REVIEW: Carbonyl Stress in Bacteria: Causes and Consequences
O. V. Kosmachevskaya, K. B. Shumaev, and A. F. Topunov*
Bach Institute of Biochemistry, Research Center of Biotechnology of the Russian Academy of Sciences, 119071 Moscow, Russia; E-mail: aftopunov@yandex.ru* To whom correspondence should be addressed.
Received May 30, 2015
Pathways of synthesis of the α-reactive carbonyl compound methylglyoxal (MG) in prokaryotes are described in this review. Accumulation of MG leads to development of carbonyl stress. Some pathways of MG formation are similar for both pro- and eukaryotes, but there are reactions specific for prokaryotes, e.g. the methylglyoxal synthase reaction. This reaction and the glyoxalase system constitute an alternative pathway of glucose catabolism – the MG shunt not associated with the synthesis of ATP. In violation of the regulation of metabolism, the cell uses MG shunt as well as other glycolysis shunting pathways and futile cycles enabling stabilization of its energetic status. MG was first examined as a biologically active metabolic factor participating in the formation of phenotypic polymorphism and hyperpersistent potential of bacterial populations. The study of carbonyl stress is interesting for evolutionary biology and can be useful for constructing highly effective producer strains.
KEY WORDS: carbonyl stress, bacteria, methylglyoxal, metabolite overproductionDOI: 10.1134/S0006297915130039
Abbreviations: AGEs, advanced glycation end products; DHAP, dihydroxyacetone phosphate; FBA, flux balance analysis; GAPD, glyceraldehyde-3-phosphate dehydrogenase; GloI/GloII and GloIII, glyoxalases I/II and III; G3P, glyceraldehyde-3-phosphate; GSH, reduced glutathione; MG, methylglyoxal; MGS, methylglyoxal synthase; Pi, inorganic phosphate; RCS, reactive carbonyl species; ROS, reactive oxygen species.
The term “carbonyl stress” that describes an imbalance
between the formation and removal of reactive carbonyl species (RCS)
first appeared in the scientific literature in the beginning of 1990s
[1]. RCS are aldehydes and ketones with a carbonyl
group electrophilic carbon atom capable of reacting with nucleophilic
nitrogen atoms in amino acids, amino peptides, and guanine bases with
the formation of N-substituted glycosamines (Shiff bases). Shiff bases
undergo Amadori rearrangement and form ketamines [2], precursors of advanced glycation end products
(AGEs) – various chemical compounds that include pyrrole,
pyrazine, imidazole, and furan derivatives. The sequence of reactions
resulting in AGE formation was first described by the French biochemist
and medic Maillard [3] (Maillard, or sugar/amino
acid reaction) and comprehensively studied by the American chemists
Hodge and Rist [4].
The Iranian scientist Rahbar [5] found glycated hemoglobin (HbA1c) in the blood of patients with diabetes, which initiated studies of nonenzymatically glycated proteins and AGEs in living organisms. More than 20 reactive aldehydes and ketones have been identified since then, the major ones being glyoxal, methylglyoxal, 3-deoxyglucosone, and malonic dialdehyde. Methylglyoxal (MG) is an α,β-dicarbonyl compound with a glycation activity that is 10,000 times higher than the corresponding activities of glucose and fructose [6]. Biosynthesis of this α-ketoaldehyde in various human and animal tissues had been discovered in the 1920s, even before the beginning of active studies of nonenzymatic glycation in biological objects. British scientists Rabbani and Thornalley introduced the term “dicarbonyl stress”, and therefore emphasized the leading role of α,β-dicarbonyl compounds in AGE formation under physiological conditions [2, 6].
Since its discovery, nonenzymatic glycation has been intensively studied in humans and other animals because of its involvement in pathogenesis of diabetes, cancer, aging, and neurodegenerative disorders. However, data on carbonyl stress in bacteria are scarce. Until recently, it even had been unclear if carbonyl stress happens in bacteria. The discovery of nonenzymatically glycated proteins in Escherichia coli cells by the Bulgarian scientists Mironova et al. provided compelling evidence that glycation reactions occur not only in eukaryotes, but also in bacteria [7]. Another researcher who noticed the presence of glycation products in bacterial cells was Pepper [8].
High levels of glycated proteins and nucleic acids observed in the exponential growth phase in bacteria with a short life cycle indicated that carbonyl compounds involved in glycation in prokaryotes are extremely reactive. Long before glycated adducts were found in the cytoplasm, bacteria had been studied for their ability to synthesize MG [9]. Later, a prokaryote-specific enzyme, methylglyoxal synthase (MGS; EC 4.2.99.11), was discovered, which catalyzes the hydrolysis of dihydroxyacetone phosphate (DHAP) into MG and inorganic phosphate (Pi) [9].
DEFENSE AGAINST NONENZYMATIC GLYCATION
Bacteria were found to synthesize MG under certain conditions; therefore, bacterial cells should possess efficient defensive mechanisms against this compound. MG toxicity is due not only to its involvement in nonenzymatic glycation, but also to its ability to generate free radical products [10-14]. Moreover, MG is capable of inhibiting both protein biosynthesis and initiation of DNA replication [15].
There are three major strategies to protect bacterial cells from nonenzymatic glycation: decreasing α-ketoaldehyde concentration, decreasing reaction activity of carbonyls or amines, and repairing or degrading RCS-modified macromolecules. The three strategies complement each other to a great extent; however, the most efficient defensive mechanism is the regulation of MG concentration by the evolutionarily conserved glyoxalase (Glo) system.
The Glo system consists of two enzymes: glyoxalase I (GloI, EC 4.4.1.5: S-D-lactoylglutathione lyase) and glyoxalase II (GloII, EC 3.1.2.6: hydroxyacylglutathione hydrolase), which are encoded by the gloA and gloB genes, respectively [16-18]. An important role of Glo is detoxification of reactive α-ketoaldehydes (predominantly, MG) [19]. The substrate for GloI is a hemithioacetal formed in a spontaneous reaction of MG with GSH. GloI catalyzes irreversible conversion of hemithioacetal into S-D-lactoylglutathione, which is then hydrolyzed to D-lactate by GloII. The D-lactate is either transported from the cell or reduced into pyruvate by D-lactate dehydrogenases (EC 1.1.2.4) [16, 19, 20]. In some bacteria, other thiols, but not glutathione, act as active carbonyl group acceptors: bacillithiol [21, 22], mycothiol [23], γ-glutamylcysteine [24], γ-glutamylcysteine peptides [21, 25]. Maximal expression of GloI is observed during the exponential growth phase, when a high glycolysis rate is essential for rapid growth of bacteria [26, 27]. Under physiological conditions, the Glo system converts MG at diffusion-controlled rate [28, 29], thus, maintaining low intracellular MG concentrations.
Escherichia coli cells contain another glyoxalase, GloIII [30, 31], whose expression is induced by heat shock and osmotic stress [32]. GloIII is identical to the Hsp31 chaperone [32] and catalyzes the direct conversion of MG with formation of intermediate products without involvement of GSH. The reaction rates for GloIII exceed the rates for the GloI and II enzymes [30]. GloIII expression is controlled by the universal stress-response regulator RpoS and reaches its maximum during the stationary phase [33]. It was suggested that GloIII is essential for the survival of noncultured strains of E. coli by providing an additional pathway for detoxification of MG and other electrophiles [33].
Alternative bacterial systems for defense against carbonyl stress have been reviewed in details in [34].
METHYLGLYOXAL SHUNT
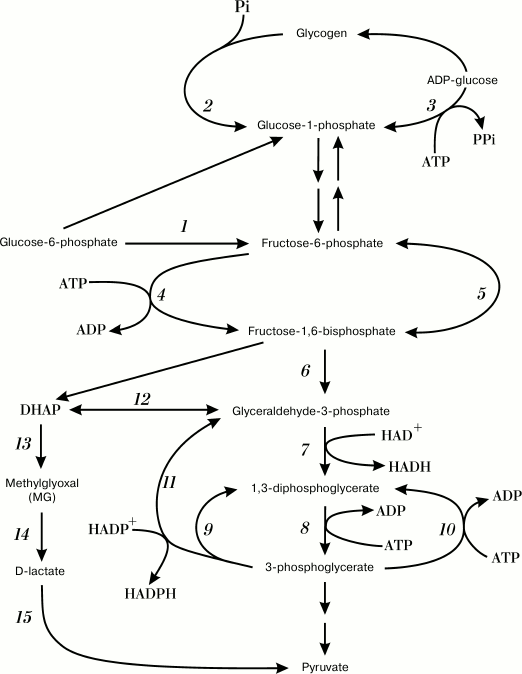
In bacterial cells, an increase in metabolic flux via glycolysis results in DHAP accumulation and redirection of the flux into an alternative pathway – the methylglyoxal (MG) shunt [35]. The MG shunt combines reactions catalyzed by MGS and GloI/II: MGS converts DHAP into MG, which is then converted into D-lactate (Fig. 1, reactions (13)-(15)). Metabolic pathways for controlled MG synthesis and degradation have been found exclusively in bacteria and in halophilic archaea [24].
Fig. 1. Futile cycles and glycolytic pathway shunts. Numbers denote enzymes catalyzing corresponding reactions: 1) glucose phosphate isomerase; 2) glycogen phosphorylase; 3) ADP-glycose pyrophosphorylase; 4) phosphofructokinase; 5) fructose diphosphatase; 6) fructose bisphosphate aldolase; 7) glyceraldehyde-3-phosphate dehydrogenase (GAPD); 8) phosphoglycerate kinase; 9) oxidized GAPD; 10) phosphoglycerate kinase; 11) nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN); 12) triosephosphate isomerase; 13) methylglyoxal synthase; 14) glyoxalases I and II; 15) D-lactate dehydrogenase.
GloI/II and MGS are constitutively expressed in E. coli cells, which indicates an importance of the MG shunt for bacterial survival. This was confirmed in experiments with mgsA-lacking E. coli strains: an inability to synthesize MG and to redirect metabolic flux via the alternative pathway resulted in inhibition of cell grown when xylose and cAMP were added to the medium [33].
It should be noted that the MG shunt cannot completely replace glycolysis. Strains with defective glycolytic enzymes were not able to grow on glucose [36]. The Km values for glyceraldehyde-3-phosphate dehydrogenase (GAPD) (0.29 mM) and MGS (0.5 mM [37] or 0.7 mM [38]) suggest that the MG shunt is inhibited under normal physiological conditions and the substrate is utilized in glycolysis [16, 39]. The MG shunt is not coupled to conservation of energy in the form of ATP [40, 41], and it represents a classical example of an inefficient pathway of glucose catabolism. However, why do most bacteria use such an inefficient and even dangerous metabolic pathway?
Under certain conditions, bacteria might utilize higher amounts of carbohydrates than is necessary for supporting basic metabolism and growth. This could happen either accidentally, e.g. due to disturbances in the regulation of energy metabolism, or purposefully (the Crabtree effect). In any case, to maintain this metabolic regime, cells are required to increase ATP turnover and, therefore, turn to dissipative hydrolysis.
Living organisms do not always use the energy of ATP efficiently, and they might even dissipate it purposefully. ATP is an energy-coupling link between anabolism and catabolism. Therefore, when the rate of catabolism exceeds the rate of anabolism, excessive amounts of catabolic intermediates and ATP are formed in cells – a state that is called “overflow metabolism” [42]. To exit this state, bacteria use energetically unfavorable metabolic pathways and futile cycles (Fig. 1) to utilize excessive metabolic intermediates and/or to provide excessive ATP hydrolysis and energy dissipation [42, 43]. Mechanisms for energy dissipation serve as cell “tools” to stabilize ATP concentration and to maintain cell energy status to support the stability of cell metabolism [43, 44]. As a rule, energy dissipation occurs via alternative or “underground” metabolic pathways [45, 46], and the MG shunt is a characteristic example of these alternative pathways in bacteria. Switching to the MG shunt allows the bacterial cell to adapt its metabolism to the imbalance of nutrients and to sustain cell growth. However, it can also result in the development of carbonyl stress if the glycation defense systems in the cell are disturbed. Elucidation of MG shunt activation mechanisms will help in understanding the causes of carbonyl stress induction in bacteria.
METHYLGLYOXAL BIOSYNTHESIS PATHWAYS
There are two independent mechanisms for carbonyl stress induction in bacteria: a decrease in the efficiency of detoxification mechanisms, and excessive metabolite production. MG can also be generated in reactions of aerobic metabolism of threonine.
Dysfunction of Methylglyoxal Detoxification Systems
As mentioned above, the major role in MG detoxification belongs to the GloI/GloII glyoxalase system. GloI is essential for defense against MG: E. coli cells can survive mutations in the gloB gene, but not in the gloA gene [47]. GloI catalyzes the limiting stage of MG detoxification, and dysfunctions or deficit of this enzyme can result in accumulation of lethal concentrations of MG.
It was reported that GloI from human endothelial cells is reversibly inhibited by nitric oxide (NO) due to nitrosylation of cysteine residues in the enzyme [48]. S-Nitrosoglutathione, a physiological NO donor, decreased the activity of human and yeast GloI [49, 50]. Glutathionylation of the Cys139 residue inhibited GloI activity [51]. These data suggest that posttranslational nitrosylation and glutathionylation of GloI might be mechanisms of regulation of glucose metabolism in cells. Human GloI was found to be phosphorylated on the Tyr106 residue by calmodulin-dependent protein kinase, which indicates that GloI could be involved in intracellular signaling [52]. Since glyoxalase is an evolutionarily conserved enzyme, we could expect that bacterial GloI might be regulated via posttranslational modifications as well (e.g. be NO-dependent).
GloI deficiency can be another cause of carbonyl stress. Thus, GloI deficit increases the sensitivity of anaerobically cultivated E. coli cells to MG [53]. The importance of GloI for normal cell growth was confirmed by decreased survival of gloA-lacking Streptococcus mutans and Salmonella typhimurium cells grown in media with a high sugar content [26, 27]. On the contrary, overexpression of Pseudomonas putida GloI increased the resistance of E. coli cells to stress [53].
Excessive Production of Glycolysis Metabolites
There are a few scenarios that can lead to excessive production of glycolysis metabolites: (i) disturbances in triosephosphate metabolism [9, 54-56]; (ii) disorders in carbohydrate metabolism regulation, caused by disorders in carbohydrate transport and incorporation into glycolysis [57-59]; (iii) deficit of intracellular inorganic phosphate (Pi) [37, 59, 60]; (iv) bacterial growth at high carbohydrate concentrations (Crabtree effect) [61, 62]; (v) deficit of amine nitrogen [16, 63]; (vi) expression of recombinant proteins [7, 64, 65]; (vii) growth in the stationary phase [7, 8, 66-68].
Disturbances in triosephosphate metabolism. The biggest contributor to the endogenous MG pool is nonenzymatic hydrolysis of the phosphate group from the triosephosphates DHAP and glyceraldehyde-3-phosphate (G3P) [69, 70]. Due to this fact, any disorders in the metabolism of these compounds will affect MG formation. Triosephosphate conversion is catalyzed by two glycolytic enzymes: triosephosphate isomerase and GAPD. Any disturbances in the biosynthesis and activity of these enzymes will unavoidably result in the excessive formation of MG.
Dysfunction or deficit of triosephosphate isomerase. The concentrations of G3P and DHAP are maintained in an equilibrium state by triosephosphate isomerase. The enzyme catalyzes the isomerization of DHAP into G3P, which is then utilized in glycolysis. The active site of triosephosphate isomerase binds endiolate phosphate – an intermediate product of G3P or DHAP conversion, capable of spontaneous loss of phosphate with the formation of MG. Moreover, both G3P and DHAP are unstable and can undergo spontaneous isomerization into endiolate phosphate under physiological conditions.
Inhibition or deficit of triosephosphate isomerases in the cytoplasm causes DHAP accumulation [71-73]. It has been shown that the absence of the tpiA gene in mutant E. coli strains results in MG accumulation [9, 74]. Mycobacterium tuberculosis strains lacking triosephosphate isomerase accumulated triosephosphates and displayed decreased virulence [54]. Hipkiss suggested that deamination of certain asparagine residues in triosephosphate isomerase during active glycolysis might cause a decrease in the enzyme activity and a corresponding increase in MG content [55]. Indeed, changing triosephosphate isomerase activity in Lactococcus lactis by only 3% caused a four-fold increase in the DHAP content [75].
Dysfunction or deficit of glyceraldehyde-3-phosphate dehydrogenase. GAPD is another enzyme whose dysfunction can result in MG formation. GAPD is redox-sensitive, containing SH-groups that can be oxidized by reactive oxygen and nitrogen species into sulfinic and sulfonic acids [76], as well as nitrosylated by NO donors [50, 77-81] or glycated by MG [82, 83]. Such posttranslational modifications decrease GAPD activity, which causes an increase of the triosephosphate pool [56]. GAPD activity also depends on the NAD+/NADH ratio. Depletion of NAD+ inhibits GAPD and results in an increase in triosephosphate concentration [84].
Disturbances in regulation of carbohydrate metabolism. Disorders in sugar transport. The first stage of carbohydrate metabolism is the transport of these compounds into cells, and very often, this stage limits the rates of carbon source utilization. Carbohydrate transport into bacterial cells is governed by the catabolic repression rule [85], which states that utilization of the preferred carbohydrate substrate suppresses utilization of other substrates. It is generally believed that, most importantly, catabolic repression prevents overload of the glycolic pathway with metabolites. This is achieved by limiting the rate of carbohydrate transport into the cell and preventing simultaneous intake of other carbohydrate substrates. An E. coli strain bearing a multicopy plasmid with the uhpT gene coding for the sugar phosphate transport system accumulated toxic concentrations of MG [57]. In contrast, Prevotella ruminicola mutants with a decreased content of glucose transporters in the membrane produced lower MG concentrations [63].
Catabolic repression is known to depend on the cAMP concentration in cells. The major function of cAMP in bacteria is regulation of transcription of inducible genes responsible for carbohydrate transport and catabolism [86]. Therefore, any changes in cAMP content or in cAMP-mediated signaling pathways might result in uncontrolled carbohydrate transport, overload of initial glycolysis stages by metabolites, and MG formation. This suggestion has been experimentally confirmed: when grown in medium containing cAMP and various sugars as carbon sources, E. coli cells accumulated toxic MG concentrations [87, 88]. In Enterobacteriaceae, a positive correlation between the presence of cAMP and the presence of MGS was observed, which probably allows the cells to simultaneously catabolize different carbohydrates and to direct excessive DHAP into the MG shunt to decrease the probability of nonenzymatic MG formation from triosephosphates [87]. In addition to glycolysis, Enterobacteriaceae possess another carbohydrate catabolism mechanism, the Entner–Doudoroff pathway, which makes it possible for them to utilize both glucose and sugar acids [89].
Excessive metabolite production can result not only from changes in cAMP content, but also from disturbances in the cAMP regulation pathways. Thus, the growth of an E. coli strain expressing a mutant cAMP receptor in minimal medium with glucose-6-phosphate was inhibited because of MG formation [90]. Disorders in cAMP signaling increased the levels of phosphofructokinase – a regulatory enzyme that determines the rate of glycolysis, and consequently, caused excessive accumulation of triosephosphates [90].
Growth on glycerol-containing media. Glycerol is a good carbon and energy source for many prokaryotic organisms. Similarly to carbohydrates, glycerol metabolism starts with its transport into the cell, where it is introduced into glycolysis at the level of DHAP. DHAP can be synthesized from glycerol via one of the two pathways. The first one is glycerol phosphorylation into glycerol-3-phosphate by ATP-dependent glycerol kinase. Glycerol-3-phosphate is then oxidized into DHAP by FAD/FMN-dependent (anaerobic) or NAD-dependent (aerobic) glycerol-3-phosphate dehydrogenases. In the second pathway, glycerol is oxidized into dihydroxyacetone by glycerol dehydrogenase and phosphorylated into DHAP by the ATP- or phosphoenolpyruvate-dependent dihydroacetone kinases. In some prokaryotes, dysfunctions in glycerol metabolism are accompanied by endogenous MG accumulation [38, 91, 92]. For example, E. coli cells that lost control over glycerol metabolism due to mutations accumulated toxic concentrations of MG [38]. Anaerobic fermentation of glycerol by transgenic E. coli was accompanied by MG accumulation [53]. High levels of glycerol kinase expression in Bacillus stearothermophilus promoted uncontrolled accumulation of glycerol-3-phosphate and caused exhaustion of the Pi pool and activation of MGS [37, 91]. Cooper and Anderson observed accumulation of high concentrations of MG upon glycogen formation from glycerol in E. coli cells lacking the triosephosphate isomerase gene [9]. Glycerol in concentrations of 5 g/liter or higher inhibited 1,3-propanediol synthesis and growth of E. coli cells under anaerobic conditions [53]. In Synechococcus sp. PCC 7002, cyanobacteria capable of efficient glycerol utilization, deletion of the mgsA gene resulted in MG formation [93]. Addition of glycerol to Synechococcus cells deficient in the sakR1 gene coding for aldo-keto reductase considerably increased endogenous MG concentration [93].
Deficit of inorganic phosphate. Phosphorus plays an important role in energy metabolism as well as in integration of carbohydrate, lipid, and protein metabolism. Inorganic phosphate is a substrate for the phosphotransferase system, phosphofructokinase, and GAPD. In facultatively anaerobic bacteria, glycolytic enzymes and ATPase compete for intracellular Pi. An increase in the availability of carbohydrates from the cultivation medium often results in depletion of internal Pi reserves due to the use of phosphate in carbohydrate phosphorylation, which follows increase in carbohydrate transport into cells. Low Pi concentrations are known to limit GAPD activity and, on the other hand, to stimulate MGS [37]. This results in DHAP accumulation and shift of metabolic flux toward the MG shunt that releases Pi in the reaction of MG synthesis (Fig. 1). Hopper and Cooper proposed that one of the physiological functions of the MG shunt is to increase Pi turnover upon cell growth in media with low phosphate content [37]. This suggestion was proved in experiments with E. coli strains lacking the mgsA gene: under physiological conditions, the E. coli cell growth was inhibited by excess of sugar phosphates and restored by Pi addition [59].
Oxidative phosphorylation was suppressed by a Pi deficit, which can be explained by competition between GAPD and ATPase for Pi [94]. Inhibition of oxidative phosphorylation promotes hyperactivation of the glycolytic flux (Crabtree effect) [84, 94] and causes related increase in MG formation.
Disturbances in GAPD activity, as well as a Pi and/or ADP deficit in the cytoplasm, might result in the substrate redirection via different (alternative to MG shunt) pathways of triosephosphate catabolism. In archaea and some Gram-positive bacteria of the genera Bacillus, Streptococcus, and Clostridium, NADP-dependent nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPDN; EC 1.2.1.9) was found that catalyzed the phosphoglycerate kinase reaction of irreversible hydrolytic oxidation of G3P into 3-phosphoglycerate without ATP formation [95, 96]. Another pathway, the glycolysis shunt, is catalyzed by oxidized GAPD (Fig. 1, reaction (9)). Mild oxidation of cysteine residues into sulfenic acid in the active site of the enzyme causes decrease in its dehydrogenase activity and induction of acyl phosphatase activity [97]. This posttranslational modification of GAPD results in the uncoupling of oxidation and phosphorylation in glycolysis and decreases ATP output [98]. Nonphosphorolytic oxidation of 1,3-diphosphoglycerate and the reverse phosphoglycerate kinase reaction constitute a futile cycle of glycolysis (Fig. 1, reactions (9) and (10)). In all cases, redirection of substrate flux via energetically unfavorable shunts and futile cycles rapidly increases pyruvate content, stabilizes the energy status of cells, and provides influx of substrates for biosynthetic reactions [98].
All these facts indicate a direct regulatory role of the intracellular Pi pool in energy metabolism, as well as indirect involvement of Pi in the development of carbonyl stress in bacteria.
High carbohydrate concentrations (Crabtree effect). As mentioned above, the amount of MG synthesized directly depends on the intensity of carbohydrate flux. Glycolysis rate might increase because of the Crabtree effect (glucose effect), which is manifested as suppression of respiration at high glucose concentrations [99]. Under aerobic conditions, some bacteria can catabolize most of the glucose into reaction byproducts (lactate, acetate, ethanol, etc.), i.e. display the Crabtree effect [100, 101]. Although the mechanism of this phenomenon remains obscure, the reason for such at the first glance absurd behavior might be the necessity to obtain an adaptive advantage that is directly determined by the number of cells in the population and their genetic diversity [102]. The Crabtree effect has been also described in yeast and in rapidly proliferating mammalian cells [103].
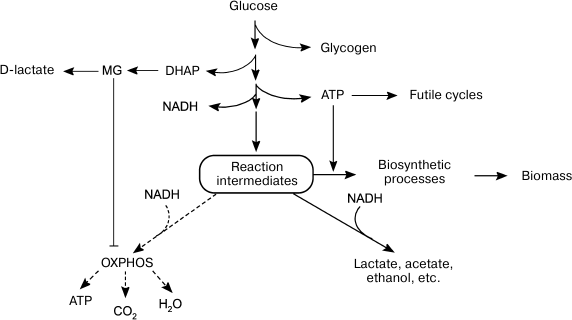
To maximize growth rate in the presence of easily available substrate, cells choose inefficient catabolism (glycolysis) over efficient catabolism (oxidative phosphorylation) (Fig. 2). The growth of microbial populations in a changing environment is a compromise between two parameters that determine metabolic strategies: the rate and the efficiency of substrate utilization [104]. Due to a number of limitations, ATP could be synthesized either at a high rate with a low yield or at a low rate with a high yield [104-107]. The ratio between the rates of ATP formation and hydrolysis (ATP turnover) determines the rate of metabolic flux. When the rate of ATP hydrolysis is lower than the rate of its biosynthesis, futile cycles, and energetically unfavorable pathways, get involved in the metabolism to maintain a high rate of glycolysis (Fig. 1). The redistribution of metabolic fluxes among these alternative pathways allows not only to stabilize the energy status of the cell, but also to accelerate substrate oxidation. As a rule, alternative pathways are induced upon amine nitrogen and/or Pi deficit, i.e. under conditions equivalent to the presence of high glucose concentrations. Obviously, such conditions also lead to activation of the MG shunt (Fig. 1, reactions (13)-(15)). Although this “high rate” strategy provides adaptive advantages, it also poses a certain danger, because damages in the systems of MG biosynthesis and utilization can cause its accumulation to lethal concentrations. In other words, the “high rate” strategy is a high-risk strategy, but bacteria are willing to risk their life to succeed in the competition for nutrition sources
Fig. 2. Alternative metabolic strategies in bacteria. Energy-saving catabolism (oxidative phosphorylation, OXPHOS) at low substrate concentrations and fast but inefficient glycolysis at high substrate concentrations.
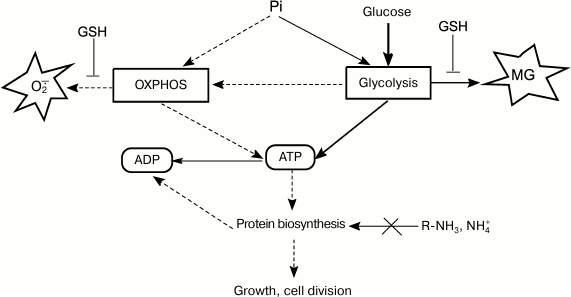
Normally, the rate of glucose consumption by cells is strictly regulated by the concentrations of Pi and nitrogen-containing substrates. Cells use global regulators to integrate information on the concentrations of glucose, nitrogen source, and Pi and to sustain their energy status by adapting metabolism to the growth conditions [108]. Disruptions in the functions of regulatory systems change the ratio between energy production and biosynthetic processes (Fig. 3). In some cases, energy metabolism imbalance might be caused by rapid changes in growth conditions. The most studied example is induction of a short-term Crabtree effect by rapid cell transition from starvation to growth in the presence of substrate excess.
Fig. 3. ATP turnover rate, an integrative parameter that reflects ratios between concentrations of major nutrients and determines intensity of metabolic flux, which, in turn, affects rates of ROS and RCS formation.
When nutrition regime changes abruptly, the cell enters a transitional period of metabolism rearrangement. An adaptive response of catabolic systems to new glucose concentrations occurs ahead of the adaptation of anabolic systems because of the necessity for gene expression induction and synthesis of protein biosynthesis system components. This imbalance results in the activation of pathways for decreasing the cell energy status. Using the FBA (flux balance analysis) method, it was predicted that in the adaptation to excessive substrate, E. coli cells will first employ futile cycles and MG and glyoxylate shunts [62], as well as use cytochrome bd-II oxidases with low electrogenic proton transfer output (H+/e– = 1) [109].
Another explanation for the short-term Crabtree effect is the fact that sugar transport systems remain active even during prolonged starvation, which allows them to immediately start consuming sugar after its appearance in the medium.
The short-term Crabtree effect is also characterized by excessive metabolite production. A typical example is triosephosphate accumulation resulting from differences in rates of metabolic pathway steps. Excessive DHAP accumulation in cells often happens because of significant increase in substrate flux at the stage of the aldolase reaction of fructose bisphosphate conversion into G3P and DHAP and simultaneous decrease in rates of subsequent reactions of phosphoenol pyruvate formation. Accumulation of sugar phosphates, fructose bisphosphate, and DHAP was first observed when glucose was added to E. coli cells grown on acetate-containing medium [110]. Pulse additions of glucose to chemostatic glucose-limited E. coli culture showed that increase in intracellular concentrations of fructose bisphosphate and DHAP happens within the first seconds after the changes in the cultivation medium composition [62, 111, 112]. Under similar conditions, mutant E. coli strains with cytochrome bd-II oxidase as the only transporter of electrons to oxygen used the pyruvate oxidase pathway and MG shunt, both of which produced less ATP than the pyruvate dehydrogenase and lactate dehydrogenase pathways [109]. These mutants accumulated MG in concentrations that exceeded by three times the MG concentrations in wild-type cells.
Streptococci that normally inhabit the human oral cavity experience short-term Crabtree effect naturally upon rapid change in food ration: a sudden increase in the concentration of sugar initiates it rapid utilization via glycolysis and conversion into an organic acid, which acts as one of the factors that determining the competitiveness and virulence of these bacteria [26].
Deficit of amine nitrogen. It is well known that carbon metabolism is regulated by the accessibility of nitrogen-containing substrates. Russell was the first to indicate the role of nitrogen source in MG synthesis during continuous cultivation of P. ruminicola from bovine rumen. When glucose was present in excessive amounts (50 mM) and nitrogen source was limited (ammonium, 3.6 mM), the number of viable cells decreased ~1000-fold [113] and MG (from 3 to 4 mM) was secreted into the cultivation medium [63]. Prevotella ruminicola cells contained polysaccharide granules, displayed high ATP concentrations, and could not maintain membrane potential [63]. A 10-fold increase in Pi concentration in the medium only slightly affected MG biosynthesis. It was suggested that when ATP could not be utilized in protein biosynthesis because of nitrogen substrate deficit, a nonphosphorylating pathway of glucose metabolism, the MG shunt, is activated [63] (Fig. 3). Similar results were obtained for E. coli cells grown in medium with excessive glucose concentration and limited ammonium concentrations [16, 114].
Expression of recombinant proteins. Expression of recombinant proteins can be another factor favoring cell transition to excessive metabolite production [115]. Because of high concentrations and high stability, recombinant proteins might accumulate stable glycated adducts in the cytoplasm. It is not surprising that the first isolated nonenzymatically glycated bacterial protein was human γ-interferon expressed in E. coli [66, 67]. We demonstrated that, when expressed in E. coli cells, soy leghemoglobin (a symbiotic hemoglobin from legumes) also undergo glycation [64, 65]. Note that our group in collaboration with the Institute of Bioorganic Chemistry of the Polish Academy of Sciences was the first to express leghemoglobin [116].
To increase the productivity of E. coli strains, it is important to use cultivation regimes that prevent cell transition to low-efficiency glycolysis, which can potentially be accompanied by development of carbonyl stress and decrease in intracellular pH [117]. However, efficient metabolism can be disturbed even at low carbohydrate substrate concentration due to the origin of fast-growing mutants with inefficient metabolism, as demonstrates by the FBA method for a model bacterial cell with recombinant protein synthesis [102].
Growth in the stationary phase. All the examples of MG shunt activation mentioned above represent cases of imbalanced growth that happens either under uncontrolled conditions or in the stationary phase in laboratory-grown bacteria. The stationary phase is accompanied by a depletion of the nutritional medium and accumulation of toxic metabolites, as well as the development of oxidative stress. Excessive formation of reactive oxygen species (ROS) can be one of the mechanisms promoting carbonyl stress in prokaryotic cells. MG is capable of generating free radical products, including O2.– in the reaction with amino acids and guanidine bases [10-14, 118, 119]. Free radical products can damage proteins and nucleic acids and initiate chain reactions of lipid peroxidation. This can cause autocatalytic increase in RCS and ROS production.
The stationary phase is characterized by the dominating presence of cells with decreased metabolic activity, i.e. cells in the resting state, which is described by the absence of cell division and low protein turnover rates. Such inert state promotes prolonged RCS contact with cell biopolymers and accumulation of intracellular glycated adducts, whereas in actively dividing cells in the exponential stage, the concentration of RCS formed during short carbonyl stress can be “diluted” in further generations. Nondividing cells are “accumulators” of glycation-induced damages, which explains higher levels of nonenzymatically glycated proteins and nucleic acids in a stationary bacterial culture as compared to exponential culture [7, 8, 66, 67, 120].
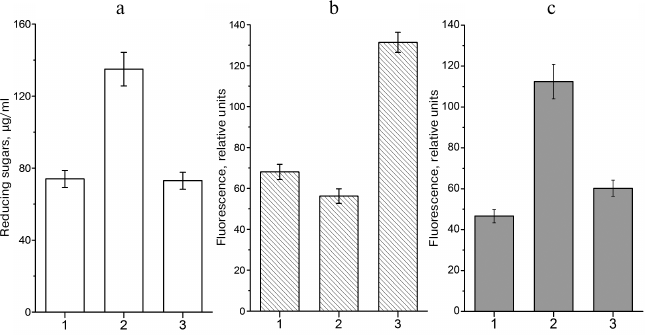
The situation at the beginning of the stationary phase is different. In the presence of excessive concentrations of a carbohydrate substrate and limited amounts of nitrogen-containing compounds (i.e. under conditions of limited cell growth), bacteria can synthesize glycogen. Glycogen synthesis is antagonistic to the MG shunt. Redirecting the carbohydrate flux into the synthesis of reserve polymer can decrease the loads on the glycolytic and MG pathways and stabilize the cell energy status (Fig. 1). There is a correlation between intracellular sugar phosphate content, amount of glycogen, and levels of nonenzymatically glycated proteins [65]. When conditions favor glycogen synthesis, the level of protein glycation in E. coli cells in the stationary phase is similar to that in cells in the exponential phase (i.e. in cells that do not accumulate glycogen) (Fig. 4).
Fig. 4. Intracellular contents of reducing sugars, glycogen, and fluorescent AGEs in leghemoglobin-expressing E. coli TB-1 culture grown in LB medium: 1) Kf , 0.14 (7 h of growth); 2) Kf, 0.14 (72 of growth); 3) Kf, 0.71 (72 of growth). Kf – filling ratio of Erlenmeyer flask (medium volume/flask volume ratio). a) Concentration of reducing sugars in the cell extract; b) glycogen content, relative units of polymer-absorbed Nile Red fluorescence (λex = 550 nm, λem = 615 nm); c) content of protein-bound AGEs expressed, relative autofluorescence intensity units (λex = 320 nm, λem = 395 nm).
The pathways for carbonyl stress induction in stationary cultures are not associated with uncontrolled carbohydrate consumption. Besides, under these conditions, nonenzymatic MG formations seems to prevail over its enzymatic synthesis.
Threonine Catabolism
α-Aminoketones (aminoacetone and 5-aminolevulinic acid) can be converted into α-ketoaldehydes by aerobic oxidation and serve as RCS in cells [121]. It is known that in the presence of oxygen, both enzymatic [122-124] and nonenzymatic [125] oxidation of aminoacetone results in MG formation. Aminoacetone is an intermediate product of threonine and alanine catabolism and can be accumulated in bacteria under certain conditions. Aminoacetone formation from threonine has been found in a number of microorganisms: E. coli [59, 126, 127], Staphylococcus aureus [128], Streptococcus faecalis, Corynebacterium erythrogenes [129], Rhodopseudomonas spheroides [130], Pseudomonas spp. [131], Arthrobacter globiformis [132], B. subtilis [133, 134], and the archaean Pyrococcus furiosus [135].
In E. coli cells, several pathways of threonine catabolism have been described. The dominating one is threonine hydroxyl group oxidation by NAD(P)+ dehydrogenase (EC 1.1. 1.103) into α-amino-β-ketobutyrate, that can be decarboxylated either spontaneously or by decarboxylase with the formation of aminoacetone and CO2 (reaction (1)) or cleaved by CoA lyase (EC 2.3.1.29) in the presence of CoA into glycine and acetyl-CoA (reaction (2)) [127, 128, 136-139]:
L-threonine + NAD(P)+ → α-amino-β-ketobutyrate → aminoacetone + CO2 + NAD(P)H, (1)
L-threonine + CoA → glycine + acetyl-CoA. (2)
These reactions are typical for both microorganisms and animals and represent the main biochemical pathways for aerobic catabolism of threonine. Whether threonine is converted into aminoacetone depends on the contents of CoA and O2 in the cell [139].
Additionally, threonine can be decarboxylated with the formation of 1-amino-2-propanol, which is then oxidized into aminoacetone (reactions (3) and (4)) [134]:
L-threonine → 1-amino-2-propanol + CO2, (3)
1-amino-2-propanol → aminoacetone + 2H+. (4)
Aminoacetone is either reduced to 1-amino-2-propanol by aminoacetone reductase, or deaminated into MG by amine oxidase (reaction (5)) [134]. It was found that in S. aureus, MG is formed from aminoacetone by nonspecific amine oxidases in a cycle of glycine or threonine oxidation [128]. A similar mechanism of MG biosynthesis was also reported for Pseudomonas [140]. In Arthrobacter, ~70% of aminoacetone from the cultivation medium could be converted into MG [132]. MG could be synthesized in reactions of threonine catabolism in E. coli cells [59].
Aminoacetone + O2 + 2H2O → MG + H2O2 + NH3 (5)
Aminoacetone accumulation could also result from activation of biosynthesis. Thus, in E. coli cells, aminoacetone was formed upon threonine biosynthesis via the glyoxylate shunt [141, 142]. Synthesis of 5-aminolevulinic acid, a precursor of tetrapyrrole, in E. coli and Rhodobacter sphaeroides cells carrying hemA genes, was accompanied by aminoacetone (0.2-0.5 g/liter) formation [143, 144]. Aminoacetone could also be generated by spontaneous decarboxylation of amino-acetoacetic acid formed by condensation of glycine with acetyl-CoA.
Similarly to MG, aminoacetone is toxic and inhibits E. coli growth [144], probably due to its damaging effect on DNA [145].
In E. coli, the absence of glucose and oxygen activates threonine deaminase (threonine dehydratase), an enzyme that catalyzes the conversion of threonine into ketobutyrate. Aminoacetone and ketobutyrate comprise the pool of ketone bodies. Accumulation of ketone bodies resulting from intense utilization of amino acids might cause the development of carbonyl stress in bacteria.
In addition to enzymatic oxidation, aminoacetone oxidation into MG could be catalyzed by Fe3+, both free or in the content of cytochrome c [124, 125, 146]. This reaction is accompanied by generation of hydrogen peroxide, superoxide radical, and aminoacetone free radical that might acerbate the damaging effect of MG on biomolecules.
REGULATORY AND SIGNALING FUNCTIONS OF METHYLGLYOXAL
Some intermediate metabolites of glycolysis (glucose 6-phosphate, fructose bisphosphate, phosphoenol pyruvate) can regulate the rates of glycolysis and related metabolic pathways. MG might also have a regulatory function. Most data on the involvement of MG in signaling were obtained in plants and yeast. Szent-Gyorgyi was the first to suggest a putative role of MG in cell growth and division when studying physiological functions of ketoaldehydes in the 1960s [147-149]. No experimental data proving the signaling role of MG in bacteria have been obtained yet, so we can only hypothesize about it.
If we change our perception of MG as a bacterial toxin exclusively, several potential physiological functions of MG can be suggested, for example, autocrine and paracrine activities inside and outside the cell, respectively. MG might be one of the glycolytic metabolites responsible for the Crabtree effect and glycolytic phenotype generation (Fig. 5). MG inhibits respiratory chains via Ca2+ signaling: adding MG to E. coli cells activated La3+-sensitive Ca2 channels, rapid increase in free cytosolic Ca2+, and consequent inhibition of F1Fo-ATPase [150]. Both glucose and fructose bisphosphate produced similar but lesser effects [103, 150, 151]. Therefore, by changing intracellular concentration of free Ca2+, MG might indirectly regulate multiple physiological processes, such as chemotaxis, spore formation, cell cycle, protein synthesis, and virulence [152].
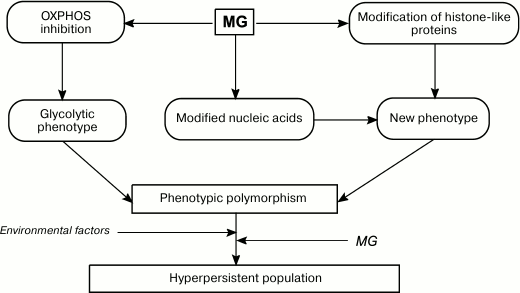
Fig. 5. Role of MG in formation of phenotypic diversity and hyperpersistence potential in bacterial populations.
Intracellular MG was found to regulate glycolysis in acidic media: GloI-deficient S. mutans mutants displayed higher glycolysis rates at pH 5.0 than the wild-type cells [26]. However, it remains unclear whether this effect is related to the MG-induced increase in the concentration of free cytosolic Ca2+.
Similarly to O2.– and H2O2, MG might be involved in cell signaling. By interacting with cysteine and/or lysine residues of transcription factors and histone-like proteins, MG can affect transcription or replication of the nucleoid. Therefore, by affecting the bacterial genome, MG serves as a mutagenic agent and epigenetic regulator and promotes generation of new phenotypes (Fig. 5). An increase in phenotypic polymorphism due to the Crabtree effect, epigenetic effects, and mutagenesis raise the adaptation capacity of bacterial population in response to environmental factors [153]. This is especially important for pathogenic and conditionally pathogenic bacteria that need to colonize the host organism. Intracellular MG might be a component of the molecular mechanism for increasing persistence of a bacterial population. Data have been obtained that support this hypothesis. An increase in intracellular MG concentration in E. coli mutants lacking GAPD and transketolase increased bacterial persistence [154]. Therefore, bacteria can shift metabolic fluxes toward biosynthesis of potentially mutagenic MG. The generated mutants undergo natural selection, e.g. toward antibiotic resistance. Hence, bacteria use controlled mutagenesis to produce populations adapted to various conditions, i.e. populations with hyperpersistence potential. An increase in persistency is important for the development of drug resistance in pathogenic bacteria and, therefore, determines the outcome of a chronic infection.
The ability of exported bacterial MG to act as a virulence factor and to produce toxic effect on neighboring cells of a host organism has been discussed in the literature. Extracellular MG was demonstrated to induce apoptosis in macrophages in M. bovis infection foci [155]. MG secreted by the periodontal pathogen Bacteroides forsythus can be a metabolic factor that favors contagion of the host organism [68].
Similar to its function in yeast cells, MG might affect transcription by modifying cysteine residues of transcription factors and changing their affinity toward DNA. When present in growth-inhibiting concentrations, MG interacts with SH-groups of the NemR transcription factor and decreases its ability to bind to the nemRA promoter, which results in the transcription of gloA [156-159].
In addition to MG, cells can have another glyoxalase reaction intermediate – S-D-lactoyl glutathione – that acts as a regulatory factor. S-D-lactoyl glutathione activates K+ channels KefGB and KefFC that transport K+ out of the cell in exchange for Na+ and H+ and cause acidification of the cytoplasm [160]. Kef channels are inhibited by GSH and activated by S-D-lactoyl glutathione [161-164]. It was suggested that the intracellular S-D-lactoyl glutathione pool determines the survival of a bacterial cell under carbonyl stress, because Kef activation can compensate for disorders in the enzymatic system of reactive aldehyde detoxification [47, 165]. Rapid conversion of S-D-lactoyl glutathione into D-lactate due to GloII overexpression increased E. coli sensitivity to MG [165]. However, we cannot exclude the existence of other intracellular targets for this compound.
CONCLUSION
Carbonyl stress is tightly related to high glucose concentrations in the bacterial environment. Glucose itself is not toxic and serves as a preferred carbon source for many microorganisms. Bacteria possess a complex system of glucose transport into the cell, and its activity correlates with the availability of nitrogen and phosphorus sources. Therefore, if bacterial regulatory systems and mechanisms for MG detoxification function normally, carbonyl stress should remain a rare event. Then why does carbonyl stress still happen? The main reason for it is occurrence of spontaneous or ROS-induced mutations that prokaryotes are extremely susceptible to. If these mutations, either beneficial or deleterious, affect enzymes of glyoxalase system and glycolysis, they can result in MG formation in concentrations toxic for the cells. As early as 1971, Freedberg discovered in a culture of E. coli cells a stable subpopulation that was able to grow in the presence of 1 mM MG as the only carbon source [38]. The activity of the glyoxalase system in these mutants was 4-8 times higher than the corresponding activity in the wild-type cells. Similar E. coli mutants with increased GloI activity were obtained in [166].
Bacteria usually encounter high sugar concentrations when grown in the laboratory. In nature, competition for limited nutrition resources is severe, and most bacteria experience starvation. For this reason, uncoupling of anabolic and catabolic processes happens only upon rapid transition from starvation to excess of food, and not in the opposite direction [62], although immediate increase in cAMP content was observed under both scenarios [62]. Such increase can cause uncontrolled transport of glucose and other carbon substrates into the cell. In nature, bacteria that often encounter high carbohydrate concentrations include microorganisms that inhabit the oral cavity or intestines of animals. Sharp changes in carbohydrate substrate concentration might disturb the regulation of metabolic pathways and result in MG biosynthesis. MG of bacterial origin has been found in thick intestine [167], gingival crevicular fluid [168], and infectious granuloma caused by tuberculosis mycobacteria [156]. The effects of bacterial MG on the host eukaryotic cells have been poorly studied so far. It was suggested that biosynthesis of high MG concentrations in the methylglyoxal synthase reaction might serve as a mechanism increasing the virulence of pathogenic bacteria [169]. Interesting results were obtained in gastroenterology studies. Campbell and coauthors presented a hypothesis on “bacterial metabolic toxins” [170], which suggests that toxic metabolites (alcohols, diols, ketones, acids, and aldehydes) produced by the intestinal microflora as a result of anaerobic fermentation of carbohydrates affect the state of this microflora and, via blood circulation, the state of various host cells, such as neurons, myocytes, and immune system cells. MG is the major toxic compound whose calculated concentration in the blood plasma might increase from micromolar to millimolar levels because of the activity of enterobacteria [170]. In eukaryotic cells, MG disrupts the mechanisms of ion-dependent signal transduction, including Ca2+-mediated signaling [150]. Such gastrointestinal disorders as irritated bowel syndrome and lactose intolerance could be caused by MG or other bacterial toxins. We hope that understanding biochemical mechanisms of carbonyl stress in bacteria will help to develop a set of measures for prevention and therapy of gastrointestinal disorders and intraperitoneal infections.
Carbonyl stress could also result from artificial modification of genomes of industrial bacteria performed to obtain highly efficient producers of compounds widely used in various industrial processes, such as 1,3-propanediol, 1,2-propanediol, and dihydroxyacetone [171, 172].
In conclusion, we would like to emphasize that understanding biochemical mechanisms of carbonyl stress is important not only for obtaining “new knowledge” for evolutionary biology, but will be useful for development of novel approaches to prevention of gastrointestinal disorders, creation of new probiotics, and solution of practical tasks of metabolic engineering.
This work was supported by the Russian Foundation for Basic Research (projects Nos. 13-04-00967, 14-04-01710, and 15-04-08891) and by the Russian Foundation for Humanities (project No. 15-36-01024).
REFERENCES
1.Baynes, J. W. (1991) Role of oxidative stress in
development of complications in diabetes, Diabetes, 40,
405-412.
2.Rabbani, N., and Thornalley, P. J. (2012) Glycation
research in amino acids: a place to call home, Amino Acids,
42, 1087-1096.
3.Maillard, L. C. (1912) Action des acids amine sur
les sucres: formation des melanoidines per voie methodique, C. R.
Acad. Sci., 154, 66-68.
4.Hodge, J. E., and Rist, C. E. (1952)
N-Glycosyl derivatives of secondary amines, J. Amer. Chem.
Soc., 74, 1494-1497.
5.Rahbar, S. (1968) An abnormal hemoglobin in red
cells of diabetics, Clin. Chim. Acta, 22, 296-298.
6.Thornalley, P. (2005) Dicarbonyl intermediates in
the Maillard reaction, Ann. N. Y. Acad. Sci., 1043,
111-117.
7.Mironova, R., Niwa, T., Hayashi, H., Dimitrova, R.,
and Ivanov, I. (2001) Evidence for nonenzymatic glycosylation in
Escherichia coli, Mol. Microbiol., 39,
1061-1068.
8.Pepper, E. (2007) Mechanisms of Long-Term
Survival in Escherichia coli: PhD thesis (Molecular
Biology), University of Southern California, USA, p. 138.
9.Cooper, R. A., and Anderson, A. (1970) The
formation and catabolism of methylglyoxal during glycolysis in
Escherichia coli, FEBS Lett., 11, 273-276.
10.Yim, H. S., Kang, S. O., Hah, Y. C., Chock, P.
B., and Yim, M. B. (1995) Free radicals generated during the glycation
reaction of amino acids by methylglyoxal. A model study of
protein-cross-linked free radicals, J. Biol. Chem., 270,
28228-28233.
11.Nohara, Y., Usui, T., Kinoshita, T., and
Watanabe, M. (2002) Generation of superoxide anions during the reaction
of guanidine compounds with methylglyoxal, Chem. Pharm. Bull.
(Tokyo), 50, 179-184.
12.Desai, K. M., and Wu, L. (2008) Free radical
generation by methylglyoxal in tissues, Drug Metab. Drug
Interact., 23, 151-173.
13.Kalapos, M. P. (2008) The tandem of free radicals
and methylglyoxal, Chem. Biol. Interact., 171,
251-271.
14.Shumaev, K. B., Gubkina, S. A., Kumskova, E. M.,
Shepel’kova, G. S., Ruuge, E. K., and Lankin, V. Z. (2009)
Mechanism for the superoxide radical formation upon L-lysine
interaction with dicarbonyl compounds, Biochemistry (Moscow),
74, 461-466.
15.Fravel, H. N., and McBrien, B. C. (1980) The
effect of methylglyoxal on cell division and the synthesis of protein
and DNA in synchronous and asynchronous cultures of Escherichia
coli B/r, J. Gen. Microbiol., 117, 127-134.
16.Ferguson, G. P., and Booth, I. R. (1998)
Importance of glutathione for growth and survival of Escherichia
coli cells: detoxification of methylglyoxal and maintenance of
intracellular K+, J. Bacteriol., 180,
4314-4318.
17.MacLean, M. J., Ness, L. S., Ferguson, G. P., and
Booth, I. R. (1998) The role of glyoxalase I in the detoxification of
methylglyoxal and in the activation of the KefB K+ efflux
system in Escherichia coli, Mol. Microbiol., 27,
563-571.
18.Kizil, G., Wilks, K., Wells, D., and
Ala’Aldeen, D. A. (2000) Detection and characterization of the
genes encoding glyoxalase I and II from Neisseria meningitides,
J. Med. Microbiol., 49, 669-673.
19.Sukdeo, N., and Honek, J. F. (2008) Microbial
glyoxalase enzymes: metalloenzymes controlling cellular levels of
methylglyoxal, Drug Metab. Drug Interact., 23, 29-50.
20.Kline, E. S., and Mahler, H. R. (1965) The lactic
dehydrogenases of E. coli, Ann. N. Y. Acad. Sci.,
119, 905-919.
21.Gaballa, A., Newton, G. L., Antelmann, H.,
Parsonage, D., Upton, H., Rawat, M., Claiborne, A., Fahey, R. C., and
Helmann, J. D. (2010) Biosynthesis and functions of bacillithiol, a
major low-molecular-weight thiol in bacilli, Proc. Natl. Acad. Sci.
USA, 107, 6482-6486.
22.Chandrangsu, P., Dusi, R., Hamilton, C. J., and
Helmann, J. D. (2014) Methylglyoxal resistance in Bacillus
subtilis: contributions of bacillithiol-dependent and independent
pathways, Mol. Microbiol., 91, 706-715.
23.Newton, G. L., Buchmeier, N., and Fahey, R. C.
(2008) Biosynthesis and functions of mycothiol, the unique protective
thiol of Actinobacteria, Microbiol. Mol. Biol. Rev., 72,
471-494.
24.Oren, A., and Gurevich, P. (1995) Occurrence of
the methylglyoxal bypass in halophilic Archaea, FEMS Microbiol.
Lett., 125, 83-87.
25.Newton, G. L., Fahey, R. C., and Rawat, M. (2012)
Detoxification of toxins by bacillithiol in Staphylococcus
aureus, Microbiology, 158, 1117-1126.
26.Korithoski, B., Levesque, C. M., and Cvitkovitch,
D. G. (2007) Involvement of the detoxifying enzyme lactoylglutathione
lyase in Streptococcus mutans aciduricity, J. Bacteriol.,
189, 7586-7592.
27.Chakraborty, S., Gogoi, M., and Chakravortty, D.
(2015) Lactoylglutathione lyase, a critical enzyme in methylglyoxal
detoxification, contributes to survival of Salmonella in the
nutrient rich environment, Virulence, 6, 50-65.
28.Marmstal, E., and Mannervik, B. (1979)
Purification, characterization and kinetic studies of glyoxalase-I from
rat liver, Biochim. Biophys. Acta, 566, 362-370.
29.Shih, M. J., Edinger, J. W., and Creighton, D. J.
(1997) Diffusion-dependent kinetic properties of glyoxalase I and
estimates of the steady-state concentrations of glyoxalase-pathway
intermediates in glycolyzing erythrocytes, Eur. J. Biochem.,
244, 852-857.
30.Misra, K., Banerjee, A. B., Ray, S., and Ray, M.
(1995) Glyoxalase-III from Escherichia coli – a single
novel enzyme for the conversion of methylglyoxal into D-lactate without
reduced glutathione, Biochem. J., 305, 999-1003.
31.Subedi, K. P., Choi, D., Kim, I., Min, B., and
Park, C. (2011) Hsp31 of Escherichia coli K-12 is glyoxalase
III, Mol. Microbiol., 81, 926-936.
32.Benov, L., Sequeira, F., and Beema, A. F. (2004)
Role of rpoS in the regulation of glyoxalase III in Escherichia
coli, Acta Biochim. Pol., 51, 857-860.
33.Szwergold, B. S. (2013) Maillard reactions in
hyperthermophilic Archaea: implications for better understanding of
non-enzymatic glycation in biology, Rejuvenation Res.,
16, 259-272.
34.Totemeyer, S., Booth, N. A., Nichols, W. W.,
Dunbar, B., and Booth, I. R. (1998) From famine to feast: the role of
methylglyoxal production in Escherichia coli, Mol.
Microbiol., 27, 553-562.
35.Cooper, R. A. (1984) Metabolism of methylglyoxal
in microorganisms, Annu. Rev. Microbiol., 38, 49-68.
37.Hopper, D. J., and Cooper, R. A. (1971) The
regulation of Escherichia coli methylglyoxal synthase: a new
control of glycolysis, FEBS Lett., 13, 213-216.
38.Freedberg, W. B., Kistler, W. S., and Lin, E. C.
(1971) Lethal synthesis of methylglyoxal by Escherichia coli
during unregulated glycerol metabolism, J. Bacteriol.,
108, 137-144.
39.Booth, I. R., Ferguson, G. P., Miller, S., Li,
C., Gunasekera, B., and Kinghorn, S. (2003) Bacterial production of
methylglyoxal: a survival strategy or death by misadventure?
Biochem. Soc. Trans., 31, 1406-1408.
40.Ferguson, G. P., Chacko, A. D., Lee, C., and
Booth, I. R. (1996) The activity of the high-affinity K+
uptake system Kdp sensitizes cells of Escherichia coli to
methylglyoxal, J. Bacteriol., 178, 3957-3961.
41.Ferguson, G. P. (1999) Protective mechanisms
against toxic electrophiles in Escherichia coli, Trends
Microbiol., 7, 242-247.
42.Russell, J. B. (2007) The energy spilling
reactions of bacteria and other organisms, J. Mol. Microbiol.
Biotechnol., 13, 1-11.
43.Igamberdiev, A. U. (1995) Logic in
Organization of Living Systems [in Russian], Voronezh University
Press, Voronezh.
44.Qian, H., and Beard, D. A. (2006) Metabolic
futile cycles and their functions: a systems analysis of energy and
control, Syst. Biol. (Stevenage), 153, 192-200.
45.D’Ari, R., and Casadesus, J. (1998)
Underground metabolism, Bioessays, 20,
181-186.
46.Notebaarta, R. A., Szappano, B., Kintses, B.,
Pal, F., Gyorkei, A., Bogos, B., Lazar, V., Spohn, R., Csorgo, B.,
Wagner, A., Ruppin, E., Pal, C., and Papp, B. (2014) Network-level
architecture and the evolutionary potential of underground metabolism,
Proc. Natl. Acad. Sci. USA, 111, 11762-11767.
47.Ozyamak, E., Black, S. S., Walker, C. A.,
Maclean, M. J., Bartlett, W., Miller, S., and Booth, I. R. (2010) The
critical role of S-lactoylglutathione formation during methylglyoxal
detoxification in Escherichia coli, Mol. Microbiol.,
78, 1577-1590.
48.Mitsumoto, A., Kim, K. R., Oshima, G., Kunimoto,
M., Okawa, K., Iwamatsu, A., and Nakagawa, Y. (1999) Glyoxalase I is a
novel nitric-oxide-responsive protein, Biochem. J., 344,
837-844.
49.Mitsumoto, A., Kim, K. R., Oshima, G., Kunimoto,
M., Okawa, K., Iwamatsu, A., and Nakagawa, Y. (2000) Nitric oxide
inactivates glyoxalase I in cooperation with glutathione, J.
Biochem., 128, 647-654.
50.Sahoo, R., Sengupta, R., and Ghosh, S. (2003)
Nitrosative stress on yeast: inhibition of glyoxalase-I and
glyceraldehyde-3-phosphate dehydrogenase in the presence of GSNO,
Biochem. Biophys. Res. Commun., 302, 665-670.
51.Birkenmeier, G., Stegemann, C., Hoffmann, R.,
Gunther, R., Huse, K., and Birkemeyer, C. (2010) Posttranslational
modification of human glyoxalase 1 indicates redox-dependent
regulation, PLoS One, 5, e10399.
52.De Hemptinne, V., Rondas, D., Toepoel, M., and
Vancompernolle, K. (2009) Phosphorylation on Thr106 and NO-modification
of glyoxalase I suppress the TNF-induced transcriptional activity of
NF-κB, Mol. Cell Biochem., 325, 169-178.
53.Zhu, M. M., Skraly, F. A., and Cameron, D. C.
(2001) Accumulation of methylglyoxal in anaerobically grown
Escherichia coli and its detoxification by expression of the
Pseudomonas putida glyoxalase I gene, Metab. Eng.,
3, 218-225.
54.Trujillo, C., Blumenthal, A., Marrero, J., Rhee,
K. Y., Schnappinger, D., and Ehrt, S. (2014) Triosephosphate isomerase
is dispensable in vitro yet essential for Mycobacterium
tuberculosis to establish infection, MBio, 5,
e00085.
55.Hipkiss, A. R. (2011) Energy metabolism and
ageing regulation: metabolically driven deamidation of triosephosphate
isomerase may contribute to proteostatic dysfunction, Ageing Res.
Rev., 10, 498-502.
56.Beisswenger, P. J., Howell, S. K., Smith, K., and
Szwergold, B. S. (2003) Glyceraldehyde-3-phosphate dehydrogenase
activity as an independent modifier of methylglyoxal levels in
diabetes, Biochim. Biophys. Acta, 1637, 98-106.
57.Kadner, R. J., Murphy, G. P., and Stephens, C. M.
(1992) Two mechanisms for growth inhibition by elevated transport of
sugar phosphates in Escherichia coli, J. Gen. Microbiol.,
138, 2007-2014.
58.Bond, D. R., and Russell, J. B. (1996) A role for
fructose 1,6-diphosphate in the ATPase-mediated energy-spilling
reaction of Streptococcus bovis, Appl. Environ.
Microbiol., 62, 2095-2099.
59.Kim, I., Kim, E., Yoo, S., Shin, D., Min, B.,
Song, J., and Park, C. (2004) Ribose utilization with an excess of
mutarotase causes cell death due to accumulation of methylglyoxal,
J. Bacteriol., 186, 7229-7235.
60.Bond, D. R., and Russell, J. B. (1998)
Relationship between intracellular phosphate, proton motive force, and
rate of nongrowth energy dissipation (energy spilling) in
Streptococcus bovis JB1, Appl. Environ. Microbiol.,
64, 976-981.
61.Cook, G. M., and Russell, J. B. (1994)
Energy-spilling reactions of Streptococcus bovis and resistance
of its membrane to proton conductance, Appl. Environ.
Microbiol., 60, 1942-1948.
62.Weber, J., Kayser, A., and Rinas, U. (2005)
Metabolic flux analysis of Escherichia coli in glucose-limited
continuous culture. II. Dynamic response to famine and feast,
activation of the methylglyoxal pathway and oscillatory behavior,
Microbiology, 151, 707-716.
63.Russell, J. B. (1993) Glucose toxicity in
Prevotella ruminicola: methylglyoxal accumulation and its effect
on membrane physiology, Appl. Environ. Microbiol., 59,
2844-2850.
64.Kosmachevskaya, O. V., and Topunov, A. F. (2009)
Existence of glycated leghemoglobin – hemoglobin of legume
plants, IMARS Highlights, 4, 10-13.
65.Kosmachevskaya, O. V., and Topunov, A. F. (2010)
Formation of glycated recombinant leghemoglobin in Escherichia
coli cells, Appl. Biochem. Microbiol., 46,
297-302.
66.Mironova, R., Niwa, T., Dimitrova, R., Boyanov,
M., and Ivanov, I. (2003) Glycation and posttranslational processing of
human interferon gamma expressed in Escherichia coli, J.
Biol. Chem., 278, 51068-51074.
67.Mironova, R., Niwa, T., Handzhiyski, Y.,
Sredovska, A., and Ivanov, I. (2005) Glycation and post-translational
processing of human interferon-gamma expressed in Escherichia
coli, J. Biol. Chem., 278, 51068-51074.
68.Maiden, M. F. J., Pham, C., and Kashket, S.
(2004) Glucose toxicity effect and accumulation of methylglyoxal by the
periodontal pathogen Bacteroides forsythus, Anaerobe,
10, 27-32.
69.Richard, J. P. (1993) Mechanism for the formation
of methylglyoxal from triosephosphates, Biochem. Soc. Trans.,
21, 549-553.
70.Phillips, S. A., and Thornalley, P. J. (1993) The
formation of methylglyoxal from triose phosphates. Investigation using
a specific assay for methylglyoxal, Eur. J. Biochem.,
212, 101-105.
71.Ationu, A., and Humphries, A. (1998) The
feasibility of replacement therapy for inherited disorder of
glycolysis: triosephosphate isomerase deficiency, Int. J. Mol.
Med., 2, 701-704.
72.Ahmed, N., Battah, S., Karachalias, N.,
Babaei-Jadidi, R., Horanyi, M., Baroti, K., Hollan, S., and Thornalley,
P. J. (2003) Increased formation of methylglyoxal and protein
glycation, oxidation and nitrosation in triosephosphate isomerase
deficiency, Biochim. Biophys. Acta, 1639, 121-132.
73.Orosz, F., Olah, J., and Ovadi, J. (2009)
Triosephosphate isomerase deficiency: new insights into an enigmatic
disease, Biochim. Biophys. Acta , 1792, 1168-1174.
74.Selvamani, V. R. S., Telaar, M., Friehs, K., and
Flaschel, E. (2014) Antibiotic-free segregational plasmid stabilization
in Escherichia coli owing to the knockout of triosephosphate
isomerase (tpiA), Microb. Cell Fact., 13, 58.
75.Solem, C., Koebmann, B., and Jensen, P. R. (2008)
Control analysis of the role of triosephosphate isomerase in glucose
metabolism in Lactococcus lactis, IET Syst. Biol.,
2, 64-72.
76.Souza, J. M., and Radi, R. (1998)
Glyceraldehyde-3-phosphate dehydrogenase inactivation by peroxynitrite,
Arch. Biochem. Biophys., 360, 187-194.
77.Mohr, S., Stamler, J. S., and Brune, B. (1994)
Mechanism of covalent modification of glyceraldehyde-3-phosphate
dehydrogenase at its active site thiol by nitric oxide, peroxynitrite
and related nitrosating agents, FEBS Lett., 348,
223-227.
78.Padgett, C. M., and Whorton, A. R. (1995)
S-Nitrosoglutathione reversibly inhibits GAPDH by S-nitrosylation,
Am. J. Physiol., 269, 739-749.
79.Galli, F., Rovidati, S., Ghibelli, L., and
Canestrari, F. (1998) S-Nitrosylation of glyceraldehyde-3-phosphate
dehydrogenase decreases the enzyme affinity to the erythrocyte
membrane, Nitric Oxide, 2, 17-27.
80.Mohr, S., Hallak, H., De Boitte, A., Lapetina, E.
G., and Brune, B. (1999) Nitric oxide-induced S-glutathionylation and
inactivation of glyceraldehyde-3-phosphate dehydrogenase, J. Biol.
Chem., 274, 9427-9430.
81.Ishii, T., Sunami, O., Nakajima, H., Nishio, H.,
Takeuchi, T., and Hata, F. (1999) Critical role of sulfenic acid
formation of thiols in the inactivation of glyceraldehyde-3-phosphate
dehydrogenase by nitric oxide, Biochem. Pharmacol., 58,
133-143.
82.Leoncini, G., Maresca, M., and Bonsignore, A.
(1980) The effect of methylglyoxal on the glycolytic enzymes, FEBS
Lett., 117, 17-18.
83.Morgan, P. E., Dean, R. T., and Davies, M. J.
(2002) Inactivation of cellular enzymes by carbonyls and protein-bound
glycation glycoxidation products, Arch. Biochem. Biophys.,
403, 259-269.
84.Diaz-Ruiz, R., Rigoulet, M., and Devin, A. (2011)
The Warburg and Crabtree effects: on the origin of cancer cell energy
metabolism and of yeast glucose repression, Biochim. Biophys.
Acta, 1807, 568-576.
85.Magasanik, B. (1961) Catabolite repression,
Cold Spring Harb. Symp. Quant. Biol., 26,
249-256.
86.Pastan, I., and Perlman, R. (1970) Cyclic
adenosine monophosphate in bacteria, Science, 169,
339-344.
87.Ackerman, R. S., Cozzarelli, N. R., and Epstein,
W. (1974) Accumulation of toxic concentrations of methylglyoxal by
wild-type Escherichia coli K-12, J. Bacteriol.,
119, 357-362.
88.Bachi, B., and Kornberg, H. L. (1975) Utilization
of gluconate by Escherichia coli. A role of adenosine
3′:5′-cyclic monophosphate in the induction of gluconate
catabolism, Biochem. J., 150, 123-128.
89.Peekhaus, N., and Conway, T. (1998) What’s
for dinner? Entner–Doudoroff metabolism in Escherichia
coli, J. Bacteriol., 180, 3495-3502.
90.Puskas, R., Fredd, N., Gazdar, C., and
Peterkofsky, A. (1983) Methylglyoxal-mediated growth inhibition in an
Escherichia coli cAMP receptor protein mutant, Arch. Biochem.
Biophys., 223, 503-513.
91.Burke, R. M., and Tempest, D. W. (1990) Growth of
Bacillus stearothermophilus on glycerol in chemostat culture:
expression of an unusual phenotype, J. Gen. Microbiol.,
136, 1381-1385.
92.Russell, J. B., and Cook, G. M. (1995) Energetics
of bacterial growth: balance of anabolic and catabolic reactions,
Microbiol. Rev., 59, 48-62.
93.Xu, D., Liu, X., Guo, C., and Zhao, J. (2006)
Methylglyoxal detoxification by an aldo-keto reductase in the
cyanobacterium Synechococcus sp. PCC 7002, Microbiology,
152, 2013-2021.
94.Koobs, D. H. (1972) Phosphate mediation of the
Crabtree and Pasteur effects, Science, 178, 127-133.
95.Iddar, A., Valverde, F., Assobhei, O., Serrano,
A., and Soukri, A. (2005) Widespread occurrence of non-phosphorylating
glyceraldehyde-3-phosphate dehydrogenase among Gram-positive bacteria,
Int. Microbiol., 8, 251-258.
96.Ito, F., Miyake, M., Fushinobu, S., Nakamura, S.,
Shimizu, K., and Wakagi, T. (2014) Engineering the allosteric
properties of archaeal non-phosphorylating glyceraldehyde-3-phosphate
dehydrogenases, Biochim. Biophys. Acta, 1844,
759-766.
97.Dan’shina, P. V., Schmalhausen, E. V.,
Arutiunov, D. Y., Pleten’, A. P., and Muronetz, V. I. (2003)
Acceleration of glycolysis in the presence of the non-phosphorylating
and the oxidized phosphorylating glyceraldehyde-3-phosphate
dehydrogenases, Biochemistry (Moscow), 68, 593-600.
98.Danshina, P. V., Schmalhausen, E. V., Avetisyan,
A. V., and Muronetz, V. I. (2001) Mildly oxidized
glyceraldehyde-3-phosphate dehydrogenase as a possible regulator of
glycolysis, IUBMB Life, 51, 309-314.
99.Crabtree, H. G. (1929) Observations on the
carbohydrate metabolism of tumors, Biochem. J., 23,
536-545.
100.Vemuri, G. N., Altman, E., Sangurdekar, D. P.,
Khodursky, A. B., and Eiteman, M. A. (2006) Overflow metabolism in
Escherichia coli during steady-state growth: transcriptional
regulation and effect of the redox ratio, Appl. Environ.
Microbiol., 72, 3653-3661.
101.Paczia, N., Nilgen, A., Lehmann, T., Gatgens,
J., Wiechert, W., and Noack, S. (2012) Extensive exometabolome analysis
reveals extended overflow metabolism in various microorganisms,
Microb. Cell Fact., 122, 1-14.
102.Molenaar, D., Van Berlo, R., De Ridder, D., and
Teusink, B. (2009) Shifts in growth strategies reflect tradeoffs in
cellular economics, Mol. Syst. Biol., 5, 323.
103.Evtodienko, Y. V., and Teplova, V. V. (1996)
Biological significance and mechanisms of Crabtree effect in rapidly
proliferating cells. Role of Ca2+ ions, Biochemistry
(Moscow), 61, 1423-1431.
104.Lele, U. N., and Watve, M. G. (2014) Bacterial
growth rate and growth yield: is there a relationship? Proc. Ind.
Natl. Sci. Acad., 80, 537-546.
105.Travisano, M., and Velicer, G. J. (2004)
Strategies of microbial cheater control, Trends Microbiol.,
12, 72-78.
106.Kreft, J. U., and Bonhoeffer, S. (2005) The
evolution of groups of cooperating bacteria and the growth rate versus
yield trade-off, Microbiology, 151, 637-641.
107.Pfeiffer, T., and Morley, A. (2014) An
evolutionary perspective on the Crabtree effect, Front. Mol.
Biosci., 21, 1-6.
108.Shimizu, K. (2014) Regulation systems of
bacteria such as Escherichia coli in response to nutrient
limitation and environmental stresses, Metabolites, 4,
1-35.
109.Sharma, P., Hellingwerf, K. J., De Mattos, M.
J. T., and Bekker, M. (2012) Uncoupling of substrate-level
phosphorylation in Escherichia coli during glucose-limited
growth, Appl. Environ. Microbiol., 78, 6908-6913.
110.Lowry, O. H., Carter, J., Ward, J. B., and
Glaser, L. (1971) The effect of carbon and nitrogen sources on the
level of metabolic intermediates in Escherichia coli, J.
Biol. Chem., 246, 6511-6521.
111.Schaefer, U., Boos, W., Takors, R., and
Weuster-Botz, D. (1999) Automated sampling device for monitoring
intracellular metabolite dynamics, Anal. Biochem., 270,
88-96.
112.Chassagnole, C., Noisommit-Rizzi, N., Schmid,
J. W., Mauch, K., and Reuss, M. (2002) Dynamic modeling of the central
carbon metabolism of Escherichia coli, Biotechnol.
Bioeng., 79, 53-73.
113.Russell, J. B. (1992) Glucose toxicity and
inability of Bacteroides ruminicola to regulate glucose
transport and utilization, Appl. Environ. Microbiol., 58,
2040-2045.
114.Emmerling, M., Dauner, M., Ponti, A., Fiaux,
J., Hochuli, M., Szyperski, T., Wuthrich, K., Bailey J. E., and Sauer,
U. (2002) Metabolic flux responses to pyruvate kinase knockout in
Escherichia coli, J. Bacteriol., 184, 152-164.
115.Sanden, A. M., Prytz, I., Tubulekas, I.,
Forberg, C., Le, H., Hektor, A., Neubauer, P., Pragai, Z., Harwood, C.,
Ward, A., Picon, A., De Mattos, T. J., Postma, P., Farewell, A.,
Nystrom, T., Reeh, S., Pedersen, S., and Larsson, G. (2003) Limiting
factors in Escherichia coli fed-batch production of recombinant
proteins, Biotechnol. Bioeng., 81, 158-166.
116.Sikorski, M. M., Topunov, A. F., Strozycki, P.,
Vorgias, C. E., Wilson, K. S., and Legocki, A. B. (1995) Cloning and
expression of plant leghemoglobin cDNA of Lupinus luteus in
Escherichia coli and purification of the recombinant protein,
Plant Sci., 108, 109-117.
117.Wang, H., Wang, F., Wang, W., Yao, X., Wei, D.,
Cheng, H., and Deng, Z. (2014) Improving the expression of recombinant
proteins in E. coli BL21 (DE3) under acetate stress: an alkaline
pH shift approach, PLoS One, 9, e112777.
118.Kosmachevskaya, O. V., Shumaev, K. B.,
Nasybullina, E. I., Gubkina, S. A., and Topunov, A. F. (2013)
Interaction of S-nitrosoglutathione with methemoglobin under conditions
of modeling carbonyl stress, Hemoglobin, 37, 205-218.
119.Kosmachevskaya, O. V., Shumaev, K. B.,
Nasybullina, E. I., and Topunov, A. F. (2014) Formation of nitri- and
nitrosylhemoglobin in systems modeling the Maillard reaction, Clin.
Chem. Lab. Med., 52, 161-168.
120.Maiden, M. F. J., Pham, C., and Kashket, S.
(2004) Glucose toxicity effect and accumulation of methylglyoxal by the
periodontal pathogen Bacteroides forsythus, Anaerobe,
10, 27-32.
121.Bechara, E. J., Dutra, F., Cardoso, V. E.,
Sartori, A., Olympio, K. P., Penatti, C. A., Adhikari, A., and
Assuncao, N. A. (2007) The dual face of endogenous
α-aminoketones: pro-oxidizing metabolic weapons, Comp.
Biochem. Physiol. C. Toxicol. Pharmacol., 146, 88-110.
122.Mathys, K. C., Ponnampalam, S. N., Padival, S.,
and Nagaraj, R. H. (2002) Semicarbazide-sensitive amine oxidase in
aortic smooth muscle cells mediates synthesis of a methylglyoxal-AGE:
implications for vascular complications in diabetes, Biochem.
Biophys. Res. Commun., 297, 863-869.
123.O’Sullivan, J., Unzeta, M., Healy, J.,
O’Sullivan, M. I., Davey, G., and Tipton, K. F. (2004)
Semicarbazide-sensitive amine oxidases: enzymes with quite a lot to do,
Neurotoxicology, 25, 303-315.
124.Sartori, A., Mano, C. M., Mantovani, M. C.,
Dyszy, F. H., Massari, J., Tokikawa, R., Nascimento, O. R., Nantes, I.
L., and Bechara, E. J. (2013) Ferricytochrome c directly
oxidizes aminoacetone to methylglyoxal, a catabolite accumulated in
carbonyl stress, PLoS One, 8, e57790.
125.Dutra, F., Knudsen, F. S., Curi, D., and
Bechara, E. J. H. (2001) Aerobic oxidation of aminoacetone, a threonine
catabolite: iron catalysis and coupled iron release from ferritin,
Chem. Res. Toxicol., 14, 1323-1329.
126.Turner, J. M. (1966) Microbial metabolism of
amino ketones. Aminoacetone formation from 1-aminopropan-2-ol by a
dehydrogenase in Escherichia coli, Biochem. J.,
99, 427-433.
127.Boylan, S. A., and Dekker, E. E. (1981)
L-threonine dehydrogenase of Escherichia coli K-12, J. Biol.
Chem., 256, 1809-1815.
128.Elliott, W. H. (1960) Aminoacetone formation by
Staphylococcus aureus, Biochem. J., 74,
478-485.
129.Green, M. L., and Elliott, W. H. (1964) The
enzymic formation of aminoacetone from threonine and its further
metabolism, Biochem. J., 92, 537-549.
130.Neuberger, A., and Tait, G. H. (1962)
Production of aminoacetone by Rhodopseudomonas spheroids,
Biochem. J., 84, 317-328.
131.Faulkner, A., and Turner, J. M. (1974)
Microbial metabolism of amino alcohols. Aminoacetone metabolism via
1-aminopropan-2-ol in Pseudomonas sp. N.C.I.B. 8858, Biochem.
J., 138, 263-276.
132.Green, M. L., and Lewis, J. B. (1968) The
oxidation of aminoacetone by a species of Arthrobacter,
Biochem. J., 106, 267-270.
133.Rahhar, D. A., Turner, J. M., and Willetts, A.
I. (1967) The role of aminoacetone in L-threonine metabolism by
Bacillus subtilis, Biochem. J., 103, 73-73.
134.Willets, A. J., and Turner, J. M. (1970).
Threonine metabolism in a strain of Bacillus subtilis: enzymic
oxidation of the intermediate DL-lactaldehyde, Biochim. Biophys.
Acta, 222, 234-236.
135.Machielsen, R., and Van der Oost, J. (2006)
Production and characterization of a thermostable L-threonine
dehydrogenase from the hyperthermophilic archaeon Pyrococcus
furiosus, FEBS J., 273, 2722-2729.
136.Bell, S. C., and Turner, J. M. (1976) Bacterial
catabolism of threonine. Threonine degradation initiated by L-threonine
NAD+ oxidoreductase, Biochem. J., 156,
449-458.
137.Newman, E. B., Kapoor, V., and Potter, R.
(1976) Role of L-threonine dehydrogenase in the catabolism of threonine
and synthesis of glycine by Escherichia coli, J.
Bacteriol., 126, 1245-1249.
138.Potter, R., Kapoor, V., and Newman, E. B.
(1977) Role of threonine dehydrogenase in Escherichia coli
threonine degradation, J. Bacteriol., 132, 385-391.
139.Tressel, T., Thompson, R., Zieske, L. R.,
Menendez, M. I., and Davis, L. (1986) Interaction between L-threonine
dehydrogenase and aminoacetone synthetase and mechanism of aminoacetone
production, J. Biol. Chem., 261, 16428-16437.
140.Higgins, I. J., and Turner J. M. (1969) Enzymes
of methylglyoxal metabolism in a pseudomonad, which rapidly metabolizes
aminoacetone, Biochim. Biophys. Acta, 184, 464-467.
141.Takors, R., Bathe, B., Rieping, M., Hans, S.,
Kelle, R., and Huthmacher, K. (2007) Systems biology for industrial
strains and fermentation processes – example: amino acids, J.
Biotechnol., 129, 181-190.
142.Lee, K. H., Park, J. H., Kim, T. Y., Kim, H.
U., and Lee, S. Y. (2007) Systems metabolic engineering of
Escherichia coli for L-threonine production, Mol. Syst.
Biol., 3, 149.
143.Nishikawa, T., Edelstein, D., Du, X. L.,
Yamagishi, S., Matsumura, T., Kaneda, Y., Yorek, M. A., Beebe, D.,
Oates, P. J., Hammes, H. P., Giardino, I., and Brownlee, M. (2000)
Normalizing mitochondrial superoxide production blocks three pathways
of hyperglycemic damage, Nature, 404, 787-790.
144.Xie, L., Eiteman, M. A., and Altman, E. (2003)
Production of 5-aminolevulinic acid by an Escherichia coli
aminolevulinate dehydratase mutant that overproduces Rhodobacter
sphaeroides aminolevulinate synthase, Biotechnol. Lett.,
25, 1751-1755.
145.Hiraku, Y., and Kawanishi, S. (1999)
Involvement of oxidative DNA damage and apoptosis in antitumor actions
of amino sugars, Free Radic. Res., 31, 389-403.
146.Hiraku, Y., Sugimoto, J., Yamaguchi, T., and
Kawanishi, S. (1999) Oxidative DNA damage induced by aminoacetone, an
amino acid metabolite, Arch. Biochem. Biophys., 365,
62-70.
147.Szent-Gyorgyi, A., Egyiid, L. G., and
McLaughlin, J. A. (1967) Ketoaldehydes and cell division,
Science, 155, 539-541.
148.Egyud, L. G. (1967) Studies on cell division:
the effect of aldehydes, ketones, and α-ketoaldehydes on the
proliferation of Escherichia coli, Curr. Mol. Biol.,
1, 14-20.
149.Egyud, L. G., and Szent-Gyorgyi, A. (1966) On
the regulation of cell division, Proc. Natl. Acad. Sci. USA,
56, 203-207.
150.Campbell, A. K., Naseem, R., Holland, I. B.,
Matthews, S. B., and Wann, K. T. (2007) Methylglyoxal and other
carbohydrate metabolites induce lanthanum-sensitive
Ca2+-transients and inhibit growth in E. coli,
Arch. Biochem. Biophys., 468, 107-113.
151.Rosas-Lemus, M., Uribe-Alvarez, C.,
Chiquete-Felix, N., and Uribe-Carvajal, S. (2014) In Saccharomyces
cerevisiae fructose-1,6-bisphosphate contributes to the Crabtree
effect through closure of the mitochondrial unspecific channel,
Arch. Biochem. Biophys., 555/556, 66-70.
152.Naseem, R., Davies, S. R., Jones, H., Wann, K.,
Holland, I. B., and Campbell, A. K. (2007) Cytosolic Ca2+
regulates protein expression in E. coli through release from
inclusion bodies, Biochem. Biophys. Res. Commun., 360,
33-39.
153.Dhar, N., and McKinney, J. D. (2007) Microbial
phenotypic heterogeneity and antibiotic tolerance, Curr. Opin.
Microbiol., 10, 30-38.
154.Girgis, H. S., Harrisa, K., and Tavazoiea, S.
(2012) Large mutational target size for rapid emergence of bacterial
persistence, Proc. Natl. Acad. Sci. USA, 109,
12740-12745.
155.Rachman, H., Kim, N., Ulrichs, T., Baumann, S.,
Pradl, L., Eddine, A. N., Bild, M., Rother, M., Kuban, R.-J., Lee, J.
S., Hurwitz, R., Brinkmann, V., Kosmiadi, G. A., and Kaufmann, S. H. E.
(2006) Critical role of methylglyoxal and AGE in mycobacteria-induced
macrophage apoptosis and activation, PLoS One, 1,
e29.
156.Rachman, H., Strong, M., Ulrichs, T., Grode,
L., Schuchhardt, J., Mollenkopf, H., Kosmiadi, G. A., Eisenberg, D.,
and Kaufmann, S. H. E. (2006) Unique transcriptome signature of
Mycobacterium tuberculosis in pulmonary tuberculosis, Infect.
Immun., 74, 1233-1242.
157.Gray, M. J., Wholey, W. Y., Parker, B. W., Kim,
M., and Jakob, U. (2013) NemR is a bleach-sensing transcription factor,
J. Biol. Chem., 288, 13789-13798.
158.Lee, C., Shin, J., and Park, C. (2013) Novel
regulatory system nemRA–gloA for electrophile reduction in
Escherichia coli K-12, Mol. Microbiol., 88,
395-412.
159.Ozyamak, E., De Almeida, C., De Moura, A. P.
S., Miller, S., and Booth, I. R. (2013) Integrated stress response of
Escherichia coli to methylglyoxal: transcriptional readthrough
from the nemRA operon enhances protection through increased expression
of glyoxalase I, Mol. Microbiol., 88, 936-950.
160.Bakker, E. P., and Mangerich, W. E. (1982)
N-ethylmaleimide induces K+-H+ antiport
activity in Escherichia coli K-12, FEBS Lett.,
140, 177-180.
161.Elmore, M. J., Lamb, A. J., Ritchie, G. Y.,
Douglas, R. M., Munro, A., Gajewska, A., and Booth, I. R. (1990)
Activation of potassium efflux from Escherichia coli by
glutathione metabolites, Mol. Microbiol., 4, 405-412.
162.Ferguson, G. P., Munro, A. W., Douglas, R. M.,
McLaggan, D., and Booth, I. R. (1993) Activation of potassium channels
during metabolite detoxification in Escherichia coli, Mol.
Microbiol., 9, 1297-1303.
163.Miller, S., Ness, L. S., Wood, C. M., Fox, B.
C., and Booth, I. R. (2000) Identification of an ancillary protein,
YabF, required for activity of the KefC glutathione-gated potassium
efflux system in Escherichia coli, J. Bacteriol.,
182, 6536-6540.
164.Roosild, T. P., Castronovo, S., Miller. S., Li,
C., Rasmussen, T., Bartlett, W., Gunasekera, B., Choe, S., and Booth,
I. R. (2009) KTN (RCK) domains regulate K+ channels and
transporters by controlling the dimer-hinge conformation,
Structure, 17, 893-903.
165.Ozyamak, E., Black, S. S., MacLean, M. J.,
Bartlett, W., Miller, S., and Booth, I. R. (2011) The critical role of
S-lactoylglutathione formation during methylglyoxal detoxification in
Escherichia coli, Mol. Microbiol., 78,
1577-1590.
166.Rekarte, U. D., Zwaig, N., and Isturiz, T.
(1973) Accumulation of methylglyoxal in a mutant of Escherichia
coli constitutive for gluconate catabolism, J. Bacteriol.,
115, 727-731.
167.Baskaran, S., Rajan, D. P., and
Balasubramanian, K. A. (1989) Formation of methylglyoxal by bacteria
isolated from human faces, J. Med. Microbiol., 28,
211-215.
168.Kashket, S., Maiden, M. F., Haffajee, A. D.,
and Kashket, E. R. (2003) Accumulation of methylglyoxal in the gingival
crevicular fluid of chronic periodontitis patients, J. Clin.
Periodontol., 30, 364-367.
169.Chakraborty, S., Karmakar, K., and
Chakravortty, D. (2014) Cells producing their own nemesis:
understanding methylglyoxal metabolism, IUBMB Life, 66,
667-678.
170.Campbell, A. K., Matthews, S. B., Vassel, N.,
Cox, C. D., Naseem, R., Chaichi, J., Holland, I. B., Greene, J., and
Wann, K. T. (2010) Bacterial metabolic “toxins”: a new
mechanism for lactose and food intolerance, and irritable bowel
syndrome, Toxicology, 278, 268-276.
171.Wendisch, V. F., Lindner, S. N., and
Meiswinkel, T. M. (2011) Use of glycerol in biotechnological
applications, Biodiesel – Quality, Emissions and
By-products (Montero, G., and Stoytcheva, M., eds.) InTech,
Rijeka.
172.Clomburg, J. M., and Gonzalez, R. (2011)
Metabolic engineering of Escherichia coli for the production of
1,2-propanediol from glycerol, Biotechnol. Bioeng., 108,
867-879.