammation, potentially leading to the development of
life-threatening complications [1]. Presumably,
activation of immune cells via mitochondrial DAMP (MTD) is caused by:
(i) mitochondrial proteins with N-terminal formyl-methionine, typical
for bacterial proteins; (ii) mitochondrial cardiolipin; (iii) ATP; and
(iv) mitochondrial DNA (mtDNA) sharing many properties with prokaryotic
DNA [1, 2]. Extracellular DNA
(eDNA) in plasma is mainly present as nuclear DNA (nDNA) and mtDNA.
Increased concentration of eDNA is observed under a variety of
pathological conditions including traumatic injury [3]. The increased mtDNA concentration in blood of
patients with trauma is a potential prognostic marker [4, 5] correlating with trauma
severity and mortality [6, 7].
The majority of investigations are operating with relative, but not absolute concentrations of mtDNA. The exact determination of mtDNA concentrations can help systematize the results obtained in different studies and understand the mechanism of action of these nucleic acids as DAMPs. In the present study, we examined this issue using quantitative determination of extracellular DNA in plasma of 34 patients with severe trauma.
It remains unclear whether mtDNA at the concentrations corresponding to the physiological values can activate the immune response. We have conducted an independent verification of the data on the activation of human neutrophils using mtDNA and found that purified mtDNA does not cause activation of neutrophils. Probably, the previously published papers were operating with unpurified mtDNA preparations, and these impurities could cause neutrophil activation.
These results confirm the predictive value of mtDNA as a prognostic factor for mortality in patients with severe trauma and at the same time raise the question concerning existence of additional factors that allow mtDNA to function as a DAMP.
MATERIALS AND METHODS
Patients’ characteristics. This research was approved by the Ethical Committee of Sklifosovsky Research Institute. We analyzed blood samples of 34 patients with hard concomitant trauma who were treated in the intensive care unit of Sklifosovsky Institute in 2012-2014. Blood samples of 10 healthy volunteers were used as control. Patients over 70 years and patients with combined traumas were not included in the study. Trauma severity was assessed by Injury Severity Score (ISS) given the localization of injury: head, thorax, abdomen, spine, pelvis, and limbs. Patients’ characteristics are presented in Table 1.
Table 1. Baseline characteristics of
patients enrolled in the study
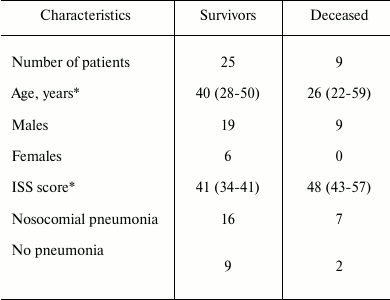
* Data are median (interquartile range).
Sample preparation and DNA extraction. Peripheral blood (3 ml) was collected from patients 24 h after injury. Before donating blood the patients and healthy donors rested for at least 10 min. EDTA-containing blood was centrifuged for 10 min at 3000g. The plasma was additionally centrifuged for 10 min at 10,000g. The upper portion of the plasma supernatant was aliquoted into clean microcentrifuge tubes (volume 0.5 ml) and stored at –20°C until DNA extraction. To monitor the efficiency of DNA extraction, each plasma sample (0.4 ml) was supplemented with 10 µl (1.7 ng/µl) exogenous 8 kb control DNA based on pBlueScriptSKII(–) plasmid. DNA from 100 µl of blood plasma was isolated using Quick-gDNA Blood MiniPrep (Zymo Research, USA) according to the manufacturer’s protocol.
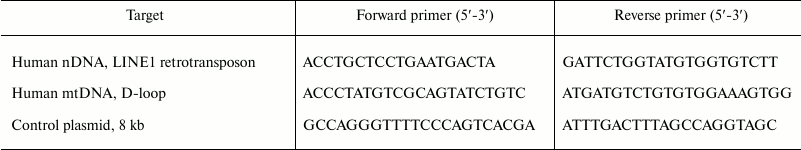
Real-time PCR. Quantitative real-time PCR was performed using an iCycler (Bio-Rad, USA). The PCR reaction system consisted of 5-µl sample under study, 10 µl of Eva Green master mix (Syntol, Russia), 0.5 µl of 10 µM each primer (sequences are presented below), 9.5 µl ddH2O. Each DNA sample was used in triplicate PCR. The targets and sequences of the primer pairs are given in Table 2. Primer specificity to mtDNA was additionally confirmed using DNA from human endothelial cells EA.hy926 lacking mtDNA (Rho0). Primer efficiency was 93-101%. The conditions of thermal cycling included initial denaturation step (94°C for 5 min), followed by 40 cycles of 94°C for 20 sec, 56°C for 20 sec, and 72°C for 20 sec; then 72°C for 5 min. PCR efficiency was calculated using serial dilutions of total DNA from HeLa cells (NEB, England) or control plasmid DNA from 10 µg/ml to 1 pg/ml. The results were normalized according to the amplification of control plasmid DNA [8].
Table 2. Characteristics of primers
Neutrophil isolation. Peripheral blood of healthy donors was collected in heparin-coated tubes. Polymorphonuclear leukocytes (neutrophils) were isolated by dextran sedimentation and subsequent centrifugation on Ficoll-Hypaque gradient (density 1.077 g/ml) as described previously [1]. The purified neutrophils were resuspended in RPMI-1640 (Paneco, Russia) supplemented with 10% low-endotoxin FBS (PAA Laboratories, Germany). Neutrophil viability (>98%) was determined by trypan blue dye exclusion.
Zymography. Isolated neutrophils (106 in 1 ml) were treated with fMLP (formyl-Met-Leu-Phe), mtDNA, or MTD in a final volume of 0.5 ml for 1 h at 37°C in 5% CO2. The quantity of MMP9 gelatinase secreted by neutrophils was measured by zymography in polyacrylamide gel containing 1 mg/ml gelatin as previously described .
Western blot. After incubation with the tested activator compounds, the neutrophils were immediately lysed in hot buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 50 mM DTT, 0.01% bromophenol blue) for 4 min at 95°C. Proteins were separated on 12% polyacrylamide gel and transferred to PVDF membrane (Amersham, USA). Antibodies to β-actin, p38, P-p38 (Cell Signaling, USA), and secondary antibodies conjugated with horseradish peroxidase (Sigma-Aldrich, USA) were used. Membranes were developed with ECL chemiluminescence reagent (Amersham). Densitometric analysis was performed by ImageJ 1.44p software.
Isolation of mitochondrial DAMPs (MTD). Mitochondrial fraction was isolated from rat liver or from human endothelial cells EA.hy926 by differential centrifugation as previously described [1]. Mitochondria were mixed with proteinase inhibitor cocktail (Amresco, USA) and disrupted using a Branson Sonifier 150 (Branson Ultrasonic Corporation, USA) at 100% amplitude, 10 times for 30 sec, intervals for 30 sec. The suspension was centrifuged at 15,000g for 10 min at 4°C, and then the supernatant was centrifuged at 100,000g for 1 h at 4°C. Protein concentration in the samples was measured by the Bradford method using the Bio-Rad protein assay kit (Bio-Rad) according to the manufacturer’s protocol.
mtDNA isolation and purification. Standard mtDNA samples were obtained from mitochondrial fraction using the DNeasy Blood & Tissue kit (Qiagen, USA). Additional purification was performed by ethanol precipitation with sodium acetate. The pellet was air-dried, dissolved in ddH2O, centrifuged at 13,000g for 30 min, and the supernatant was used for the further experiments. Concentration and purity of the samples were determined by measuring OD230, OD260, and OD280 with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, USA).
Data analysis. Results are presented indicating mean values and standard deviation, quartile, and median values or a box plot. Intergroup differences were analyzed by the Mann–Whitney test.
RESULTS
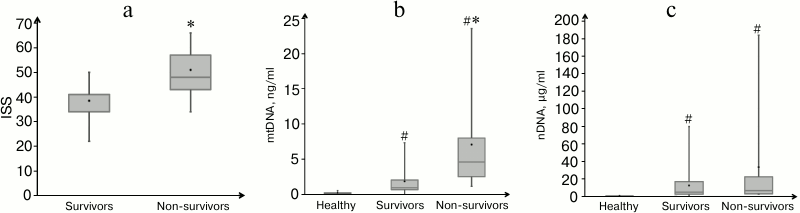
Patients’ characteristics and measurement of nDNA and mtDNA concentration. Trauma severity indicated as ISS score differed significantly between the group of survivors and deceased patients (p = 0.02) (Table 1 and Fig. 1a). In the blood of healthy donors (control group, n = 10), the median concentration of nDNA in plasma was 0.1 µg/ml (interquartile range 0.08-0.16 µg/ml), and the median concentration of mtDNA – 0.19 ng/ml (range 0.15-0.26 ng/ml). The median concentrations of nDNA and mtDNA of surviving patients (n = 25) were 4.5 µg/ml (range 2.2-16.6 µg/ml) and 0.9 ng/ml (range 0.7-2.0 ng/ml), respectively (Fig. 1, b and c). For non-survivors (n = 9), these values were 6.3 µg/ml (range 2.6-22.1 µg/ml) and 4.6 ng/ml (range 2.5-8.0 ng/ml), respectively (Fig. 1, b and c). Statistically significant difference between the groups of survivors and deceased patients was observed only at mtDNA level (p = 0.02).
Fig. 1. Characteristics of trauma patients and their nDNA and mtDNA concentrations in plasma. a) Trauma severity defined as ISS score in the groups of survivors (n = 25) or non-survivors (n = 9). Concentration of mtDNA (b) and nDNA (c) in the plasma of healthy donors (n = 10), and in survivor and non-survivor patients with severe trauma. Mean values are shown as black dots. # p < 0.001, difference between healthy and traumatic patients by Mann–Whitney test; * p < 0.02, difference between deceased and survived patients by Mann–Whitney test.
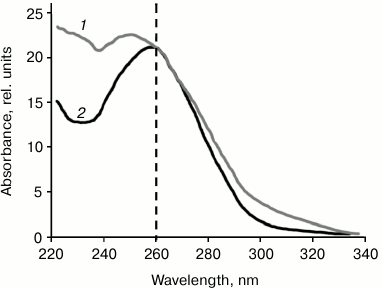
Isolation and purification of mtDNA. mtDNA was isolated from a culture of human EA.hy926 endothelial cells or from rat liver. Standard methods of mtDNA isolation used in [1, 10] resulted in DNA samples with ratio A260/A280 = 1.68 and A260/A230 = 0.95, thus indicating the presence of impurities. An additional purification step (see “Materials and Methods”) changed these ratios to the values typical for pure DNA samples: A260/A280 = 1.91 and A260/A230 = 1.67 (Fig. 2). In further experiments on human neutrophil activation, we compared mtDNA samples after standard isolation and after additional purification.
Fig. 2. UV spectrum of mtDNA isolated from rat liver by standard procedure (1) and after additional purification step (2).
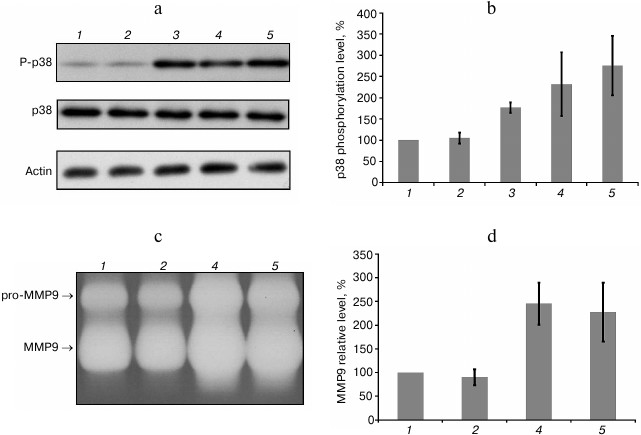
Influence of MTD and mtDNA on neutrophil activation. MAPK p38 phosphorylation (P-p38) can serve as a marker of early human neutrophil activation. MAPK is rapidly activated by stimulation of neutrophils by various DAMPs [11, 12]. In our study, as well as in the study of C. J. Hauser and coworkers [1], MTD and fMLP treatment led to increase in P-p38 level after 5 min, reached a maximum after 10 min, and gradually declined over the next 30 min (data not shown). DNA samples isolated by the standard protocol induced p38 phosphorylation (Fig. 3a). However, purified mtDNA samples obtained from either human endothelial cells, or rat liver, did not increase level of p38 phosphorylation in none of the 11 experiments (Fig. 3, a and b).
Fig. 3. Additionally purified mtDNA does not cause activation of human neutrophils: 1) negative control; 2) additionally purified mtDNA preparation (5 mg/ml); 3) mtDNA isolated by the standard procedure (5 mg/ml); 4) MTD preparation (50 mg/ml); 5) fMLP (10 nM) (positive control). Mean values and standard deviation are shown. a) P-p38 level after 10 min (result of a typical experiment); b) results of densitometric analysis of Western blots (n = 11); c) zymography of culture medium samples after 45 min of incubation (result of a typical experiment); pro-MMP9 is a proenzyme form of MMP9; d) results of densitometric analysis of zymograms (n = 4).
Neutrophil activation also leads to the secretion of matrix metalloproteinase-9 (MMP9) involved in extracellular matrix breakdown under normal and inflammatory conditions. Incubation of neutrophils with fMLP and MTD increased MMP9 in the culture medium as detected by gelatin zymography (Fig. 3, c and d). At the same time, incubation of neutrophils with purified mtDNA at concentrations several orders of magnitude higher than the actual mtDNA concentration in healthy blood and after injuries did not cause any increase in MMP9 secretion (Fig. 3, c and d).
DISCUSSION
Initially, the concept of mtDNA action as a DAMP was based on the works of C. J. Hauser with coworkers [1, 10]. These widely cited studies showed that mtDNA is detected in the blood of traumatic patients at high concentrations (median value 2.7 mg/ml), and mtDNA is able to induce activation of the isolated human neutrophils. The exact mtDNA concentration in blood of patients with severe trauma was identified only in a few studies. Initially, plasma mtDNA concentration was measured in 2004 with the median value of 8,586,300 copies/ml (~0.15 ng/ml) in patients with trauma and 1,607,000 copies/ml (~0.03 ng/ml) in the control group [7]. According to the recent publications, the median value of mtDNA concentrations in patients with severe trauma varied from 1.7 ng/ml [4] to 200 ng/ml [5]. It should be noted that direct methods for estimation of total extracellular DNA not using PCR yield values not exceeding 2-3 mg/ml [13, 14]. Since one diploid cell usually contains from several hundred to several thousand mtDNA molecules, it is easy to calculate that the weight of nDNA several hundred or thousand times exceeds cell mtDNA content. Thus, assuming that traumatic injury leads to equal release of mtDNA and nDNA proportions into the bloodstream, the amount of nuclear DNA in plasma should be two or three orders of magnitude higher than any mtDNA, thus corresponding to the data obtained in our work. However, it should be noted that nDNA/mtDNA ratio may be influenced by many factors, such as removal of extracellular nucleic acid, presence of pathogens in the body, type of damaged tissue, relative mtDNA content therein, and the type of cell death.Considerable variation in the absolute mtDNA quantification may be associated with technical aspects rarely deserving the researcher’s attention. First, quantifying mtDNA requires use of an exogenous control – a double-stranded circular DNA molecule added to the sample prior to isolation of total DNA [15]. In our work, we used a recombinant plasmid as an exogenous control, thus improving the accuracy of mtDNA determination. Second, it is extremely important to use primers specific only to the mtDNA sequence and not to the numerous mitochondrial pseudogenes present in the nuclear genome. Some investigations provide no information concerning this issue [1, 5], potentially leading to the overestimation of mtDNA concentration. We consider that the median plasma mtDNA concentration in the patients with trauma do not exceed 10 ng/ml (Fig. 1a). However, this issue should be examined in independent experiments recruiting more patients. It should also be noted that mtDNA circulating in blood is present not only in plasma, but also on the surface of cells, as well as in microsomes. The concentration and the role of these nucleic acids in the development of the immune response have not yet been determined.
We observed a significant correlation between increased mtDNA level and mortality (Fig. 1b), consistent with other studies [4, 8]. However, it remains unclear whether mtDNA participates in the life-threatening excessive activation of innate immunity, or just represents a marker of cell injury. Earlier, C. J. Hauser’s group demonstrated that mtDNA acts as a DAMP, activating human neutrophils [1, 10]. They showed that mtDNA released after trauma can activate human neutrophils via TLR9. We found that mtDNA purified by standard procedures used in these studies contains unidentified impurities (Fig. 2) that probably cause activation of human neutrophils. None of the 11 experiments with properly purified mtDNA used in concentrations of up to 5 µg/ml resulted in p38 phosphorylation in neutrophils, whereas MTD, fMLP, and “unpurified” mtDNA caused this effect (Fig. 3). Likewise, purified mtDNA did not cause an increase in MMP9 secretion (Fig. 3).
Our work confirmed data that mitochondrial fragments activate inflammatory cascades in neutrophils and stimulate their degranulation. However, we found that pure mtDNA does not cause the activation of neutrophils. Nucleic acids induce neutrophil inflammatory responses by interaction with TLR9 localized mainly in endosomes [16]. Perhaps mtDNA itself in the absence of additional factors is not able to penetrate the cells to interact with TLR9. Also, one cannot exclude the possibility that Hauser et al. [1, 10] dealt with the donors carrying other TLR9 allelic variants, or increased expression of TLR9 led to their exposure at the plasma membrane of neutrophils [17].
It is not new to observe the putative activator/modulator of immune function after more thorough purification appears to have either low or no activity. For instance, nuclear protein HMGB1 (high-mobility group protein B1), after cell damage, functions as powerful DAMP, causing a strong immune response. However, pure preparation of recombinant HMGB1 is virtually inactive [18]. We used additionally purified mtDNA, thus allowing us to define its role in immune response activation more precisely.
There are several options of mtDNA action as an activator of the immune response. First, mtDNA nitrogenous bases may be oxidized in vivo, thus significantly enhancing its activity as a DAMP. Intra-articular injection of mtDNA and oxidized oligonucleotides results in joint inflammation [19]. Besides, it was shown that oxidized mtDNA activates NLRP3 inflammasome [20]. Second, activation of neutrophils may require additional factor(s) (coDAMP). mtDNA inside the cell is associated with nucleoprotein complexes containing helicase TWINKLE [21] and transcription factor TFAM [22], the major component of these complexes. Protein TFAM, as well as HMGB1, contains conserved high-mobility group domains, potentially causing activation of neutrophils. It has been shown that TFAM in combination with formylated mitochondrial proteins enhances activation of human monocytes [23]. Synergistic TFAM action also takes place in combination with DNA, leading to the activation of plasmacytoid dendritic cells [24]. Third, other immune system cells, such as monocytes and macrophages, may serve as mtDNA target and can be activated by unmethylated CpG-rich regions [25] found in prokaryotic and mitochondrial genomes [26].
According to the concept of phenoptosis proposed by V. P. Skulachev, excessive immune activation to either injury or infection may be considered as an execution of the self-destructive program [27]. Identification of exogenous and endogenous “danger patterns” causing septic and sterile inflammation appears to be an important problem. At the same time, despite significant progress in this area [28], the role of the particular mitochondrial components in the activation of innate immunity is still not well understood.
The authors are very grateful to Prof. V. P. Skulachev, without whom this work would not have been done. The authors are also extremely grateful to G. F. Sud’ina (A. N. Belozersky Institute of Physico-Chemical Biology) for her inestimable help with neutrophils and discussing the results.
This work was supported by the Russian Foundation for Basic Research (14-04-01057; neutrophil experiments) and the Russian Science Foundation (14-24-00107; plasma DNA measurement).
REFERENCES
1.Zhang, Q., Raoof, M., Chen, Y., Sumi, Y., Sursal,
T., Junger, W., Brohi, K., Itagaki, K., and Hauser, C. J. (2010)
Circulating mitochondrial DAMPs cause inflammatory responses to injury,
Nature, 464, 104-107.
2.Krysko, D. V., Agostinis, P., Krysko, O., Garg, A.
D., Bachert, C., Lambrecht, B. N., and Vandenabeele, P. (2011) Emerging
role of damage-associated molecular patterns derived from mitochondria
in inflammation, Trends Immunol., 32, 157-164.
3.Tsang, J. C., and Lo, Y. M. (2007) Circulating
nucleic acids in plasma/serum, Pathology, 39,
197-207.
4.Gu, X., Yao, Y., Wu, G., Lv, T., Luo, L., and Song,
Y. (2013) The plasma mitochondrial DNA is an independent predictor for
post-traumatic systemic inflammatory response syndrome, PLoS
One, 8, e72834.
5.Yamanouchi, S., Kudo, D., Yamada, M., Miyagawa, N.,
Furukawa, H., and Kushimoto, S. (2013) Plasma mitochondrial DNA levels
in patients with trauma and severe sepsis: time course and the
association with clinical status, J. Crit. Care, 28,
1027-1031.
6.Lo, Y. M., Rainer, T. H., Chan, L. Y., Hjelm, N.
M., and Cocks, R. A. (2000) Plasma DNA as a prognostic marker in trauma
patients, Clin. Chem., 46, 319-323.
7.Lam, N. Y., Rainer, T. H., Chiu, R. W., Joynt, G.
M., and Lo, Y. M. (2004) Plasma mitochondrial DNA concentrations after
trauma, Clin. Chem., 50, 213-216.
8.Khubutia, M. Sh., Shabanov, A. K., Skulachev, M.
V., Bulava, G. V., Savchenko, I. M., Grebenchikov, O. A., Sergeev, A.
A., Zorov, D. B., and Zinovkin, R. A. (2013) Mitochondrial and nuclear
DNA in patients with severe polytrauma, Gen. Reanimatol.,
9, 30-35.
9.Popova, E. N., Pletjushkina, O. Y., Dugina, V. B.,
Domnina, L. V., Ivanova, O. Y., Izyumov, D. S., Skulachev, V. P., and
Chernyak, B. V. (2010) Scavenging of reactive oxygen species in
mitochondria induces myofibroblast differentiation, Antioxid. Redox
Signal., 13, 1297-1307.
10.Zhang, Q., Itagaki, K., and Hauser, C. J. (2010)
Mitochondrial DNA is released by shock and activates neutrophils via
p38 map kinase, Shock, 34, 55-59.
11.Alvarez, M. E., Bass, J. I. F., Geffner, J. R.,
Calotti, P. X. F., Costas, M., Coso, O. A., Gamberale, R., Vermeulen,
M. E., Salamone, G., and Martinez, D. (2006) Neutrophil signaling
pathways activated by bacterial DNA stimulation, J. Immunol.,
177, 4037-4046.
12.Zu, Y.-L., Qi, J., Gilchrist, A., Fernandez, G.
A., Vazquez-Abad, D., Kreutzer, D. L., Huang, C.-K., and Ramadan, I.
(1998) p38 mitogen-activated protein kinase activation is required for
human neutrophil function triggered by TNF-α or FMLP stimulation,
J. Immunol., 160, 1982-1989.
13.Laktionov, P. P., Tamkovich, S. N., Rykova, E.
Y., Bryzgunova, O. E., Starikov, A. V., Kuznetsova, N. P., Sumarokov,
S. V., Kolomiets, S. A., Sevostianova, N. V., and Vlassov, V. V. (2004)
Extracellular circulating nucleic acids in human plasma in health and
disease, Nucleosides Nucleotides Nucleic Acids, 23,
879-883.
14.Shaked, G., Douvdevani, A., Yair, S., Zlotnik,
A., and Czeiger, D. (2014) The role of cell-free DNA measured by a
fluorescent test in the management of isolated traumatic head injuries,
Scand. J. Trauma Resusc. Emerg. Med., 22, 21.
15.Myers, M. B., Mittelstaedt, R. A., and Heflich,
R. H. (2009) Using phiX174 DNA as an exogenous reference for measuring
mitochondrial DNA copy number, Biotechniques, 47,
867-869.
16.Leifer, C. A., Kennedy, M. N., Mazzoni, A., Lee,
C., Kruhlak, M. J., and Segal, D. M. (2004) TLR9 is localized in the
endoplasmic reticulum prior to stimulation, J. Immunol.,
173, 1179-1183.
17.Lindau, D., Mussard, J., Wagner, B. J., Ribon,
M., Ronnefarth, V. M., Quettier, M., Jelcic, I., Boissier, M. C.,
Rammensee, H. G., and Decker, P. (2013) Primary blood neutrophils
express a functional cell surface Toll‐like receptor 9, Eur.
J. Immunol., 43, 2101-2113.
18.Rouhiainen, A., Tumova, S., Valmu, L., Kalkkinen,
N., and Rauvala, H. (2007) Pivotal advance: analysis of proinflammatory
activity of highly purified eukaryotic recombinant HMGB1 (amphoterin),
J. Leukoc. Biol., 81, 49-58.
19.Collins, L. V., Hajizadeh, S., Holme, E.,
Jonsson, M., and Tarkowski, A. (2004) Endogenously oxidized
mitochondrial DNA induces in vivo and in vitro
inflammatory responses, J. Leukoc. Biol., 75,
995-1000.
20.Shimada, K., Crother, T. R., Karlin, J.,
Dagvadorj, J., Chiba, N., Chen, S., Ramanujan, V. K., Wolf, A. J.,
Vergnes, L., Ojcius, D. M., Rentsendorj, A., Vargas, M., Guerrero, C.,
Wang, Y., Fitzgerald, K. A., Underhill, D. M., Town, T., and Arditi, M.
(2012) Oxidized mitochondrial DNA activates the NLRP3 inflammasome
during apoptosis, Immunity, 36, 401-414.
21.Spelbrink, J. N., Li, F. Y., Tiranti, V., Nikali,
K., Yuan, Q. P., Tariq, M., Wanrooij, S., Garrido, N., Comi, G.,
Morandi, L., Santoro, L., Toscano, A., Fabrizi, G. M., Somer, H.,
Croxen, R., Beeson, D., Poulton, J., Suomalainen, A., Jacobs, H. T.,
Zeviani, M., and Larsson, C. (2001) Human mitochondrial DNA deletions
associated with mutations in the gene encoding Twinkle, a phage T7 gene
4-like protein localized in mitochondria, Nature Genet.,
28, 223-231.
22.Parisi, M. A., and Clayton, D. A. (1991)
Similarity of human mitochondrial transcription factor 1 to high
mobility group proteins, Science, 252, 965-969.
23.Crouser, E. D., Shao, G., Julian, M. W., Macre,
J. E., Shadel, G. S., Tridandapani, S., Huang, Q., and Wewers, M. D.
(2009) Monocyte activation by necrotic cells is promoted by
mitochondrial proteins and formyl peptide receptors, Crit. Care
Med., 37, 2000-2009.
24.Julian, M. W., Shao, G., Bao, S., Knoell, D. L.,
Papenfuss, T. L., VanGundy, Z. C., and Crouser, E. D. (2012)
Mitochondrial transcription factor A serves as a danger signal by
augmenting plasmacytoid dendritic cell responses to DNA, J.
Immunol., 189, 433-443.
25.Hartmann, G., and Krieg, A. M. (1999) CpG DNA and
LPS induce distinct patterns of activation in human monocytes, Gene
Ther., 6, 893-903.
26.Pollack, Y., Kasir, J., Shemer, R., Metzger, S.,
and Szyf, M. (1984) Methylation pattern of mouse mitochondrial DNA,
Nucleic Acids Res., 12, 4811-4824.
27.Skulachev, V. P. (1999) Phenoptosis: programmed
death of an organism, Biochemistry (Moscow), 64,
1418-1426.
28.Hill, S., and Van Remmen, H. (2014) Mitochondrial
stress signaling in longevity: a new role for mitochondrial function in
aging, Redox Biol., 2, 936-944.