Association of High Light-Inducible HliA/HliB Stress Proteins with Photosystem 1 Trimers and Monomers of the Cyanobacterium Synechocystis PCC 6803
D. V. Akulinkina1, Yu. V. Bolychevtseva1, I. V. Elanskaya2, N. V. Karapetyan1, and N. P. Yurina1*
1Bach Institute of Biochemistry, Russian Academy of Sciences, 119071 Moscow, Russia; fax: +7 (495) 954-2732; E-mail: nyurina@inbi.ras.ru2Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia; fax: +7 (495) 939-4309; E-mail: ivelanskaya@mail.ru
* To whom correspondence should be addressed.
Received February 19, 2015; Revision received April 14, 2015
Hlip (high light-inducible proteins) are important for protection of the photosynthetic apparatus of cyanobacteria from light stress. However, the interaction of these proteins with chlorophyll–protein complexes of thylakoids remains unclear. The association of HliA/HliB stress proteins with photosystem 1 (PS1) complexes of the cyanobacterium Synechocystis PCC 6803 was studied to understand their function. Western blotting demonstrated that stress-induced HliA/HliB proteins are associated with PS1 trimers in wild-type cells grown under moderate light condition (40 µmol photons/m2 per sec). The content of these proteins increased 1.7-fold after light stress (150 µmol photons/m2 per sec) for 1 h. In the absence of PS1 trimers (ΔpsaL mutant), the HliA/HliB proteins are associated with PS1 monomers and the PS2 complex. HliA/HliB proteins are associated with PS1 monomers but not with PS1 trimers in Synechocystis PS2-deficient mutant grown at 5 µmol photons/m2 per sec; the content of Hli proteins associated with PS1 monomers increased 1.2-fold after light stress. The HliA/HliB proteins were not detected in wild-type cells of cyanobacteria grown in glucose-supplemented medium at 5 µmol photons/m2 per sec, but light stress induces the synthesis of stress proteins associated with PS1 trimers. Thus, for the first time, the association of HliA/HliB proteins not only with PS1 trimers, but also with PS1 monomers is shown, which suggests a universal role of these proteins in the protection of the photosynthetic apparatus from excess light.
KEY WORDS: high light-inducible proteins HliA/HliB, light stress, photosystem 1, cyanobacteriaDOI: 10.1134/S0006297915100053
Abbreviations: β-DM, n-dodecyl-β-D-maltoside; HliA/HliB proteins, high light-inducible proteins; PS1(2), photosystem 1(2).
Non-photochemical quenching of excess absorbed energy under high light
intensity protects the plant and cyanobacterial photosynthetic
apparatus from destruction [1-4]. Light-induced stress proteins Hlip (high
light-inducible proteins) or SCPs (small chlorophyll a/b
Cab-like proteins) play an important role in protecting the
photosynthetic apparatus of cyanobacteria from destruction [5, 6]. These proteins, required
for the survival of organisms under high light intensity, are found to
be similar to light-harvesting chlorophyll a/b-binding proteins
(Cab) of plants and appear to be their evolutionary predecessors [7, 8]. Hli proteins are located in
the thylakoid membrane and contain one transmembrane helix and a
chlorophyll-binding domain. They are characterized by a low molecular
weight of 6-10 kDa [5-7, 9]. In cyanobacteria, these proteins are coded by
light-inducible hli genes, which are found in all currently
sequenced genomes of cyanobacteria; the number of hli gene
copies depends on the species and the ecotype of cyanobacteria [9, 10].
Five Hli proteins have been identified for the Synechocystis PCC 6803 bacterium (hereinafter referred to as Synechocystis), four of which are low molecular weight proteins HliA/HliB, HliC/HliD; the fifth protein is the C-terminal fragment of ferrochelatase [11, 12]. The genes encoding HliA-HliD are induced by various stress conditions, including not only high intensity light, but also low temperature as well as nitrogen and sulfur starvation [11, 13], which complicates the elucidation of mechanisms behind the induction of the synthesis of these proteins.
Mutant cyanobacteria with inactivated hliA-hliD genes were shown to be sensitive to high intensity light and to differ from wild-type cells by their pigment content, and they are incapable of non-photochemical dissipation of absorbed light energy [14]. Hli proteins can prevent generation of reactive oxygen species by binding free chlorophyll molecules formed under stress, which generate singlet oxygen [15]. Besides that, Hli proteins appear to participate in tetrapyrrole biosynthesis regulation, therefore regulating chlorophyll synthesis [15]. HliD was found to bind chlorophyll a and β-carotene and to dissipate absorbed energy by transferring energy from chlorophyll a (state Gy) to β-carotene (state S1) [16].
Of particular interest are two proteins from this family, HliA and HliB, since they are critical for the survival of Synechocystis cells under light stress [17]. The hliA and hliB genes were found to be functionally complementary [15] and to be characterized by a high degree of nucleotide sequence similarity (87%) [11], which indicates a relatively recent duplication of the gene [12, 18]. The expression of hliA and hliB genes and a high degree of sequence similarity suggests that they play a similar role in the cell, revealing close co-regulation [12].
The data concerning binding these important proteins to chlorophyll–protein complexes of cyanobacterial thylakoid membranes is varied. HliA and HliB in Synechocystis were shown to be associated with PS1 trimers, but not monomers, and to be necessary for their stabilization [17]. On the other hand, HliA and HliB proteins in Synechocystis were found to be bound to the CP47 protein in PS2, but not PS1 [19]. For a better understanding of HliA/HliB functions, it was necessary to determine which chlorophyll–protein complexes in cyanobacterial thylakoids are associated with them. The aim of this study was to assess the association of light-induced stress proteins HliA and HliB with PS1 trimers and monomers in wild-type Synechocystis cells, in a PS2-deficient mutant, and in a mutant unable to form PS1 trimers.
MATERIALS AND METHODS
Cyanobacteria strains and growth conditions. We studied wild-type cells of the cyanobacterium Synechocystis PCC 6803, as well as a PS2-deficient (ΔpsbDI, ΔpsbDII, ΔpsbC) mutant [20] and the ΔpsaL mutant, which is unable to form PS1 trimers due to a psaL gene disruption [21]. The cyanobacteria were grown in BG-11 liquid medium [22] at 30°C under constant fluorescent daylight illumination lamps and ambient aeration by a magnetic stirrer until mid-log phase of growth. Wild-type cells and the cells of mutant unable to form PS1 trimers were grown at medium light intensity of 40 µmol photons/m2 per sec. The PS2-deficient mutant cells were grown at low light intensity (5 µmol photons/m2 per sec) supplemented with 5 mM glucose and antibiotics (chloramphenicol 20 µg/ml, spectinomycin 20 µg/ml). The ΔpsaL mutant was grown in the presence of kanamycin (80 µg/ml). To create light stress, wild-type cells and PS2-deficient mutants grown under the aforementioned conditions were illuminated by high-intensity light (150 µmol photons/m2 per sec) for 1 h. The use of not very high light stress intensity was intended to prevent photodestruction and cell death, especially for the PS2-deficient mutant. In comparative studies of wild-type cells and PS2-deficient mutant cells, the wild-type cells were pre-adapted to the mutant growth conditions.
Thylakoid membrane isolation and chlorophyll–protein complex fractionation. Thylakoid membranes were isolated by the method described in [23, 24]. For extraction of native photosystem complexes from the thylakoid membrane, the mild nonionic n-dodecyl-β-D-maltoside (or β-DM) detergent was used. It was added at chlorophyll/detergent ratio of 1 : 15. After incubation at 4°C for 30 min, the lysate was centrifuged at 18,000g for 10 min. Chlorophyll–protein complexes were extracted by anion-exchange chromatography on a DEAE-Toyopearl-650 column [23, 24].
Evaluation of chlorophyll content and PS1 complex activity. Chlorophyll a content in samples was evaluated in ethanol extract [25].
PS1 trimer fraction activity was determined by the capability of P700 (primary electron donor in PS1 reaction center) photooxidation as a light-induced change in absorption at 810 nm (against 870 nm) under illumination by 730-nm light. The measurements were performed using a DUAL-PAM-101 fluorimeter with the ED-P700 DW-101 device (Walz, Effelrich, Germany). The PS2 activity was estimated by the variable fluorescence, which was measured with the PAM-101 fluorimeter with 680-nm light.
Isolation of proteins, SDS-PAGE, and Western blotting. Proteins were fractionated using PAGE in the presence of SDS [26]. Protein content was measured by the Bradford method [27]. Samples containing 20 µg of protein were applied to a lane. The samples were incubated at 95°C for 10 min before application, and then centrifuged at 18,000g for 10 min.
Protein electrophoresis was performed in Tris-glycine buffer (25 mM Tris, 250 mM glycine, 0.1% SDS, pH 7.5). The proteins were transferred from gel to a nitrocellulose membrane in a blotting chamber with Tris-glycine transfer buffer (25 mM Tris, 250 mM glycine, 20% ethanol, 0.02% SDS, pH 7.5) over 1 h at 200 mA. Then the membrane with the transferred proteins was put for 1 h at 4°C in TBST blocking buffer (50 mM Tris-HCl, 200 mM NaCl, 0.1% Tween 20, pH 7.5) supplemented with 5% dry skim milk, and then primary antibodies were added. We used polyclonal rabbit antibodies to HliA/HliB (1 : 4000) (Abcam, USA). The membrane was incubated with the antibodies overnight at 4°C under constant stirring. As secondary antibodies, we used goat anti-rabbit IgG conjugated to horseradish peroxidase (1 : 10,000) (AgriSera, Sweden). Each step was accompanied by repeated washing of membranes with TBST buffer. Immune complexes on a membrane were detected with the ECL fluorescent detection system (GE Healthcare, England), and the signals were registered on X-ray film (Retina, Germany). The film was scanned and the data were processed with the Image J program (http://rsbweb.nih.gov/ij/).
RESULTS
Solubilization of thylakoid membranes and extraction of chlorophyll–protein complexes. Thylakoid membranes were lysed with the mild nonionic detergent β-DM, using 1 : 15 chlorophyll/detergent ratio. In preliminary experiments, various β-DM concentrations were used for thylakoid membrane solubilization (with chlorophyll/detergent ratio of 1 : 15; 1 : 18; and 1 : 20). For further work, the chlorophyll/detergent ratio of 1 : 15 was chosen since this concentration allowed a sufficient yield of chlorophyll–protein complexes together with the lowest formation of free pigments. The same β-DM concentration was used in [17].
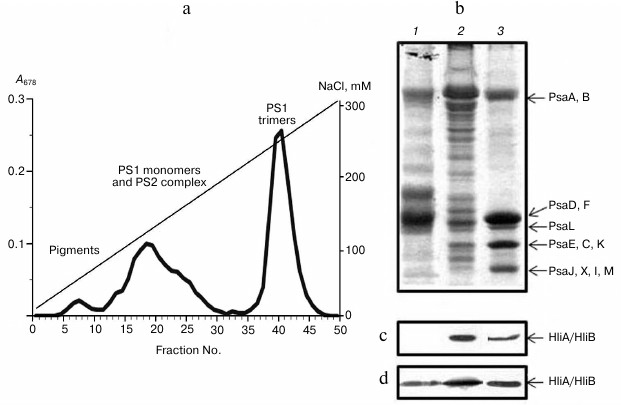
Fractionation of chlorophyll–protein complexes from Synechocystis cyanobacterium thylakoid membranes with anion-exchange chromatography yielded three separate peaks, characterized by absorption spectroscopy as well as by protein composition and PS1 activity. We isolated a PS1 trimer fraction, a fraction containing PS1 monomers together with PS2 complex, and a fraction of free chlorophylls and carotenoids (Fig. 1a). Chlorophyll content in PS1 trimers of wild-type cells averaged 68 ± 4% of the total chlorophyll, meaning that most of thylakoid membrane chlorophyll of this cyanobacterium is located in PS1 trimers, like in Arthrospira platensis studied earlier [28].
The comparison of light-induced change in P700 absorption in PS1 trimers and photosynthetic membranes from wild-type cells indicated activity of isolated PS1 trimers. PAGE revealed the fact that the PS1 trimer fraction from wild-type cells is intact according to their protein content, since it contains all typical protein components of PS1 (Fig. 1b): high molecular weight PsaA and PsaB and low molecular weight proteins (PsaD,F; PsaL; PsaE,C,K; and PsaJ,X,I,M). Fractions containing PS1 monomers and PS2 complex were represented by proteins from both photosystems. From PS2-deficient Synechocystis cells, PS1 monomers were isolated completely free of PS2.
Fig. 1. Association of stress-induced proteins HliA/HliB with chlorophyll–protein complexes from thylakoid membranes of Synechocystis wild-type cells. a) Chromatographic profile of chlorophyll–protein complexes fractionation on a DEAE-Toyopearl 650M anion-exchange column; b) electrophoretogram of photosystem proteins after anion-exchange chromatography in fractions 17-20 (1), 23-26 (2), and 40-43 (3). Proteins HliA/HliB in the fractions were detected by Western blotting in cells grown under normal conditions (c) or in cells exposed to light stress (d).
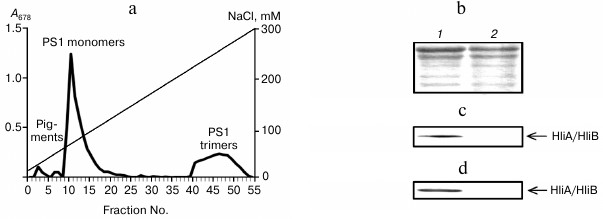
Association of HliA/HliB proteins with PS1 monomers in PS2-deficient mutant. To demonstrate the association of HliA/HliB with PS1 monomer, a PS2-deficient Synechocystis mutant containing only PS1 in thylakoid membranes was used. Fractionation of the mutant chlorophyll–protein complexes on an anion-exchange column yielded three peaks: PS1 trimers, PS1 monomers, and free pigments. As indicated in Fig. 2a, PS1 content in the mutant is decreased as compared to PS1 monomer content in wild-type cells grown under normal conditions (Fig. 1a). Western blotting showed HliA/HliB proteins to be associated only with the PS1 monomer fraction and to be absent in the PS1 trimer fraction (Fig. 2, b and c). PS2-deficient mutant cells were exposed to light stress (150 µmol photons/m2 per sec, 1 h). After light stress, the HliA/HliB proteins were also detected in the fraction containing PS1 monomers (fractions 10-14), with HliA/HliB content increased by 1.2-fold (Fig. 2d) compared to cells grown under normal illumination (40 µmol photons/m2 per sec). HliA/HliB proteins were not detected in mutant PS1 trimer fraction after light stress (Fig. 2d). Therefore, the study conducted on PS2-deficient mutant Synechocystis cells showed that HliA/HliB proteins can be associated with PS1 monomers and that their synthesis is induced by moderate light stress.
Fig. 2. Association of HliA/HliB proteins with PS1 monomers in PS2-deficient mutant Synechocystis cells. a) Fractionation profile of chlorophyll–protein complexes of mutant cells; b) electrophoretogram of photosystem proteins after anion-exchange chromatography and staining with Coomassie R-250, used for application of equal protein aliquots. Lanes: 1) fractions 10-14; 2) fractions 45-49. HliA/HliB proteins in the fractions were detected by Western blotting in cells grown under normal conditions (c) or in cells exposed to light stress (d).
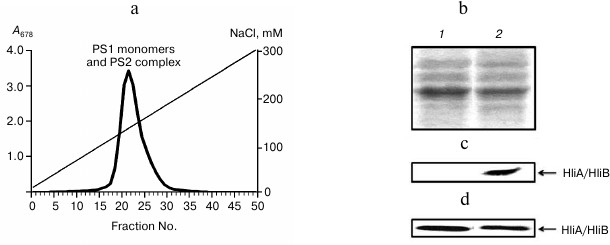
Fig. 3. Association of HliA/HliB proteins with PS1 monomers in ΔpsaL mutant cells of Synechocystis lacking PS1 trimers. a) Fractionation profile of mutant cell chlorophyll–protein complexes; b) electrophoregram of photosystem proteins after anion-exchange chromatography and staining with Coomassie R-250, used for application of equal protein aliquots. Lanes: 1) fractions 22-24; 2) fractions 26-30. HliA/HliB proteins in the fractions were detected by Western blotting in cells grown under normal conditions (c) or in cells exposed to light stress (d).
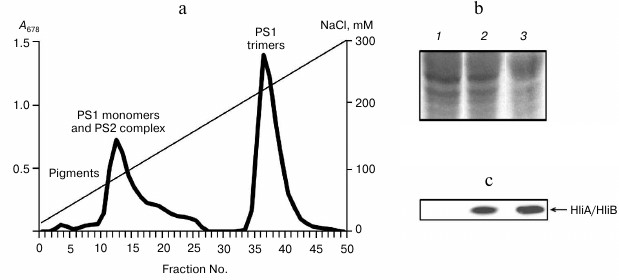
Fig. 4. Association of HliA/HliB proteins with chlorophyll–protein complexes of wild-type Synechocystis cells grown in glucose medium under low light (5 µmol photons/m2 per sec). a) Fractionation profile of chlorophyll–protein complexes from cells exposed to light stress; b) electrophoregram of proteins stained with Coomassie R-250, used for application of equal protein aliquots. Lanes: 1) fractions 10-14; 2) fractions 17-22; 3) fractions 36-40. c) HliA/HliB proteins in the fractions were detected by Western blotting in cells exposed to light stress.
In contrast to higher plants, up to 80% of chlorophyll in cyanobacterial cells is located in PS1 complex [28], which is present in thylakoids predominantly in the form of trimers [20, 29-31]. The presence of PS1 trimers is supposed to be essential for higher complex stability and protection from photodestruction [17-30]. Chlorophyll–protein complexes from thylakoid membranes contain, in addition to such core proteins as PsaA, PsaB, and low molecular weight proteins, also additional proteins, such as IsiA and Hli. In this study, we analyzed HliA/HliB localization in chlorophyll–protein complexes of PS1 trimers and monomers in Synechocystis PCC 6803 cyanobacteria thylakoid membranes. Most articles devoted to studies of Hli proteins localization in chlorophyll–protein complexes are associated with analyzing their localization in PS2. Light-induced HliA/HliB proteins were shown to be localized in cyanobacterial PS2 [19, 32-35]. HliA/HliB proteins are supposed to participate in chlorophyll stabilization for its re-use in repairing damaged PS2 complex and in biogenesis of newly synthesized PS2 [16, 33, 34]. Contradictory data was obtained on HliA/HliB localization in PS1. For example, HliA/HliB proteins were found not to be associated with Synechocystis PS1 [33]. On the other hand, HliA/HliB proteins were shown to be present in PS1 trimers and to be absent in cyanobacterial PS1 monomers [17]. We determined that HliA/HliB proteins are associated with PS1 trimers from cells grown under normal conditions and that their concentration increases 1.7-fold under light stress. Our results correlate with data of Wang et al. [17] on the presence of Hli proteins in PS1 trimers from wild-type cells under stress. Wang et al. [17] did not detect Hli proteins in the fraction containing PS1 monomers and PS2 complex, which may be caused by photodestruction under severe light stress (200-400 µmol photons/m2 per sec for 12 h). Data similar to ours on the presence of HliA/HliB proteins in cells grown under low intensity light was obtained in an earlier study [19].
Our study of PS2-deficient Synechocystis cells has shown for the first time that HliA/HliB proteins in cyanobacterial cells grown under normal conditions are associated with PS1 monomers and that their concentration increases under light stress. This data correlate with the study carried out on higher plant cells. The α-helical protein Ohp from Arabidopsis thaliana, which is evolutionarily homologous to Hlip, was shown to be bound to a PS1 monomer [10].
Conflicting data may be explained by the fact that the PS1 protein turnover time greatly exceeds that of PS2 proteins [36]. Because of this, the concentration of additional HliA/HliB proteins participating in chlorophyll salvaging and associated with PS1 is less than in PS2. Their concentration may be lower than the detection limit for the methods used in those studies. Moreover, the differences may be caused by different cell growth conditions [36]. The connection of Hli proteins with PS1 monomers and trimers does not seem surprising since they may apparently assist in chlorophyll accumulation and storage, and function as carrier proteins for chlorophyll in cases of PS1 breakdown and the synthesis of nascent PS1 complexes, as proposed for PS2. Hli proteins are believed to participate in the coordinated pigment and newly synthesized apoprotein delivery system during the biogenesis of PS1 and PS2 photosynthetic complexes, decreasing the risk of phototoxic unbound chlorophyll accumulation [37].
As our data shows, HliA/HliB proteins can be associated with both PS1 monomers and trimers. This suggests that the binding site for these proteins is located on the surface of PS1 monomer and is not shielded during oligomerization. A similar localization on the PS1 surface is typical for the additional protein IsiA [38]. Our studies of PS2-deficient mutants determined that HliA/HliB proteins are not associated with PS1 trimers. The fact that the PS1 trimer relative content in PS2-deficient mutant cells is decreased compared to wild-type cells (Figs. 1a and 2a) supports the hypothesis on the importance of the presence of Hli proteins for PS1 trimer stabilization [17]. PS1 trimers isolated from wild-type cells and PS2-deficient mutants were photochemically active.
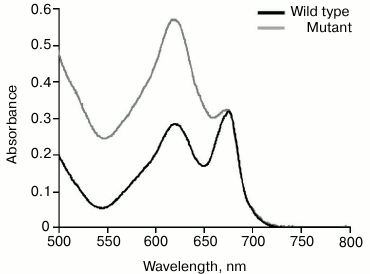
It is possible that significant alterations in structure of chlorophyll–protein complexes from PS2-deficient mutant thylakoid membrane structure [39] prevent HliA/HliB binding to PS1 trimers. Apparently, PS2 deletion causes a disruption in the structure of thylakoid membrane chlorophyll–protein complexes and/or an alteration in PS1 trimer structure. This causes HliA/HliB to no longer associate with PS1 trimers in PS2-deficient mutant. Changes in the functional megacomplex consisting of phycobilisome antenna complex, PS1, and PS2 [40] are demonstrated by data on phycobilisome content (absorption maximum at 625 nm). According to absorption spectra of wild-type cyanobacterial cells and PS2-deficient mutant cells normalized at the absorption maximum of chlorophyll, phycobilisome content in PS2-deficient mutant is two times greater than that in wild-type cyanobacterial cells (Fig. 5). Apparently, in the case of PS2 deletion, the structure of thylakoid membrane megacomplex is severely disrupted, which may also cause alterations in PS1 trimer structure, preventing association of HliA/HliB proteins.
Fig. 5. Absorption spectra of wild-type Synechocystis cells and PS2-deficient mutant. The spectra are normalized at the maximum absorption of chlorophyll (680 nm).
To reveal possible interdependence of HliA/HliB association with PS1 monomers and trimers, the ΔpsaL mutant was studied, which lacks PS1 trimers. Absence of PS1 trimers was found to have no effect on HliA/HliB binding to PS1 monomers and PS2 complexes. Apparently, alterations in thylakoid membrane cause by the absence of PS1 trimers are not so great and do not cause changes in HliA/HliB protein binding to other thylakoid chlorophyll–protein complexes.
Thus, our results show that HliA/HliB proteins can bind to basic chlorophyll–protein complexes of cyanobacterial thylakoid membranes: to PS1 monomers, PS2 complex [19], and PS1 trimers. The association of HliA/HliB with PS1 as well as with PS2 indicates their universal role in protection of the cyanobacterial photosystems.
The authors thank Prof. W. F. J. Vermaas (Arizona State University, USA) for providing cyanobacteria mutants.
This study was conducted with financial support from the Program 1.7P of the Presidium of the Russian Academy of Sciences “Molecular and Cell Biology” and the Russian Foundation for Basic Research (project Nos. 13-04-00533a and 12-04-00603a).
REFERENCES
1.Horton, P., Ruban, A., and Walters, R. G. (1996)
Regulation of light harvesting in green plants, Annu. Rev. Plant
Physiol. Plant Mol. Biol., 47, 655-684.
2.Niyogi, K. (1999) Photoprotection revisited:
genetic and molecular approaches, Annu. Rev. Plant Physiol. Plant
Mol. Biol., 50, 333-359.
3.Karapetyan, N. V. (2007) Non-photochemical
quenching of fluorescence in cyanobacteria, Biochemistry
(Moscow), 72, 1127-1135.
4.Karapetyan, N. V. (2008) Protective dissipation of
excess absorbed energy by photosynthetic apparatus of cyanobacteria:
role of antenna terminal emitters, Photosynth. Res., 7,
195-204.
5.Dolganov, N. A., Bhaya, D., and Grossman, A. R.
(1995) Cyanobacterial protein with similarity to the chlorophyll
a/b-binding proteins of higher plants: evolution and
regulation, Proc. Natl. Acad. Sci. USA, 92,
636-640.
6.Funk, C., and Vermaas, W. (1999) A cyanobacterial
gene family coding for single-helix proteins resembling part of the
light-harvesting proteins from higher plants, Biochemistry,
38, 9397-9404.
7.Kilian, O., Steunou, A. S., Grossman, A. R., and
Bhaya, D. A. (2008) A novel two domain-fusion protein in cyanobacteria
with similarity to the CAB/ELIP/HLIP superfamily: evolutionary
implications and regulation, Mol. Plant, 1, 155-166.
8.Yurina, N. P., Mokerova, D. V., and Odintsova, M.
S. (2013) Light-inducible stress plastid proteins of phototrophs,
Russ. J. Plant Physiol., 60, 577-588.
9.Muramatsu, M., and Hihara, Y. (2012) Acclimation to
high-light conditions in cyanobacteria: from gene expression to
physiological responses, J. Plant Res., 125, 11-39.
10.Jansson, S., Andersson, J., Kim, S. J., and
Jackowski, G. O. (2000) An Arabidopsis thaliana protein
homologous to cyanobacterial high light-inducible proteins, Plant
Mol. Biol., 42, 345-351.
11.He, Q., Dolganov, N., Bjorkman, O., and Grossman,
A. R. (2001) The high light-inducible polypeptides in
Synechocystis PCC6803: expression and function in high light,
J. Biol. Chem., 276, 306-314.
12.Kufryk, G., Hernandez-Prieto, M. A., Kieselbach
T., Miranda, H., Vermaas, W., and Funk, C. (2008) Association of small
Cab-like proteins (SCPs) of Synechocystis sp. PCC 6803 with
photosystem II, Photosynth. Res., 95, 135-145.
13.Mikami, R., Kanesaki, Y., Suzuki, I., and Murata,
N. (2002) The histidine kinase Hik33 perceives osmotic stress and cold
stress in Synechocystis sp. PCC 6803, Mol. Microbiol.,
46, 905-915.
14.Havaux, M., Guedeney, G., He, Q., and Grossman,
A. R. (2003) Elimination of high light-inducible polypeptides related
to eukaryotic chlorophyll a/b-binding proteins results in
aberrant photoacclimation in Synechocystis PCC 6803, Biochim.
Biophys. Acta, 1557, 21-33.
15.Xu, H., Vavilin, D., Funk, C., and Vermaas, W.
(2004) Multiple deletions of small Cab-like proteins in the
cyanobacterium Synechocystis sp. PCC 6803: consequences for
pigment biosynthesis and accumulation, J. Biol. Chem.,
279, 27971-27979.
16.Staleva, H., Komenda, J., Shukla, M. K., Slouf,
V., Kana, R., Polivka, T., and Sobotka, R. (2015) Mechanism of
photoprotection in the cyanobacterial ancestor of plant antenna
proteins, Nat. Chem. Biol., 11, 287-291.
17.Wang, Q., Jantaro, S., Lu, B., Majeed, W.,
Bailey, M., and He, Q. (2008) The high light-inducible polypeptides
stabilize trimeric photosystem I complex under high light conditions in
Synechocystis PCC 6803, Plant Physiol., 147,
1239-1250.
18.Bhaya, D., Dufresne, A., Vaulot, D., and
Grossman, A. (2002) Analysis of the hli gene family in marine
and freshwater cyanobacteria, FEMS Microbiol. Lett., 215,
209-219.
19.Yao, D., Kieselbach, T., Komenda, J., Promnares,
K., Hernandez-Prieto, M., Tichy, M., Vermaas, W., and Funk, C. (2007)
Localization of the small Cab-like proteins in photosystem II, J.
Biol. Chem., 282, 267-276.
20.Shen, G., Boussiba, S., and Vermaas, W. F. J.
(1993) Synechocystis sp. PCC 6803 strains lacking photosystem I
and phycobilisome function, Plant Cell, 12,
1853-1863.
21.Chitnis, V. P., and Chitnis, P. R. (1993) PsaL
subunit is required for the formation of photosystem I trimers in the
cyanobacterium Synechocystis sp. PCC 6803, FEBS Lett.,
336, 330-334.
22.Rippka, R., Deruelles, J., Waterbury, J. B.,
Herdmann, M., and Stanier, R. Y. (1979) Generic assignments, strain
histories, and properties of pure cultures of cyanobacteria, J. Gen.
Microbiol., 111, 1-61.
23.Shubin, V. V., Bezsmertnaya, I. N., and
Karapetyan, N. V. (1992) Isolation from Spirulina membranes of
two photosystem I-type complexes one of which contains chlorophyll
responsible for the 77 K fluorescence band at 760 nm, FEBS
Lett., 309, 340-342.
24.Shubin, V. V., Tsuprun, V. L., Bezsmertnaya, I.
N., and Karapetyan, N. V. (1993) Trimeric forms of the photosystem I
reaction center complex pre-exist in the membranes of the
cyanobacterium Spirulina platensis, FEBS Lett.,
334, 79-82.
25.Lichtenthaler, H. K. (1987) Chlorophylls and
carotenoids: pigments of photosynthetic biomembranes, Methods
Enzymol., 148, 350-382.
26.Laemmli, U. K. (1970) Cleavage of structural
proteins during the assembly of the head of bacteriophage T4,
Nature, 227, 680-685.
27.Bradford, M. M. (1976) A rapid and sensitive
method for the quantitation of microgram quantities of protein
utilizing the principle of protein–dye binding, Anal.
Biochem., 72, 248-254.
28.Rakhimberdieva, M. G., Boichenko, V. A.,
Karapetyan, N. V., and Stadnichuk, I. N. (2001) Interaction of
phycobilisomes with photosystem 2 dimers and photosystem 1 monomers and
trimers of the cyanobacterium Spirulina platensis,
Biochemistry, 40, 15780-15788.
29.Kruip, J., Karapetyan, N. V., Terekhova, I. V.,
and Rogner, M. (1999) In vitro oligomerization of a membrane
protein complexes: liposome based reconstitution of trimeric
photosystem I from isolated monomers, J. Biol. Chem.,
274, 18181-18188.
30.Karapetyan, N. V., Holzwarth, A. R., and Rogner,
M. (1999) The photosystem I of cyanobacteria: molecular organization,
excitation dynamics and physiological significance, FEBS Lett.,
460, 395-400.
31.Karapetyan, N. V., Bolychevtseva, Yu. V., Yurina,
N. P., Terekhova, I. V., Shubin, V. V., and Brecht, M. (2014)
Long-wavelength chlorophylls in photosystem I of cyanobacteria: origin,
localization, and functions, Biochemistry (Moscow), 79,
213-220.
32.Promnares, K., Komenda, J., Bumba, L.,
Nebesarova, J., Vacha, F., and Tichy, M. (2006) Cyanobacterial small
chlorophyll-binding protein ScpD (HliB) is located on the periphery of
photosystem II in the vicinity of PsbH and CP47 subunits, J. Biol.
Chem., 281, 32705-32713.
33.Hernandez-Prieto, M., Tibiletti, T., Abasova, L.,
Kirilovsky, D., Vass, I., and Funk, C. (2011) The small Cab-like
proteins of the cyanobacterium Synechocystis sp. PCC 6803: their
involvement in chlorophyll biogenesis for photosystem II, Biochim.
Biophys. Acta, 1807, 1143-1151.
34.Yao, D. C. I., Brune, D. C., Vavilin, D., and
Vermaas, W. F. J. (2012) Photosystem II component lifetimes in the
cyanobacterium Synechocystis sp. strain PCC 6803: small Cab-like
proteins stabilize biosynthesis intermediates and affect early steps in
chlorophyll synthesis, J. Biol. Chem., 287, 682-692.
35.Sinha, R. K., Komeda, J., Knoppova, J.,
Sedlarova, M., and Pospisil, P. (2012) Small Cab-like proteins prevent
formation of singlet oxygen in the damaged photosystem II complex of
the cyanobacterium Synechocystis sp. PCC 6803, Plant Cell
Environ., 35, 806-818.
36.Yao, D. C. I., Brune, D. C., and Vermaas, W. F.
J. (2012) Lifetimes of photosystem I and II proteins in the
cyanobacterium Synechocystis sp. PCC 6803, FEBS Lett.,
586, 169-173.
37.Chidgrey, J. W., Linhartova, M., Komenda, J.,
Jackson, P. J., Dickman, M. J., Canniffe, D. P., Konik, P., Pilny, J.,
Hunter, C. N., and Sobotka, R. (2014) A cyanobacterial chlorophyll
synthase–HliD complex associates with the Ycf39 protein and the
YidC/Alb3 insertase, Plant Cell, 26, 1267-1279.
38.Boekema, E. J., Hifney, A., Yakushevska, A. E.,
Piotrowski, M., Keegstra, W., Berry, S., Michel, K.-P., Pistorius, E.
K., and Kruip, J. (2001) A giant chlorophyll–protein complex
induced by iron deficiency in cyanobacteria, Nature, 412,
745-748.
39.Van de Meene, A. M. L., Sharp, W. P., McDaniel,
J. H., Friedrich, H., Vermaas, W. F. J., and Roberson, R. W. (2012)
Gross morphological changes in thylakoid membrane structure are
associated with photosystem I deletion in Synechocystis sp. PCC
6803, Biochim. Biophys. Acta, 1818, 1427-1434.
40.Liu, H., Zhang, H., Niedzwiedzki, D. M., Prado,
M., He, G., Gross, M. L., and Blankenship, R. E. (2013) Phycobilisomes
supply excitations to both photosystems in a megacomplex in
cyanobacteria, Science, 342, 1104-1107.